Abstract
Human rhinovirus species C (HRV-C) was the second most common HRV species detected in hospitalized patients in Italy with acute respiratory disease during a 1-year surveillance period. Sequencing of the picornavirus VP4/VP2 region allowed molecular typing of HRV-A and HRV-B and provisional typing of HRV-C.
In addition to human rhinovirus (HRVs) species A and B, a third HRV species, HRV-C, has been recently discovered. HRV-C has been repeatedly detected in association with lower respiratory tract infections (LRTI) (3, 5-8, 10, 11, 13, 18), and this finding has raised a great amount of interest regarding its potential pathological role in respiratory infections. Furthermore, several issues remain to be addressed, including the epidemiology of HRV-C infections in different regions of the world and the most suitable method for determining HRV-C species and type.
In this study, we examined the molecular signatures of the three HRV species circulating inside a hospital population during a prospective 1-year study in Northern Italy. Sequencing of VP4/VP2 coding genes was used for HRV typing (17) along with cis-acting replication element (cre) detection (2).
During the period 1 October 2008 to 30 September 2009, 1,500 respiratory samples were collected from 985 individuals, including both pediatric and adult patients, admitted to or staying in the hospital with a diagnosis of acute respiratory infection. Respiratory samples were aliquoted and handled as reported previously (14, 16). A panel of 17 respiratory viruses was investigated as reported previously (13), including HRVs (9) and human enteroviruses (HEVs) (21). Two methods were used for HRV molecular typing. The first allowed amplification of a 400-nucleotide (nt) fragment within the 5′ noncoding region (NCR) (4). The second method (17) amplified a region encompassing part of the 5′ NCR and the VP2 gene, thus yielding a 549-nt fragment (nt 534 to 1083). For HEV typing, two methods were used (12, 17). Primer sets utilized for amplification were also used for sequencing with the ABI PRISM Big Dye Terminator cycle sequencing reaction kit. Phylogenetic analysis was performed with MEGA 4.1 software (20); trees were constructed using the neighbor-joining method, and the Kimura-2 parameter was used for simultaneously estimating distance among sequences. Bootstrap values included 1,000 replicates.
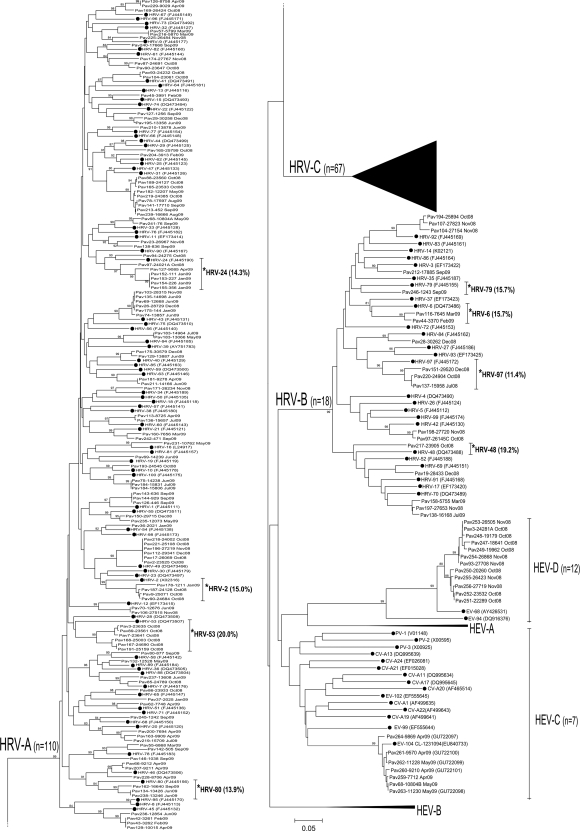
Overall, 835 of 1,500 samples (55.6%) were positive for one or more respiratory viruses, and 405 (27.0%) were positive for HRVs. HEVs were detected in 30 samples (2.0%). HRV infection occurred in 296 (30.1%) patients, for a total of 336 episodes. An episode was defined as a single respiratory event, whose duration was defined by the presence of the virus along with clinical symptoms. In parallel, 21 HEV strains from as many patients (2.1%) were identified. Of these, 12 were enterovirus (EV)-68, 7 were EV-104, 1 was echovirus-13, and 1 was not typed. The 336 episodes of HRV infection were investigated by phylogenetic analysis, which was performed by evaluating sequences of the VP4/VP2 region (Fig. 1), and, when the VP4/VP2 product was insufficient for sequencing, by examining the sequences of the 5′ NCR (5′ untranslated region [UTR]; data not shown). In the latter case, only the HRV species, and not the type, was reported. By using the VP4/VP2 sequencing method, 110 HRV episodes were attributed to species A (32.7%), 18 (5.4%) to species B, and 67 (19.9%) to species C, for a total of 195 episodes. As a general rule, we were able to sequence the VP4/VP2 region only in samples with viral load greater than 103 to 104 RNA copies/ml respiratory sample. In addition, by using 5′NCR sequencing, 43 episodes (12.8%) were attributed to species A, 13 (3.9%) to species B, and 26 (7.7%) to species C, for a total of 82 episodes. Thus, a total of 277/336 (82.4%) HRV episodes could be attributed to a single species.
