Abstract
“Mycobacterium avium subsp. hominissuis” often causes cervical lymphadenitis in children; its prompt and accurate identification enables adequate therapy, tracing, and prevention. The aims of this study were to determine the causative agent of lymphadenitis using culture, PCR, and triplex quantitative real-time PCR (qPCR) methods with DNA directly isolated from tissue, as well as to identify possible sources of infection from the environment. We confirmed the diagnoses by detecting M. avium subsp. hominissuis using qPCR with DNA directly isolated from lymph node biopsy specimens of two patients. In order to trace the source of infection from the environment, a method of DNA isolation from soil and other environmental samples, such as dust, cobwebs, and compost, was developed. The triplex qPCR examination revealed the presence of M. avium subsp. hominissuis in a high proportion of the environmental samples (42.8% in the first patient's house and 47.6% in the second patient's house). Both patients were also exposed to M. avium subsp. avium, which was present due to the breeding of infected domestic hens. The high infectious dose of M. avium subsp. hominissuis or the increased susceptibility of humans to M. avium subsp. hominissuis compared to M. avium subsp. avium could be the reason why the children were infected with M. avium subsp. hominissuis.
Lymphadenitis in children occurs as a result of inflammation of the cervical, submaxillary, or submandibular lymph nodes. Cases usually present as unilateral enlargement of the lymph nodes. Awareness of the condition has increased in recent years due to the consistent upward trend in the incidence of the disease (13, 27). The identification of the causative agent of cervical lymphadenitis is essential for the administration of correct treatment and subsequent follow-up. Differentiation between enlargement caused by the Mycobacterium tuberculosis complex and that caused by nontuberculous mycobacteria (NTM) is crucial for determining the appropriate therapy.
The most prevalent causative agent in children between the ages of 1 and 5 in developed countries with a low incidence of human tuberculosis is NTM or potentially pathogenic mycobacteria (PPM) (3, 5, 22, 53), especially “Mycobacterium avium subsp. hominissuis” (50). The routes of M. avium subsp. hominissuis infection have not yet been clearly identified, but bearing in mind that human-to-human transmission has not been demonstrated, infection from an environmental source is considered to be most likely. M. avium subsp. hominissuis is ubiquitous in the environment and has been isolated from different types of samples such as soil, water, aerosols, dust, protozoa, and even small invertebrates (6, 12, 19, 22, 25).
The diagnosis of PPM causing cervical lymphadenitis is difficult and is often delayed due to the long incubation period required to grow the mycobacteria in vitro. Other methods of diagnosing the etiological agent include microscopic examination after Ziehl-Neelsen (ZN) staining, cytopathological analysis, DNA isolation from fine-needle aspirates (FNA), and subsequent PCR identification (37, 55). So far, culture-independent methods lack the specificity required to identify members of the M. avium complex (MAC), which consists of 28 different serotypes (54).
The most widely used tool for molecular identification of the members of the MAC is the detection of the insertion sequences IS1245 and IS901 (1, 10, 32). M. avium subsp. avium strains of serotypes 1, 2, and 3, and M. avium subsp. hominissuis strains of serotypes 4 to 6, 8 to 11, and 21 are closely related but differ in the presence and number of copies of these insertion sequences. M. avium subsp. hominissuis is characterized by multiple copies of IS1245 and the absence of IS901 (9, 20, 31, 51), whereas M. avium subsp. avium possesses multiple IS901 copies and a single copy of IS1245 (23). The principal source of human infection is infected birds, especially domestic hens (25, 35).
The aims of this study were to determine the causative agent of lymphadenitis through the use of conventional culture and triplex quantitative real-time PCR (qPCR) methods with DNA directly isolated from lymph node tissue. In order to identify the source of the infection from the environment, a method for DNA isolation from soil and other environmental samples, such as dust, cobwebs, and compost, was introduced, and its DNA yield based upon the model of M. avium subsp. paratuberculosis was determined. PCR results were compared to results of conventional microbiological culture.
MATERIALS AND METHODS
Sample origin.
Two patients, ages 7 and 2 years, were included in this study. Neither of them had any history of health problems. The time taken to develop an enlarged neck lymph node was several months; both were diagnosed with cervical lymphadenitis. A typical symptom that both of the patients shared was unilateral enlargement of the lymph node. Normal X-ray scans and no history of tuberculosis were also noted. Before the samples were obtained, informed consent was obtained from the parents of the children, and the samples were used according to the regulations of the respective hospital.
(i) Patient 1.
A 7-year-old boy from the Czech Republic with no history of health problems developed an enlarged lymph node in the neck area on the left side in a period of 6 months. The isolate was cultured from a tissue sample after surgical excision of the enlarged lymph node. To identify possible sources of infection, seven samples from the patient's environment, i.e., the patient's house and garden, were collected (Table 1). An additional 32 soil samples were examined only by microscopy and mycobacterial culture. These samples were collected from the garden where the patient spent most of his time in the few months before the enlargement of the lymph node.
