Abstract
We determined the association between DNA load and mortality in patients with Vibrio vulnificus infection. Real-time PCR performed on sera of 27 culture-positive patients showed a significantly higher median DNA load in nonsurvivors than in survivors. Hence, real-time PCR can be used as an early prognostic factor in V. vulnificus septicemia.
Vibrio vulnificus is a halophilic, virulent, Gram-negative bacillus that can cause primary septicemia, wound infection, and/or gastroenteritis and is acquired by consumption of raw fish or shellfish or exposure of wounds to contaminated seawater (5, 10, 13). The mortality rate in cases of V. vulnificus infection due to shellfish consumption is approximately 53%, but it is even higher (67%) in patients with liver disease (1). The mortality rate may increase to 100% in patients with septicemia if treatment is delayed by 72 h (11).
DNA hybridization, enzyme assays, and various PCR methods targeting V. vulnificus-specific genes have been used for diagnosis. Real-time PCR technology enables quantification of microorganisms (9, 16). An association between disease severity/mortality and bacterial DNA load measured by quantitative real-time PCR has been demonstrated for Neisseria meningitidis and Staphylococcus aureus infections (2, 6), but there has been no such study reported for patients with Vibrio septicemia. Our aim was to determine the association between disease-related mortality and bacterial DNA load in patients with V. vulnificus infection and whether bacterial load can be used as an early prognostic factor to predict the severity of disease.
One hundred forty-five patients aged ≥18 years with skin and soft-tissue infections (e.g., cellulitis, necrotizing fasciitis) at four university hospitals were included between 2006 and 2008. Blood sera of 22 of the V. vulnificus-infected patients in our previous study (9) were included in this study. Blood, skin, and soft tissues were cultured on admission.
Clinical isolates were initially identified with a Vitek II automated system (bioMérieux, Marcy l'Etoile, France). Serum was stored at −70°C, and DNA was extracted by using Qiagen blood mini kit, followed by real-time PCR targeting the V. vulnificus-specific toxR gene. The toxR gene was cloned as described by Takahashi et al. (14), using sequences in GenBank (accession no. AF170883). The primers used were tox-130 (TGTTCGGTTGAGCGCATTAA) and tox-200 (GCTTCAGAGCTGCGTCATTC), and the probe was (6-carboxyfluorescein-CGCTCCTGTCAGATTCAACCAACAACG-Black Hole Quencher-1) (9, 14). V. vulnificus infection was confirmed by identifying V. vulnificus in blood, skin, or soft tissue. This study was approved by the Ethics in Human Research Committee of each participating university hospital.
All statistical analyses were performed with SPSS version 17.0. Discrete variables were expressed as percentages of counts, and continuous variables were expressed as medians and interquartile ranges (IQRs). For continuous data, comparisons between groups were made with the Mann-Whitney test, with the association between bacterial DNA load and mortality being evaluated by the nonparametric Mann-Whitney U test. The correlation between bacterial DNA load and APACHE II score was assessed using the Spearman correlation coefficient. A P value of <0.05 was considered statistically significant.
V. vulnificus was identified by culture in 32 of the 145 patients. Real-time PCR was performed on sera obtained from 27 of these 32 patients on admission; the mean age of the 27 patients was 54.2 years, 22 were male (81.5%), and 13 died of septicemia (48.1% mortality). Additionally, 18 (66.7%) of the 27 had fever, and altered mentation was observed in 5, all of whom died.
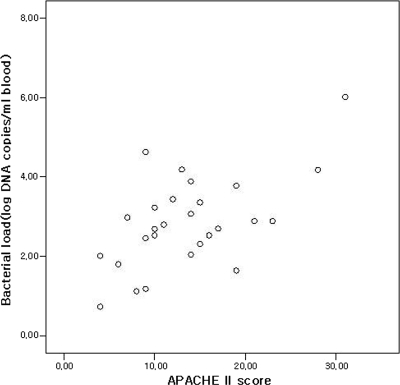
On admission, the median V. vulnificus DNA load in the 27 culture-positive patients was 625 copies per ml serum (IQR, 109 to 2,740). It was 2,300 copies/ml (IQR, 558 to 11,350; average of ranks, 18.04) in those who died and 316.5 copies/ml in the survivors (IQR, 51 to 705; average of ranks, 10.25). These data yielded a Mann-Whitney U value of 38.5 and a Wilcoxon W value of 143.5 (P = 0.011). On admission, the mean APACHE II score was 15 (IQR, 12.5 to 22) for those who died and 9 (IQR, 6.75 to 14) for the survivors (P = 0.002). Bacterial DNA load and disease severity showed a continuous positive correlation (Spearman's rank correlation, r = 0.450; P = 0.019; Fig. 1).