FIG. 1.
A neighbor-joining phylogenetic tree analysis of the VP4/VP2 region of HRV (n = 195, accession numbers, HM366725 to HM366919) and HEV (n = 21, accession numbers HM370281 to HM370295; Pav260-Pav264, previously published accession numbers, GU722101 to GU722097) strains identified in Pavia, Italy. HEV branches were used to root the tree. Reference strains for HRV-A, HRV-B, HEV-C, and HEV-D were included in the tree reconstruction; these are indicated by black dots and the relevant accession numbers are in brackets. Strains showing more than 10% nucleotide divergence with respect to the reference strain (but less than 7% amino acid divergence) were marked with asterisks. Details of the HRV-C strains are shown in Fig. 2.
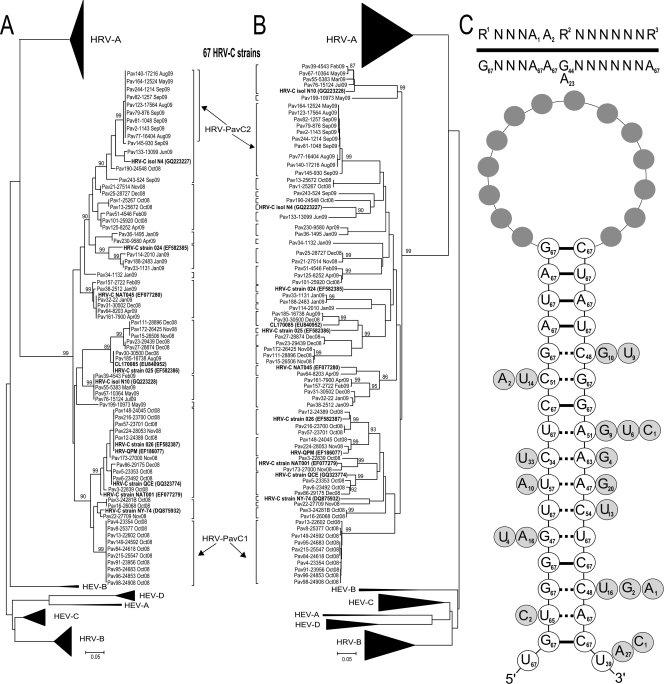
As for HRV-Cs, no definitive type identification has thus far been achieved. However, currently in GenBank the entire genome sequence is available for 11 strains. In this study (Fig. 2), the 67 HRV-C strains analyzed by sequencing of the VP4/VP2 region were classified using as a molecular cutoff both the 10% nucleotide (nt) divergence, as proposed by Wisdom et al. (23), and the 7% amino acid (aa) divergence, as proposed by Arden et al. (1). According to the former (nucleotide sequence) approach, the 67 HRV-C strains of this study clustered into 29 types (Fig. 2B), whereas according to the latter (amino acid) approach the same strains clustered into 13 types (Fig. 2A). In both trees, two clusters were clearly distinct, each including 10 different HRV-C strains. These clusters are referred to as HRV-PavC1 and HRV-PavC2 (Fig. 2). HRV-PavC1 includes 10 strains from as many patients, all circulating in October 2008. The second cluster (HRV-PavC2) included 10 HRV strains from 10 patients that circulated between May and September 2009. All HRV-C strains from this study were found to fall within the putative types already reported in GenBank. In our series, several HRV-A and HRV-B strains showed a nucleotide divergence greater than 10% with respect to the reference strain and an amino acid divergence of less than 7% (Fig. 1).