TABLE 1.
Distribution of mycobacteria in patients' environmental samples as determined by microscopy with Ziehl-Neelsen staining, culture, and triplex qPCR
| Patient | Area | Sample |
Hen contact | ZN staining | qPCR (copies/g) |
Isolate identification | ||
|---|---|---|---|---|---|---|---|---|
| Location | Origin | IS901 | IS1245 | |||||
| 1 | Residence | Kitchen | Potting soil from supermarket | − | − | − | 5.88 × 103 | − |
| Hennery | Coop with young (6-wk-old) chickens | Cobwebs by window | − | − | − | − | − | |
| Coop with old hens | Dust and cobwebs | + | − | 1.00 × 100 | −a | − | ||
| Drinking water from vessel | + | − | − | − | − | |||
| Greenhouse | Soil | Soil without hen and chicken droppings | − | − | − | 5.64 × 102 | − | |
| Iceberg lettuce | Leaf without soil contamination | − | − | − | − | − | ||
| Garden | Compost | Material without hen and chicken droppings | − | − | − | 1.42 × 104 | − | |
| 2 | Residence | Children's room | Potting soil fertilized with chicken droppings | + | + | 7.14 × 101 | 2.25 × 107 | M. colombiense |
| Living room | Soil from store-bought cactus | − | − | − | 4.42 × 104 | |||
| Bedroom | Dust from a vacuum cleaner | − | − | − | 6.21 × 103 | M. engbaekii | ||
| Hennery | Coop with old hens | Cobwebs above nests | + | − | 1.00 × 100 | 2.60 × 104 | Mycobacterium sp. | |
| Dry droppings from hens | + | + | 1.00 × 100 | 3.93 × 103 | Mycobacterium sp. | |||
| Sand from pen | + | + | − | 4.47 × 105 | ||||
| Greenhouse | Patch with vegetable | Soil fertilized with chicken droppings | + | + | 3.28 × 101 | 2.67 × 107 | ||
| Tagetes patula | Soil fertilized with chicken droppings | + | + | 1.52 × 101 | 7.20 × 106 | |||
| Thuja | Soil fertilized with chicken droppings | + | + | 5.46 × 100 | 2.06 × 107 | |||
| Garden | Pen for dog | Soil with dog feces and urine | − | + | − | 3.11 × 101 | ||
| Grass | Soil | + | + | − | 1.50 × 107 | |||
| Pelargonium | Soil fertilized with chicken droppings | + | + | − | − | M. kumamotonense | ||
| Cobwebs with dust on plants | − | − | − | 7.19 × 104 | ||||
| Hedge plants | Soil | + | − | 4.14 × 101 | 1.34 × 106 | M. kumamotonense | ||
| Street | Near residence | Soil next to road | + | + | − | 1.57 × 105 | Mycobacterium sp. | |
| Garden of neighboring farm I | Shaded corner | Moss | − | − | − | 2.22 × 101 | M. chelonae | |
| Compost | Material without chicken droppings | − | + | − | 1.00 × 100 | |||
| Spruce tree | Soil under tree | + | + | − | 2.17 × 105 | |||
| Grass | Soil | + | + | 1.05 × 102 | 1.59 × 104 | |||
| Hennery of neighboring farm II | Coop with old hens | Dry chicken droppings from pen | + | + | − | − | Mycobacterium sp. | |
| Cobwebs | + | + | 2.60 × 102 | 9.68 × 103 | ||||
IS1245 qPCR was negative due to the low number of M. avium subsp. avium cells in the examined matrix.
(ii) Patient 2.
A girl from Slovakia, admitted to the Faculty Hospital in Bratislava, was 2 1/2 years old at the time of diagnosis. Samples for culture and qPCR examination were taken directly from the enlarged neck lymph node, which was situated on the left side. Sampling was done immediately after surgical excision. Altogether, samples from three parts (peripheral, internal, and caseous center) were collected. Approximately 2 g of each tissue was collected in a sterile container, immediately chilled, and transported to the laboratory. To assess the prevalence of mycobacteria in the environment as a potential source of infection, a total of 21 environmental samples from the patient's immediate environment were examined, including samples from their home, garden, greenhouse, and hen house, as well as the backyards of their closest neighbors (Table 1). Most of the samples examined were from places in the house and garden where the child played. Environmental samples were taken using a sterile scraper and placed into sterile plastic bags. After transportation to the laboratory, the tissue and environmental samples were aliquoted into separate boxes to avoid possible cross contamination between samples.
Microscopy and culture examinations. (i) Microscopy.
Smears were prepared from each sample after homogenization and decontamination and were stained using the ZN technique for the presence of acid-fast bacilli (AFB). At least 200 fields of view were examined for each sample using a light microscope at a magnification of ×1,000 under oil immersion (B17; Olympus, Tokyo, Japan).
(ii) Examination by culture.