FIG. 1.
Correlation between V. vulnificus DNA loads and APACHE II scores at admission.
We used the area under the receiver operating characteristic curve as an overall prognostic measure for parameters. The accuracy of a fatal prognosis at the time of admission was 0.79 (95% confidence interval [CI], 0.59 to 0.92). At an optimal DNA cutoff value of 625, the sensitivity and specificity of a fatal prognosis were 77% (95% CI, 46 to 95%) and 79% (95% CI, 49 to 95%), respectively, with corresponding positive and negative likelihood ratios of 3.59 and 0.29. At a DNA cutoff value of 500, the corresponding values were 77% (95% CI, 46 to 95%) and 71% (95% CI, 42 to 91%), respectively. Ten of the 14 patients with >500 copies/ml died, whereas 10 of the 13 patients with ≤500 copies/ml survived. Only 2 of the 10 patients with >1,000 copies/ml survived, whereas 12 of the 17 patients with ≤1,000 copies/ml survived.
When the eight patients who were real-time PCR positive but culture negative were included, the median DNA load was 774 copies/ml (IQR, 136 to 4,340), i.e., 2,175 copies/ml (IQR, 504.7 to 9,267.5; average of ranks, 22.3) in those who died and 342 copies/ml (IQR, 82.5 to 977.5; average of ranks, 13.5) in the survivors. The median APACHE II score was 18 (IQR, 13.0 to 23.5) for those who died and 9 (IQR, 6.0 to 12.5) for the survivors (P = 0.0001).
Since V. vulnificus septicemia rapidly progresses with a subsequent mortality rate of ≥50% within a few days of disease onset, early diagnosis and treatment are mandatory (8). Thus, it is important to predict disease severity and outcome at the time of patient presentation.
There have been many studies of the association between the extent of bacteremia and the severity of the clinical picture (4, 17). Hall and Gold (4) reported that all of 20 patients with septic shock who had ≥100 CFU/ml of blood died, compared with only 41% of those who had <100 CFU/ml of blood. High-level bacteremia has been shown to correlate with a poor prognosis in Gram-negative septicemia (3, 15). Advances in molecular techniques have led to the use of real-time PCR for predicting disease severity and death (2, 6, 12). Ho et al. (6) found that the mecA DNA level among patients with methicillin-resistant S. aureus bacteremia was significantly higher in nonsurvivors than in survivors. Also, Iwaya et al. (7) demonstrated in a mouse infection model that real-time PCR was useful for accurately estimating Serratia marcescens cell numbers in blood and for predicting the severity of septicemia. However, no previous studies on the association between the bacterial DNA load and the severity of V. vulnificus septicemia or mortality have been done. In our study, we found a significant difference in the median DNA load at presentation between patients with V. vulnificus septicemia who died versus those who survived. In addition, we found that the V. vulnificus DNA load correlated with the APACHE II score. Hence, real-time PCR is useful for predicting the prognosis of V. vulnificus septicemia. A limitation of this study is that it included few clinical cases because of the rarity of this infection.
In conclusion, in this study, it was found that in patients with V. vulnificus septicemia, the V. vulnificus DNA level was significantly higher in nonsurvivors than in survivors. The results of this study suggest that real-time PCR can be used for early prediction of death due to V. vulnificus septicemia.
Acknowledgments
We do not have any commercial interest or other association that might pose a conflict of interest.
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Centers for Diseases Control and Prevention. 1993. Vibrio vulnificus infections associated with raw oyster consumption—Florida, 1981-1992. Centers for Diseases Control and Prevention, Atlanta, GA.