FIG. 2.
Phylogenetic tree based on amino acid (A) or nucleotide (B) HRV and HEV sequences. Eleven complete HRV-C genome sequences previously published with relevant accession numbers are in bold. HRV-PavC1 and HRV-PavC2 are two strain clusters temporally related to each other. (C) Predicted VP2 cre secondary structure of 67 HRV-C strains identified in Pavia. In the stem-loop, the nucleotide pairs joined by the thick line were detected in all 67 HRV-C strains (100%), as indicated in each circle. The other nucleotide pairs containing one variable nucleotide show the type(s) and, within the closed circle, the number of nucleotide substitutions detected in different strains. Above the loop, the conserved and variable nucleotides among the 14 nucleotides forming the loop are indicated along with the frequency of the variable nucleotide present in different strains.
In addition, as reported in Fig. 2C, the 67 HRV-C strains were assayed for the presence of the cre element. In all 67 strains, the cre motif was detected in the VP2 region, thus confirming that all these strains belong to the HRV-C species (2, 13). Thus, cre provides a distinctive molecular signature for differentiating HRV-C from HEVs (cre located in the 2C region), HRV-A (cre in the 2A region), and HRV-B (cre in the VP1 region).
By using a VP4/VP2 sequence divergence threshold of approximately 11% for HRV-A and 10% for HRV-B prototypes for distinguishing between intraserotype and interserotype strains, results of molecular typing of HRV-A and HRV-B strains entirely corresponded with those of serological typing (74 HRV-A and 25 HRV-B serotypes) (23). However, to date, HRV-C strains are not taxonomically classified into types, even though a similar level of molecular diversity (equal or greater than HRV-A and HRV-B) seems to exist among HRV-C strains or variants. Using a 10% nucleotide divergence as a threshold, in October 2009, 47 hypothetical HRV-C types were identified (23), and in December 2009 the number of potential types grew to 60 (22). In our study, all 67 HRV-C strains possessed sequences already reported in GenBank, as of 30 April 2010. In addition, while recombination events are known to be frequent among HEVs (15), analysis of the 5′ NCR of HRVs is consistent with multiple interspecies recombination events also in HRV evolution (22).
In conclusion, HRVs are the most common virus population circulating within a hospital-based population, and HRV-Cs circulate at a rate close to that of HRV-As. Sequencing of VP4/VP2 genes is required for HRV-C identification as well as cre detection in the VP2 region. However, VP1 gene sequencing is needed to genotypically assign types, as recently reported by Simmonds et al. (19).
Nucleotide sequence accession numbers.
HRV and HEV VP4/VP2 sequences have been submitted to GenBank under accession numbers HM366725 to HM366919 to HM366919 and HM370281 to HM370295, respectively.
Acknowledgments
We thank all the technical staff of the Virology Unit for careful preparation and handling of specimens. We are also indebted to Daniela Sartori for careful preparation of the manuscript and to Laurene Kelly for correction of the English.
This work was supported by the Ministero della Salute, Fondazione IRCCS Policlinico San Matteo, Ricerca Corrente (grants no. 80622 and 80557).