The homogenization and decontamination of the samples were performed as previously described (18). Briefly, approximately 1 g of each sample was suspended in 10 ml of sterile water, shaken for 30 min at 1,400 rpm, and then left standing at room temperature for 30 min. After decontamination of the centrifuged pellet, 80 μl of the suspension was inoculated onto two solid media and one liquid medium: Herrold's egg yolk medium, Stonebrink's egg-based medium, and Sula's liquid serum medium (26). Incubation of the cultures was carried out at 25 and 37°C. The cultures were monitored for the first week in order to rule out contamination and then every second week for up to 3 months.
(iii) Identification of isolates.
AFB-positive isolates were further identified to species level by multiplex conventional PCR as described previously (32). If they did not belong to the MAC, they were sequenced (16S rRNA gene) as described by Harmsen et al. (21). The sequences obtained were compared to the available sequences deposited in the RIDOM (21) and NCBI databases.
DNA isolation from lymph nodes.
DNA isolation from 50 mg of tissue samples obtained from the patient after surgery was based on a slightly modified DNeasy blood and tissue kit protocol (Qiagen, Hilden, Germany) as described by Slana et al. (45). Every isolation run of up to 12 samples included a negative isolation control, which contained corresponding amount of water.
PCR methods.
In order to classify the bacterial species in the extracted lymph node samples, several PCR methods were used. Isolated DNA was tested for the presence of mycobacterial DNA using the conventional multiplex PCR system (32). To exclude the possibility of infection by M. tuberculosis, an IS6110 PCR detection method (Mycobacterium tuberculosis PCR kit; Geneproof, Brno, Czech Republic) was used. Triplex qPCR with an internal amplification control was used for the sensitive detection of M. avium subsp. hominissuis and M. avium subsp. avium in all tissue and environmental samples (45). All samples were analyzed with experimental replicates. Each PCR run included both positive (plasmid standard) and negative (water instead of DNA) PCR controls.
DNA isolation from environmental samples.
The DNA isolation protocol was based on the MoBio PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) with slight modifications. The principle of extraction is based on mechanical and chemical cell lyses followed by immobilization of the DNA on a silica spin column and subsequent washing and elution. Briefly, 0.25 g of soil was added to the PowerSoil bead beating tubes containing cell lysis solution, followed by the addition of 60 μl of the C1 solution and 6.25 μg of fish sperm DNA (Amresco, Solon, OH). The samples were homogenized using a MagNA Lyser instrument (Roche Molecular Diagnostics, Mannheim, Germany) at 6,400 rpm for 60 s.
The following steps were performed in accordance with the manufacturer's instructions. After the DNA binding solution was added to the cleared supernatant, the mixture was loaded onto the spin column. Washing of the spin column was performed according to the manufacturer's instructions with an additional centrifugation step to remove any remaining ethanol from the column. The DNA was eluted in 100 μl of preheated Tris-EDTA (TE) buffer (Amresco) after a 3-min incubation period on the column. After centrifugation, the filtrate was reapplied onto the spin column in order to improve the yield. As for DNA isolation from tissue, the negative isolation control was included in all isolation runs.
Efficiency of DNA isolation from soil and limit of detection.
The efficiency of the described protocol for DNA isolation from soil was tested by artificial contamination of four different types of soil: field soil, sand, peat, and pot soil. A model described previously for M. avium subsp. paratuberculosis was used (46). Briefly, a 1-ml aliquot of liquid culture of M. avium subsp. paratuberculosis was washed twice in TE buffer. Twenty 1-mm zirconia silica beads (Biospec, Bartlesville, OK) were added before mechanical disruption using the MagNA Lyser instrument (Roche). The suspension was centrifuged at 0.1 × g for 30 s to remove the largest clumps. The supernatant was transferred to a new tube and diluted to six different concentrations, which were quantified and used to spike negative soil samples. After DNA isolation, M. avium subsp. paratuberculosis was quantified using the F57 qPCR assay (46). Each M. avium subsp. paratuberculosis concentration was analyzed with eight physical replicates. The limit of detection was determined as the lowest cell concentration that could be detected in all replicates of the isolated samples. The DNA isolation yield was determined by calculating the median of all replicates (within the respective M. avium subsp. paratuberculosis artificial contamination concentrations).
IS1245 RFLP analysis.
Tissue and soil isolates from patient 2 were subjected to restriction fragment length polymorphism (RFLP) analysis according to the standardized method described previously (29, 51). DNA was isolated according to the method described by van Soolingen et al. (52) and subsequently digested with the restriction endonuclease PvuII. DNA fragments were separated by agarose gel electrophoresis and transferred to a nylon membrane by vacuum blotting. DNA was hybridized with a labeled probe, and IS1245 profiles were analyzed according to the number and position of the bands.
RESULTS
Patient diagnosis. (i) Patient 1.
