Abstract
HIV clinics in Canada provide care to an increasing number of patients born outside of Canada with HIV-1 non-B subtype infections. Because the Easy Q HIV-1 v1.2 assay (EQ; bioMérieux) failed to detect some non-B subtype infections, a multiassay HIV-1 viral load (VL) study was conducted with patients with diverse HIV subtype infections. Patients were enrolled from the Southern Alberta HIV Clinic (SAC), Calgary, Alberta, Canada (n = 349) and the McGill HIV Clinic (MHC), Montreal, Quebec, Canada (n = 20) and had four or five tubes of blood drawn for testing by EQ and three other commercial HIV VL assays: (i) the Versant 3.0 HIV-1 test, with the Versant 440 instrument (branched DNA [bDNA]; Siemens), (ii) the RealTime HIV-1 test, with the m2000rt instrument (m2000rt; Abbott Molecular Diagnostics), and (iii) the COBAS AmpliPrep TaqMan HIV-1 48 test (CAP-CTM; Roche Molecular Diagnostics). Blood was processed according to the individual manufacturer's requirements and stored frozen at −86°C. The HIV subtype was known for patients who had undergone HIV genotypic resistance testing (Virco, Belgium). Data analyses were done using standard statistical methods within Stata 9.0 (StataCorp, College Station, TX). A total of 371 samples were tested on 369 patients, of whom 291 (81%) had a Virco genotype result of B (195; 53%) or non-B (96; 26%) subtypes A to D and F to K, as well as circulating recombinant forms (CRFs) (i.e., CRF01_AE and CRF02_AG). Most (58/78; 74%) patients of unknown subtype were recent African emigrants who likely have non-subtype B infection. Overall bias was small in pairwise Bland-Altman plots, but the limits of agreement between assays were wide. Discordant viral load results occurred for 98 samples and were due to missing values, false negatives, and significant underquantification that varied by HIV subtype. Results were obtained for all 371 samples with m2000rt, but for only 357 (97%) with CAP-CTM, 338 (92%) with EQ, and 276 (75%) with bDNA due to errors/equipment failures. False-negative results (nondetection of viral RNA versus other assay results) occurred for all platforms, as follows: for m2000rt, 8 (2%) [B(4) and non-B(4) subtypes], CAP-CTM, 9 (2.5%) [B(6) and non-B(3) subtypes]; EQ, 20 (6%) [B(7) and non-B(13) subtypes]; bDNA, 5 (2%) [B(1) and C(4)]. EQ and bDNA had the highest rates of underquantification by ≥1.0 log10 copies/ml, mainly for HIV non-B subtypes. Performance significantly varied between HIV VL platforms according to subtype. HIV viral diversity in the population being tested must be considered in selection of the viral load platform.
Human immunodeficiency virus type 1 (HIV-1) plasma RNA quantitative viral load (VL) assays have become an essential tool not only in HIV care but also in understanding HIV pathogenesis (22, 23, 28, 37). Precision and reproducibility of measured plasma HIV-1 RNA levels and the absolute CD4 T-lymphocyte count are critical diagnostic tests for guiding patient care decisions (16-18, 22, 23). HIV viral load measurements are used to assess an individual's infectivity (25), to gauge their risks for disease progression (28), to monitor their response to highly active antiretroviral therapy (HAART) (47), and to assess the likelihood of emergence of viral resistance (5, 18, 34). Successful HAART is defined as suppression of plasma viremia and is clinically documented based on two sequential quantitative HIV-1 plasma RNA measurements that are below the lower quantification limit of an approved viral load assay (18). Clinical trials of novel antiretroviral drugs and regimens also use the difference between the pretreatment and end point HIV-1 plasma RNA level or the rate of decline of viral load to assess potency (27, 35, 43, 46).
Until the recent development of quantitative real-time PCR (qRT-PCR) commercial assays there was limited standardization of the performance and reporting of the results from different commercial manufacturers' HIV-1 viral load tests. Diagnostic limitations of the earlier assays included target detection of primarily HIV group M, subtype B viruses, variation between different molecular methods for performing quantification (i.e., nucleic acid signal branch amplification [NASBA], PCR end point detection, and branched DNA [bDNA] signal amplification), variable assay dynamic ranges, and nonstandardization of the HIV viral load level against an international quantification standard, which all resulted in the reporting of results in a nonstandardized format (i.e., in copies/ml versus log10 IU/ml) (26, 29). For these reasons, results of earlier versions of HIV-1 viral load assays for a given patient may not be readily comparable between different methods. Indeed, clinically significant (i.e., ≥0.5- to 1.0-log) differences in results have at times been reported between individual assay platforms across the dynamic ranges of these assays for the same samples (3, 4, 7, 8, 12, 13, 21, 31).