- 2.Darton, T., M. Guiver, S. Naylor, D. L. Jack, E. B. Kaczmarski, R. Borrow, and R. C. Read. 2009. Severity of meningococcal disease associated with genomic bacterial load. Clin. Infect. Dis. 48:587-594. [DOI] [PubMed] [Google Scholar]
- 3.DuPont, H. L., and W. W. Spink. 1969. Infections due to gram-negative organisms: an analysis of 860 patients with bacteremia at the University of Minnesota Medical Center, 1958-1966. Medicine (Baltimore) 48:307-332. [DOI] [PubMed] [Google Scholar]
- 4.Hall, W. H., and D. Gold. 1955. Shock associated with bacteremia; review of thirty-five cases. AMA Arch. Intern. Med. 96:403-412. [DOI] [PubMed] [Google Scholar]
- 5.Haq, S. M., and H. H. Dayal. 2005. Chronic liver disease and consumption of raw oysters: a potentially lethal combination—a review of Vibrio vulnificus septicemia. Am. J. Gastroenterol. 100:1195-1199. [DOI] [PubMed] [Google Scholar]
- 6.Ho, Y. C., S. C. Chang, S. R. Lin, and W. K. Wang. 2009. High levels of mecA DNA detected by a quantitative real-time PCR assay are associated with mortality in patients with methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol. 47:1443-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwaya, A., S. Nakagawa, N. Iwakura, I. Taneike, M. Kurihara, T. Kuwano, F. Gondaira, M. Endo, K. Hatakeyama, and T. Yamamoto. 2005. Rapid and quantitative detection of blood Serratia marcescens by a real-time PCR assay: its clinical application and evaluation in a mouse infection model. FEMS Microbiol. Lett. 248:163-170. [DOI] [PubMed] [Google Scholar]
- 8.Kim, D. M., Y. Lym, S. J. Jang, H. Han, Y. G. Kim, C. H. Chung, and S. P. Hong. 2005. In vitro efficacy of the combination of ciprofloxacin and cefotaxime against Vibrio vulnificus. Antimicrob. Agents Chemother. 49:3489-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, H. S., D. M. Kim, G. P. Neupane, Y. M. Lee, N. W. Yang, S. J. Jang, S. I. Jung, K. H. Park, H. R. Park, C. S. Lee, and S. H. Lee. 2008. Comparison of conventional, nested, and real-time PCR assays for rapid and accurate detection of Vibrio vulnificus. J. Clin. Microbiol. 46:2992-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klontz, K. C., S. Lieb, M. Schreiber, H. T. Janowski, L. M. Baldy, and R. A. Gunn. 1988. Syndromes of Vibrio vulnificus infections. Clinical and epidemiologic features in Florida cases, 1981-1987. Ann. Intern. Med. 109:318-323. [DOI] [PubMed] [Google Scholar]
- 11.Neupane, G. P., and D. M. Kim. 2010. In vitro time-kill activities of ciprofloxacin alone and in combination with the iron chelator deferasirox against Vibrio vulnificus. Eur. J. Clin. Microbiol. Infect. Dis. 29:407-410. [DOI] [PubMed] [Google Scholar]
- 12.Rolain, J. M., M. N. Mallet, P. E. Fournier, and D. Raoult. 2004. Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother. 54:538-541. [DOI] [PubMed] [Google Scholar]
- 13.Starks, A. M., T. R. Schoeb, M. L. Tamplin, S. Parveen, T. J. Doyle, P. E. Bomeisl, G. M. Escudero, and P. A. Gulig. 2000. Pathogenesis of infection by clinical and environmental strains of Vibrio vulnificus in iron-dextran-treated mice. Infect. Immun. 68:5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi, H., Y. Hara-Kudo, J. Miyasaka, S. Kumagai, and H. Konuma. 2005. Development of a quantitative real-time polymerase chain reaction targeted to the toxR for detection of Vibrio vulnificus. J. Microbiol. Methods 61:77-85. [DOI] [PubMed] [Google Scholar]
- 15.Vollberg, C. M., and J. L. Herrera. 1997. Vibrio vulnificus infection: an important cause of septicemia in patients with cirrhosis. South. Med. J. 90:1040-1042. [DOI] [PubMed] [Google Scholar]
- 16.Wolffs, P., R. Knutsson, B. Norling, and P. Radstrom. 2004. Rapid quantification of Yersinia enterocolitica in pork samples by a novel sample preparation method, flotation, prior to real-time PCR. J. Clin. Microbiol. 42:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagupsky, P., and F. S. Nolte. 1990. Quantitative aspects of septicemia. Clin. Microbiol. Rev. 3:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]