The tissue sample was negative under microscopy with ZN staining but positive through culture examination for mycobacteria. The isolate M. avium subsp. hominissuis was identified through the presence of the specific 16S rRNA gene for mycobacteria and the IS1245 sequence as well as the absence of IS901, as analyzed by triplex qPCR (Table 2).
TABLE 2.
Examined tissue samples from patients with neck lymphadenitis
| Patient | Country | Age (yr)/sex | Specimen | Test result |
||||
|---|---|---|---|---|---|---|---|---|
| AFBa | Culture | PCR, IS6110 | qPCR |
|||||
| IS901 | IS1245 | |||||||
| 1 | Czech Republic | 7/male | Extirpated tissue with caseous lesions | − | + | − | − | +b |
| 2 | Slovakia | 2/female | Extirpated tissue with caseous lesion, periphery | − | − | − | − | − |
| Tissue without caseous lesions | − | − | − | − | − | |||
| Extirpated tissue with caseous lesion, caseated center of 1 cm in diam | − | − | − | − | +c | |||
AFB, acid-fast bacilli visible after Ziehl-Neelsen staining.
Identification was carried out with isolated DNA from the mycobacterial isolate.
Quantification was not possible.
(ii) Patient 2.
The tissue samples were negative under microscopy and culture examinations. However, after direct DNA isolation from the tissue, the presence of IS1245 was confirmed by triplex qPCR testing, which revealed that the lymphadenopathy was in fact due to M. avium subsp. hominissuis infection (Table 2).
Limit of detection and DNA isolation efficiency.
The detection limit varied for the soil types tested in the range of 2.5 × 103 to 8.9 × 103 copies per g of soil. Specifically, detection limits of 4.2 × 103 copies per 1 g of field soil, 2.5 × 103 copies per 1 g of sand, and 8.9 × 103 copies per 1 g of peat and potting soil were established. At these concentrations in all eight replicates, positive results using qPCR was obtained. Samples with lower numbers of mycobacteria were still considered to be positive, but the probability of the detection in repeated experiments is lower. The median DNA isolation efficiency was 59.1% for field soil, 53.8% for sand, 30.2% for peat, and 36.9% for pot soil samples (Table 3).
TABLE 3.
DNA isolation efficiency and limit of detection of Mycobacterium avium subsp. paratuberculosis in different types of soil tested
| Theoretical no. of MAP cells | Field soil |
Sand |
Peat |
Potting soil |
||||
|---|---|---|---|---|---|---|---|---|
| Signal ratioa | DNA recovery (%) | Signal ratio | DNA recovery (%) | Signal ratio | DNA recovery (%) | Signal ratio | DNA recovery (%) | |
| 5.5 × 104 | 8/8 | 54.9 | 8/8 | 52.8 | NTb | NT | NT | NT |
| 1.3 × 104 | 8/8 | 57.8 | 8/8 | 44.3 | NT | NT | NT | NT |
| 6.1 × 103 | 8/8 | 60.5 | 8/8 | 54.9 | 8/8 | 26.2 | 8/8 | 31.5 |
| 4.2 × 103 | 8/8 | 44.0 | 8/8 | 49.6 | NT | NT | NT | NT |
| 2.5 × 103 | 7/8 | 148.0 | 8/8 | 65.3 | 7/8 | 30.2 | 6/8 | 36.9 |
| 7.9 × 102 | 5/8 | 92.1 | 7/8 | 75.0 | 3/8 | 56.7 | 1/8 | 48.9 |
Number of positive samples/total number of samples tested.
NT, not tested.
Environmental samples. (i) Residence of patient 1.
None of the seven environmental samples examined by microscopy, culture, and triplex qPCR were positive for AFB by ZN staining. The samples were also negative under culture examination. Three of the samples examined by triplex qPCR were positive for M. avium subsp. hominissuis DNA (two samples of soil and one sample of dunghill), and one sample was positive for M. avium subsp. avium DNA (dust and cobwebs from the hen house; Table 1).
There were 32 additional samples of pot soil or soil from the garden examined using only the culture method, 5 of which resulted in M. avium subsp. hominissuis isolates. They were all subjected to RFLP analysis and showed the same RFLP profile as the isolate from the lymph node of the patient (Fig. 1).
FIG. 1.