Newer qRT-PCR HIV-1 viral load assays have now been developed and are increasingly replacing these older assay methods in clinical laboratories. Evaluations of these qRT-PCR assays reported to date have shown an improved quantification both across a broader dynamic range (0 to >7.0 log10 IU/ml) and for a wider variety of HIV groups and subtypes (11, 36, 40). HIV viral load assays that use qRT-PCR methods have incorporated primers and probes to detect and quantify group M non-B subtype as well as group N and O viruses (11, 15, 19, 36, 39, 45), due to previously recognized assay problems stemming from the sequence diversity of such strains (1, 6, 20, 42). Only the Abbott Laboratories qRT-PCR HIV assay has successfully detected an HIV-1 group P infection (32). Limited clinical evaluations of currently available HIV-1 viral load assays have been performed with patients with diverse HIV subtype infections (11, 15, 19, 33, 36, 38, 40, 41), but there have been several recent reports on the clinical impact of underquantification of group M, non-subtype B infections (10, 14, 44).
HIV clinics in Canada are caring for an increasing number of patients born outside Canada with non-B subtype infections. The expansion of the HIV epidemic in countries where Canada has historic connections, as well as a change in immigration policy in 2001, has led to this increasing HIV viral diversity within Canada (1, 24). Collaboration between our laboratory and HIV clinicians recently identified several cases attending our clinic with non-subtype B infections where HIV-1 viral load levels were false negative or grossly underquantified based on the Easy Q v1.2 assay (bioMèrieux, Laval, Quebec, Canada) (21, 30). Due to concerns about the performance of our current HIV-1 viral load assay, we undertook a large multiassay comparison of the three FDA-approved qRT-PCR HIV viral load manufacturers' platforms and included bDNA for comparison of an earlier assay version still used by many laboratories in order to determine the optimal testing platform for our increasingly diverse patient population.
(This work was presented at the Conference on Retroviruses and Opportunistic Infections, 16 to 19 February 2010, San Francisco, CA.)
MATERIALS AND METHODS
Patient population.
This study was approved by the Conjoint Health Ethics Research Board, University of Calgary, and the McGill University Bioethics Committee. Patients were enrolled between October 2007 and 2008 in the Southern Alberta HIV Clinic, Calgary, Alberta, Canada, or the McGill HIV Clinic, Montreal, Quebec, Canada. We prioritized recruitment of patients with detectable viremia and with either known non-subtype B infection or birth outside Canada. Data on age, gender, country of birth, date of HIV seropositive results, absolute CD4 count at the time of the HIV viral load determination, HIV clinical status, current HIV antiretroviral medications, and HIV subtype were collected.
Sample collection.
After signing consent forms, patients had four or five EDTA tubes of blood drawn for HIV-1 viral load measurements by the four methods outlined below. Blood was transported within 4 h of collection to the Molecular Microbiology Laboratory at Calgary Laboratory Services (CLS) and immediately processed to obtain plasma according to each manufacturers' instructions. Plasma aliquots (1 to 2 ml) were stored frozen (−86°C) in labeled racks until testing to minimize the freeze-thawing of sample tubes, according to the manufacturer's instructions.
HIV-1viral load determinations.
A senior molecular laboratory technologist (T. Lloyd) with company training and certification on each platform performed all of the assays strictly in accordance with each manufacturers' instructions. All of the required equipment to perform all of the assays was either already in-house (EasyMAG/EasyQ) or brought on-site for sample testing. An HIV-1 RNA quantification panel from Health Canada that included standardized samples which contained a small, middle, and large amount of viral RNA representative of the dynamic range was run on each assay platform. No assay repeatability studies were performed, due to the limited amount of available plasma.
(i) EQ assay.
The NucliSens Easy Q v.1.2 assay (EQ; bioMérieux, Laval, Quebec, Canada) assay uses a molecular beacon targeted to the gag p24 gene to a perform qRT-PCR isothermal nucleic acid amplification of the sample RNA for comparison to internal quantification calibrators added to the reaction mixture. This assay detects HIV-1 group M A to D and F to K viruses. Purified RNA was obtained from a 1.0-ml plasma sample using the EasyMAG automated extractor, and qRT-PCR amplification and detection were done using the automated EasyQ instrument. The assay dynamic range is 1.60 to 6.7 log10 copies/ml for this amount of plasma.
(ii) m2000rt.
The RealTime HIV-1 assay (m2000rt; Abbott Molecular Diagnostics, Mississauga, Ontario, Canada) targets a conserved region of the pol-int genes and amplifies HIV-1 RNA using a partially double-stranded qRT-PCR method. A partial sequence of the pumpkin polymerase gene acts as the internal control. This assay detects HIV-1 group M A to D and F to K viruses and several circulating recombinant forms (CRFs), as well as group N, O, and P viruses. Purified RNA was obtained from a 0.6-ml plasma sample by using the m2000sp automated extractor, and qRT-PCR amplification and detection were done using the fully automated m2000rt instrument. The assay dynamic range is 1.60 to 7.0 log10 copies/ml for this amount of plasma.
(iii) CAP-CTM assay.