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Arden, K. E., C. E. Faux, N. T. O'Neill, P. McErlean, A. Nitsche, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2010. Molecular characterization and distinguishing features of a novel human rhinovirus (HRV) C, HRVC-QCE, detected in children with fever, cough and wheeze during 2003. J. Clin. Virol. 47:219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordey, S., D. Gerlach, T. Junier, E. M. Zdobnov, L. Kaiser, and C. Tapparel. 2008. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA 14:1568-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khetsuriani, N., X. Lu, W. G. Teague, N. Kazerouni, L. J. Anderson, and D. D. Erdman. 2008. Novel human rhinoviruses and exacerbation of asthma in children. Emerg. Infect. Dis. 14:1793-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiang, D., I. Kalra, S. Yagi, J. K. Louie, H. Boushey, J. Boothby, and D. P. Schnurr. 2008. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J. Clin. Microbiol. 46:3736-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamson, D., N. Renwick, V. Kapoor, Z. Liu, G. Palacios, J. Ju, A. Dean, K. St George, T. Briese, and W. I. Lipkin. 2006. Mass Tag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J. Infect. Dis. 194:1398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau, S. K., C. C. Yip, A. W. Lin, R. A. Lee, L. Y. So, Y. L. Lau, K. H. Chan, P. C. Woo, and K. Y. Yuen. 2009. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J. Infect. Dis. 200:1096-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau, S. K., C. C. Yip, H. W. Tsoi, R. A. Lee, L. Y. So, Y. L. Lau, K. H. Chan, P. C. Woo, and K. Y. Yuen. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 45:3655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linsuwanon, P., S. Payungporn, R. Samransamruajkit, N. Posuwan, J. Makkoch, A. Theanboonlers, and Y. Poovorawan. 2009. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J. Infect. 59:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu, X., B. Holloway, R. K. Dare, J. Kuypers, S. Yagi, J. V. Williams, C. B. Hall, and D. D. Erdman. 2008. Real-time RT-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46:533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McErlean, P., L. A. Shackelton, E. Andrews, D. R. Webster, S. B. Lambert, M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2008. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C). PLoS One 3:e1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, E. K., K. M. Edwards, G. A. Weinberg, M. K. Iwane, M. R. Griffin, C. B. Hall, Y. Zhu, P. G. Szilagyi, L. L. Morin, L. H. Heil, X. Lu, and J. V. Williams, New Vaccine Surveillance Network. 2009. A novel group of rhinoviruses is associated with asthma hospitalizations. J. Allergy Clin. Immunol. 123:98-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nix, W. A., M. S. Oberste, and M. A. Pallansch. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piralla, A., F. Rovida, G. Campanini, V. Rognoni, A. Marchi, F. Locatelli, and G. Gerna. 2009. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J. Clin. Virol. 45:311-317. [DOI] [PubMed] [Google Scholar]
- 14.Rovida, F., E. Percivalle, M. Zavattoni, M. Torsellini, A. Sarasini, G. Campanini, S. Paolucci, F. Baldanti, M. G. Revello, and G. Gerna. 2005. Monoclonal antibodies versus reverse transcription-PCR for detection of respiratory viruses in a patient population with respiratory tract infections admitted to hospital. J. Med. Virol. 75:336-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarasini, A., E. Percivalle, F. Rovida, G. Campanini, E. Genini, M. Torsellini, S. Paolucci, F. Baldanti, A. Marchi, M. G. Revello, and G. Gerna. 2006. Detection and pathogenicity of human metapneumovirus respiratory infection in pediatric Italian patients during a winter-spring season. J. Clin. Virol. 35:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83:333-340. [DOI] [PubMed] [Google Scholar]
- 18.Savolainen-Kopra, C., S. Blomqvist, T. Kilpi, M. Roivainen, and T. Hovi. 2009. Novel species of human rhinoviruses in acute otitis media. Pediatr. Infect. Dis. J. 28:59-61. [DOI] [PubMed] [Google Scholar]
- 19.Simmonds, P., C. L. McIntyre, C. Savolainen-Kopra, C. Tapparel, I. M. Mackay, and T. Hovi. 21 July 2010, posting date. Proposals for the classification of human rhinovirus species C into genotypically-assigned types. J. Gen. Virol. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed]
- 20.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 21.Van Doornum, G. J., M. Schutten, J. Voermans, G. J. J. Guldemeester, and H. G. M. Niesters. 2007. Development and implementation of real-time nucleic acid amplification for the detection of enterovirus infections in comparison to rapid culture of various clinical specimens. J. Med. Virol. 79:1868-1876. [DOI] [PubMed] [Google Scholar]
- 22.Wisdom, A., A. E. Kutkowska, E. C. McWilliam Leitch, E. Gaunt, K. Templeton, H. Harvala, and P. Simmonds. 2009. Genetics, recombination and clinical features of human rhinovirus species C (HRV-C) infections; interactions of HRV-C with other respiratory viruses. PLoS One 4:e8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wisdom, A., E. C. McWilliam Leitch, E. Gaunt, H. Harvala, and P. Simmonds. 2009. Screening respiratory samples for detection of human rhinoviruses (HRVs) and enteroviruses: comprehensive VP4-VP2 typing reveals high incidence and genetic diversity of HRV species C. J. Clin. Microbiol. 47:3958-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]