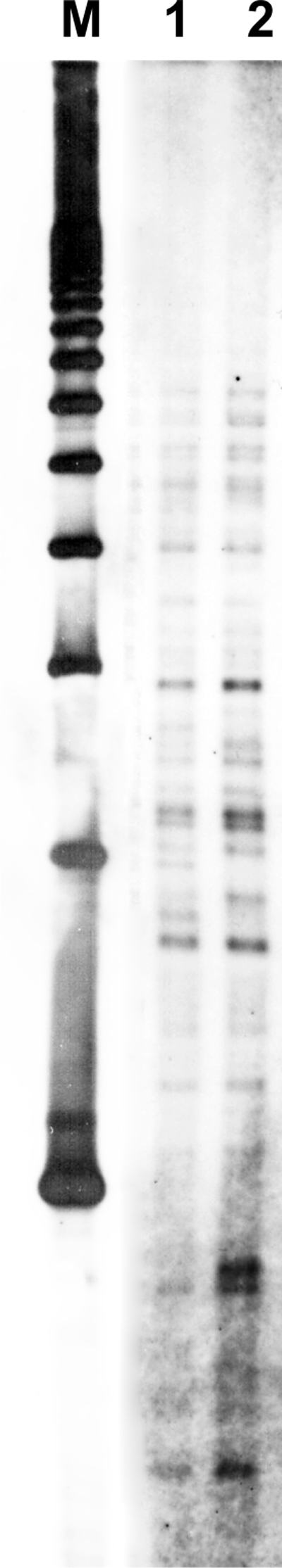
Comparison of IS1245 RFLP types of the isolate obtained from patient 1 (lane 1) and an isolate obtained from pot soil in his house (lane 2). A 1-kb ladder was used as a marker (lane M).
Residence of patient 2 and neighboring houses.
The ZN microscopy examination of the decontaminated samples resulted in 16 (69.5%) out of 21 samples being positive for AFB. Fourteen of these samples were also positive for either M. avium subsp. avium or M. avium subsp. hominissuis by triplex qPCR, of which two were positive by culture; these were identified by 16S rRNA sequencing as M. kumamotonense and a Mycobacterium strain that could not be further identified. Culture examination resulted in seven positive isolates, none of which belonged to the M. avium species as confirmed by conventional multiplex PCR (32). The 16S rRNA gene was sequenced, and the following strains were obtained: M. colombiense, M. engbaekii, M. chelonae, two isolates of M. kumamotonense, and two isolates of a Mycobacterium species that could not be identified using the available deposited sequences. From the 21 environmental samples examined by triplex qPCR, 10 (47.6%) were positive for M. avium subsp. hominissuis and 9 (42.8%) were positive for M. avium subsp. avium. The M. avium subsp. hominissuis-positive samples were present in the house of the patient (potting soil and dust) as well as in the soil from the garden and greenhouse. M. avium subsp. avium was found predominantly in the hennery and neighbor's garden (Table 1).
DISCUSSION
Although NTM isolates have been differentiated using real-time qPCR (28, 39, 44, 47), the majority of research is still devoted to detecting and identifying members of the M. tuberculosis complex (14). Real-time PCR for the detection of MAC and M. tuberculosis complex in fine-needle aspirates and tissue biopsy samples has been confirmed as a useful tool for the rapid diagnosis of mycobacterial lymphadenitis (49). Recently, a triplex qPCR system for the simultaneous detection of M. avium subsp. avium and M. avium subsp. hominissuis in pig tissues has been described (45). We have used the same triplex qPCR system, and it has proven to be an efficient and rapid tool for the identification of M. avium subsp. hominissuis from tissue samples of two patients (Table 2) and their environments (Table 1).
The recovery of mycobacteria by culture is a standard method, although due to limitations encountered (i.e., their slow growth), in recent times culture-independent methods have been preferred (25). Our results show that qPCR is a much more sensitive method for the detection of mycobacteria, as shown with patient 2, whose diagnosis was not confirmed by culture. The diagnosis of M. avium subsp. hominissuis as a causative agent in neck lymphadenitis in children by using qPCR has been performed also with patients in Germany, where the amount of IS1245 ranged between 101 and 102 copies (data not shown). The reliability of the culture method was evaluated in a study analyzing diagnostic methods for detecting mycobacteria in pig tissue samples. A high number of tuberculous lesions present in lymph nodes were ZN positive but not confirmed by culture (43). This and other studies suggest that correlation among different diagnostic methods would be complicated (24, 45).
The infectious route of mycobacteria (other than in tuberculosis) has been studied on several occasions. Some studies implicate water as the source of infection, especially in HIV-infected individuals (16, 17, 36). Van Coppenraet et al. (50) could not find a link between pet birds and the incidence of mycobacterial lymphadenitis in children, concluding that infection can occur from any environmental source, while other authors have found a strong correlation between soil and mycobacterial infection (7, 38, 56). Our results suggest that soil contaminated with M. avium subsp. hominissuis was the main source of infection for both children. Surprisingly, both children were in close contact with soil and dust contaminated with M. avium subsp. avium in connection with flocks of infected hens reared near their residences (Table 1). Using the triplex qPCR system, we are not able to differentiate between M. avium subsp. avium and mixed M. avium subsp. avium/M. avium subsp. hominissuis infections. In some samples we have detected a larger amount of IS1245 than of IS901; however, we cannot conclude that there was a mixed infection solely on the basis of this result. These results suggest a higher sensitivity of humans to M. avium subsp. hominissuis infection than to infections caused by M. avium subsp. avium (25, 35, 50).
Although M. avium subsp. hominissuis has often been isolated from soil using the culture method, studies of microbial soil ecology suggest that a large percentage of microbial cells present in the environment are not able to be isolated in vitro; i.e., they could be viable but noncultivable, making their detection using molecular techniques necessary (2, 48). Furthermore, mycobacteria present in soil samples are easily outgrown by other, faster-growing microorganisms (15). Their strong adherence to soil particles and their viability can also present difficulties in their isolation by culture (4, 8). Due to these obstacles, the use of molecular methods for detection and identification of mycobacteria in the environment is favorable. Mycobacterial DNA has been isolated from soil in previous studies for the purpose of an ecological study or genus-specific identification, although no further species identification was done (30, 33, 34).
Previous studies indicate that M. avium subsp. hominissuis is the most common etiological agent of NTM lymphadenitis in children (50). The patients in our study were exposed to an infectious environment containing M. avium subsp. avium due to the breeding of domestic hens. In such small flocks, old hens (3 years or older) could be present. The number of positive M. avium subsp. avium DNA samples was similar to that of positive M. avium subsp. hominissuis DNA samples (Table 1). The reason why the children were infected with M. avium subsp. hominissuis and not with M. avium subsp. avium is not clear. It has been described that M. avium subsp. hominissuis is pathogenic mainly to humans (31, 35), while infections with M. avium subsp. avium are more typical for birds (11, 35, 40-42), perhaps indicating why there was an increased susceptibility to M. avium subsp. hominissuis in these patients. Moreover, M. avium subsp. hominissuis DNA was found in a higher quantity in the environment, indicating an exposure of the patients to a higher infectious dose.
In conclusion, the source of infection was confirmed as soil for one of the patients by RFLP of the IS1245 sequence, and this has also been suggested for the second patient, using direct triplex qPCR and culture methods.
Acknowledgments
We thank R. Stanik (Faculty Hospital, Bratislava, Slovakia) for supplying the tissue samples and J. Svobodova (Regional Institute of Public Health, Brno, Czech Republic) for providing the mycobacterial isolate.
This work was supported by the Ministry of Agriculture of the Czech Republic (project no. MZE0002716202) and the Ministry of Education, Youth and Sports of the Czech Republic (project AdmireVet no. CZ.1.05/2.1.00/01.0006-ED 0006/01/01).
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Bartos, M., et al. 2006. Identification of members of Mycobacterium avium species by Accu-Probes, serotyping, and single IS900, IS901, IS1245 and IS901-flanking region PCR with internal standards. J. Microbiol. Methods 64:333-345. [DOI] [PubMed] [Google Scholar]
- 2.Beran, V., M. Havelkova, J. Kaustova, L. Dvorska, and I. Pavlik. 2006. Cell wall deficient forms of mycobacteria: a review. Veterinarni Medicina 51:365-389. [Google Scholar]
- 3.Blyth, C. C., et al. 2009. Nontuberculous mycobacterial infection in children: a prospective national study. Pediatr. Infect. Dis. J. 28:801-805. [DOI] [PubMed] [Google Scholar]
- 4.Brooks, R. W., K. L. George, B. C. Parker, J. O. Falkinham, and H. Gruft. 1984. Recovery and survival of nontuberculous mycobacteria under various growth and decontamination conditions. Can. J. Microbiol. 30:1112-1117. [DOI] [PubMed] [Google Scholar]
- 5.Choi, P., et al. 2009. Polymerase chain reaction for pathogen identification in persistent pediatric cervical lymphadenitis. Arch. Otolaryngol. Head Neck Surg. 135:243-248. [DOI] [PubMed] [Google Scholar]
- 6.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Groote, M. A., N. R. Pace, K. Fulton, and J. O. Falkinham. 2006. Relationships between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl. Environ. Microbiol. 72:7602-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhand, N. K., J. A. L. M. Toribio, and R. J. Whittington. 2009. Adsorption of Mycobacterium avium subsp. paratuberculosis to soil particles. Appl. Environ. Microbiol. 75:5581-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingos, M., A. Amado, and A. Botelho. 2009. IS1245 RFLP analysis of strains of Mycobacterium avium subspecies hominissuis isolated from pigs with tuberculosis lymphadenitis in Portugal. Vet. Rec. 164:116-120. [DOI] [PubMed] [Google Scholar]
- 10.Dvorska, L., M. Bartos, G. Martin, W. Erler, and I. Pavlik. 2001. Strategies for differentiation, identification and typing of medically important species of mycobacteria by molecular methods. Veterinarni Medicina 46:309-328. [Google Scholar]
- 11.Dvorska, L., et al. 2007. Avian tuberculosis in naturally infected captive water birds of the Ardeideae and Threskiornithidae families studied by serotyping, IS901 RFLP typing, and virulence for poultry. Vet. Microbiol. 119:366-374. [DOI] [PubMed] [Google Scholar]
- 12.Dvorska, L., et al. 2004. Study of Mycobacterium avium complex strains isolated from cattle in the Czech Republic between 1996 and 2000. Vet. Microbiol. 99:239-250. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson, M., R. Bennet, and N. Danielsson. 2001. Non-tuberculous mycobacterial lymphadenitis in healthy children: another “lifestyle disease”? Acta Paediatr. 90:1340-1342. [DOI] [PubMed] [Google Scholar]
- 14.Espy, M. J., et al. 2006. Real-time PCR in clinical microbiology: applications for a routine laboratory testing. Clin. Microbiol. Rev. 19:165-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkinham, J. O., III. 2002. Nontuberculous mycobacteria in the environment. Clin. Chest Med. 23:529-551. [DOI] [PubMed] [Google Scholar]
- 16.Falkinham, J. O. 2009. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J. Appl. Microbiol. 107:356-367. [DOI] [PubMed] [Google Scholar]
- 17.Falkinham, J. O., M. D. Iseman, P. de Haas, and D. van Soolingen. 2008. Mycobacterium avium in a shower linked to pulmonary disease. J. Water Health 6:209-213. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, O., et al. 2001. Diptera as vectors of mycobacterial infections in cattle and pigs. Med. Vet. Entomol. 15:208-211. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, O. A., et al. 2004. Beetles as possible vectors of infections caused by Mycobacterium avium species. Vet. Microbiol. 102:247-255. [DOI] [PubMed] [Google Scholar]
- 20.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium-avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmsen, D., et al. 2003. RIDOM: comprehensive and public sequence database for identification of Mycobacterium species. BMC Infect. Dis. 3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inderlied, C. B., C. A. Kemper, and L. E. M. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansen, T. B., et al. 2007. New probes used for IS1245 and IS1311 restriction fragment length polymorphism of Mycobacterium avium subsp. avium and Mycobacterium avium subsp. hominissuis isolates of human and animal origin in Norway. BMC Microbiol. 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaevska, M., I. Slana, P. Kralik, and I. Pavlik. 2010. Examination of Mycobacterium avium subsp. avium distribution in naturally infected hens by culture and triplex quantitative real time PCR. Veterinarni Medicina 55:325-330. [Google Scholar]
- 25.Kazda, J., I. Pavlik, J. O. Falkinham III, and K. Hruska. 2009. The ecology of mycobacteria: impact on animal's and human's health. Springer, Bertlin, Germany.
- 26.Kubin, M., E. Wisingerova, J. Pekarek, and B. Prochazka. 1986. The patterns of complex and partially purified mycobacterial antigens in macrophage-migration inhibition testing. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 261:362-369. [DOI] [PubMed] [Google Scholar]
- 27.Kuth, G., J. Lamprecht, and G. Haase. 1995. Cervical lymphadenitis due to mycobacteria other than tuberculosis—an emerging problem in children. ORL J. Otorhinolaryngol. Relat. Spec. 57:36-38. [DOI] [PubMed] [Google Scholar]
- 28.Lim, S. Y., B. J. Kim, M. K. Lee, and K. Kim. 2008. Development of a real-time PCR-based method for rapid differential identification of Mycobacterium species. Lett. Appl. Microbiol. 46:101-106. [DOI] [PubMed] [Google Scholar]
- 29.Matlova, L., et al. 2004. Impact of sawdust and wood shavings in bedding on pig tuberculous lesions in lymph nodes, and IS1245 RFLP analysis of Mycobacterium avium subsp. hominissuis of serotypes 6 and 8 isolated from pigs and environment. Vet. Microbiol. 102:227-236. [DOI] [PubMed] [Google Scholar]
- 30.Mendum, T. A., B. Z. Chilima, and P. R. Hirsch. 2000. The PCR amplification of non-tuberculous mycobacterial 16S rRNA sequences from soil. FEMS Microbiol. Lett. 185:189-192. [DOI] [PubMed] [Google Scholar]
- 31.Mijs, W., et al. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505-1518. [DOI] [PubMed] [Google Scholar]
- 32.Moravkova, M., et al. 2008. Strategy for the detection and differentiation of Mycobacterium avium species in isolates and heavily infected tissues. Res. Vet. Sci. 85:257-264. [DOI] [PubMed] [Google Scholar]
- 33.Nieminen, T., et al. 2006. 16S rRNA targeted sandwich hybridization method for direct quantification of mycobacteria in soils. J. Microbiol. Methods 67:44-55. [DOI] [PubMed] [Google Scholar]
- 34.Pakarinen, J., et al. 2007. Proliferation of mycobacteria in a piggery environment revealed by Mycobacterium-specific real-time quantitative PCR and 16S rRNA sandwich hybridization. Vet. Microbiol. 120:105-112. [DOI] [PubMed] [Google Scholar]
- 35.Pavlik, I., P. Svastova, J. Bartl, L. Dvorska, and I. Rychlik. 2000. Relationship between IS901 in the Mycobacterium avium complex strains isolated from birds, animals, humans, and the environment and virulence for poultry. Clin. Diagn. Lab. Immunol. 7:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Primm, T. P., C. A. Lucero, and J. O. Falkinham. 2004. Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 17:98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy, V. C. K., et al. 2008. Mycobacterial culture of fine needle aspirate—a useful tool in diagnosing tuberculous lymphadenitis. Indian J. Med. Microbiol. 26:259-261. [DOI] [PubMed] [Google Scholar]
- 38.Reed, C., et al. 2006. Environmental risk factors for infection with Mycobacterium avium complex. Am. J. Epidemiol. 164:32-40. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes, G., et al. 2008. Detection of Mycobacterium immunogenum by real-time quantitative Taqman PCR. J. Microbiol. Methods 73:266-268. [DOI] [PubMed] [Google Scholar]
- 40.Shitaye, E. J., et al. 2009. Comparison of the conventional culture, the manual fluorescent MGIT system and the automated fluorescent MGIT 960 culture system for the detection of Mycobacterium avium ssp avium in tissues of naturally infected hens. Folia Microbiol. 54:137-141. [DOI] [PubMed] [Google Scholar]
- 41.Shitaye, J. E., et al. 2008. Diagnostic testing of different stages of avian tuberculosis in naturally infected hens (Gallus domesticus) by the tuberculin skin and rapid agglutination tests, faecal and egg examinations. Veterinarni Medicina 53:101-110. [Google Scholar]
- 42.Shitaye, J. E., et al. 2008. Mycobacterium avium subsp. avium distribution studied in a naturally infected hen flock and in the environment by culture, serotyping and IS901 RFLP methods. Vet. Microbiol. 127:155-164. [DOI] [PubMed] [Google Scholar]
- 43.Shitaye, J. E., et al. 2006. Mycobacterial and Rhodococcus equi infections in pigs in the Czech Republic between the years 1996 and 2004: the causal factors and distribution of infections in the tissues. Veterinarni Medicina 51:497-511. [Google Scholar]
- 44.Shrestha, N. K., et al. 2003. Detection and differentiation of Mycobacterium tuberculosis and nontuberculous mycobacterial isolates by real-time PCR. J. Clin. Microbiol. 41:5121-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slana, I., M. Kaevska, P. Kralik, A. Horvathova, and I. Pavlik. 2010. Distribution of Mycobacterium avium subsp. avium and M. a. hominissuis in artificially infected pigs studied by culture and IS901 and IS1245 quantitative real time PCR. Vet. Microbiol. 144:437-443. [DOI] [PubMed] [Google Scholar]
- 46.Slana, I., P. Kralik, A. Kralova, and I. Pavlik. 2008. On-farm spread of Mycobacterium avium subsp. paratuberculosis in raw milk studied by IS900 and F57 competitive real time quantitative PCR and culture examination. Int. J. Food Microbiol. 128:250-257. [DOI] [PubMed] [Google Scholar]
- 47.Tell, L. A., et al. 2003. Real-time polymerase chain reaction testing for the detection of Mycobacterium genavense and Mycobacterium avium complex species in avian samples. Avian Dis. 47:1406-1415. [DOI] [PubMed] [Google Scholar]
- 48.Trevors, J. T. 1996. Nucleic acids in the environment. Curr. Opin. Biotechnol. 7:331-336. [DOI] [PubMed] [Google Scholar]
- 49.van Coppenraet, E. S. B., et al. 2004. Real-time PCR assay using fine-needle aspirates and tissue biopsy specimens for rapid diagnosis of mycobacterial lymphadenitis in children. J. Clin. Microbiol. 42:2644-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Coppenraet, L. E. S. B., P. E. W. de Haas, J. A. Lindeboom, E. J. Kuijper, and D. van Soolingen. 2008. Lymphadenitis in children is caused by Mycobacterium avium hominissuis and not related to ‘bird tuberculosis’. Eur. J. Clin. Microbiol. Infect. Dis. 27:293-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Soolingen, D., et al. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 53.Wagner, D., and L. S. Young. 2004. Nontuberculous mycobacterial infections: a clinical review. Infection 32:257-270. [DOI] [PubMed] [Google Scholar]
- 54.Wolinsky, E., and W. B. Schaefer. 1973. Proposed numbering scheme for mycobacterial serotypes by agglutination. Int. J. Syst. Bacteriol. 23:182-183. [Google Scholar]
- 55.Wright, C. A., K. G. Hoek, B. J. Marais, P. van Helden, and R. M. Warren. 2010. Combining fine-needle aspiration biopsy (FNAB) and high-resolution melt analysis to reduce diagnostic delay in mycobacterial lymphadenitis. Diagn. Cytopathol. 38:482-488. [DOI] [PubMed] [Google Scholar]
- 56.Yajko, D. M., et al. 1995. Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:176-182. [PubMed] [Google Scholar]