The COBAS AmpliPrep-COBAS TaqMan 48 HIV-1 assay (CAP-CTM; Roche Molecular Diagnostics, Laval, Quebec, Canada) targets a conserved region of the gag p41 gene and uses TaqMan differential fluorescence-tagged primers to amplify HIV-1 RNA. Additional fluorescence primers and probes detect quantification standards, and a partial sequence of the long terminal repeat region acts as the internal control. This assay detects HIV-1 group M A to D and F to K viruses and several CRFs. Purified RNA was obtained from a 1.0-ml plasma sample using the COBAS AmpliPrep and qRT-PCR amplification, and detection was performed using the fully automated COBAS TaqMan 48 instrument. The assay dynamic range is 1.6 to 7.0 log10 copies/ml for this amount of plasma.
(iv) bDNA assay.
The Versant HIV-1 RNA 3.0 assay (bDNA; Siemens Medical Solutions, Mississauga, Ontario, Canada) targets a highly conserved region of the pol gene, and several primers and probes are used to perform signal amplification of branched DNA across this region. This assay detects HIV-1 group M A to D and F to K viruses and several CRFs. Purified RNA was obtained from a 1.0-ml plasma sample, and qPCR amplification and detection in the bDNA assay were performed using the Versant 440 v.3.0 instrument. This assay does not use an internal control. The assay dynamic range is 1.7 to 5.7 log10 copies/ml for this amount of plasma.
Comparison of HIV-1 viral load tests.
The log10-transformed viral loads obtained using the four methods were assessed for agreement using pairwise and matrix Bland-Altman plots by employing the Stata 9.0 program (StataCorp, College Station, TX). The viral load level was considered to be zero where no viral RNA target was detected (TND). The viral load level was considered to be the limit of detection (LDL) when the amount of viral RNA detected was lower than the assay's LDL. False negatives were defined as a TND or no amplification result for a qRT-PCR assay where all of the other assays detected HIV-1 RNA above the LDL or where an assay gave a result that was below the LDL while all of the other assays gave an HIV-1 VL of ≥1.0 log10 copies/ml above this level. Falsely high results were defined as a viral load that was ≥1.0 log10 copies/ml higher than all other assay results. Poor performance occurred if there was more than one missing value, if one or more assay results were false negative, or if the viral load differed by two or more methods by 1 log10 copies/ml, because a 0.5-log10 or 3-fold increase or decrease in viral load is considered to be a minimal change in plasma viremia (16, 17). A P value of <0.05 was considered statistically significant.
RESULTS
Patient population.
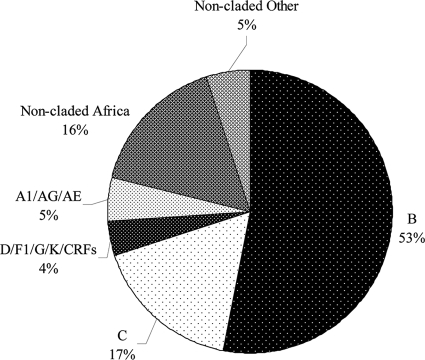
A total of 369 patients were enrolled, 347 (94%) from SAC and another 22 (6%) from the MHC. One-third of the patients were female (135; 37%). Patients had a mean age of 40.3 ± 10.9 years (± standard deviation) and a mean CD4 count of 424 ± 217 cells/mm3. Figure 1 outlines the distribution of HIV diversity for enrolled patients. The HIV-1 genotype resistance testing was available for 291 (79%) cases and included 195 (66%) B subtype and 96 (33%) non-B subtype infections. Of the 177 Canadian-born patients, 174 were confirmed to have HIV subtype B infections, while 2 had subtype C and 1 had a subtype AG infection. The other 21 patients with HIV subtype B infections were originally from European countries (15 patients), the United States (2 patients), South America/Mexico (2 patients), Southeast Asia (5 patients), Africa/Middle East (4 patients), or their origin was unknown (3 patients). Of the 96 documented non-B infections, 74 were from individuals originally from African countries, and other countries of origin (number of individuals in parentheses) were Southeast Asia (4), Canada (3), and Europe (1); these individuals had highly diverse subtype infections, including A1 (7), AG (11), AE (1), C (62), D (4), F1 (2), G (5), K (1), CRF02_AG (2), and CRF01_AE (1).
FIG. 1.
HIV-1 group M infections among study patients, by subtype.
A total of 78 (21%) cases had an unknown genotype because viral load was undetectable on HAART without any preceding testing. Fifty-eight (74%) were presumed to have non-subtype B infection because they had recently emigrated from Africa.
Viral load distributions.
Overall, the population tested was representative of the entire dynamic range, and the distribution of the quantitative values fit a normal curve for all of the assays (data not shown). HIV-1 viral load values for both the m2000rt (mean, 4.0 ± 1.1 log10 copies/ml) and CAP-CTM (mean, 4.0 ± 1.2 log10 copies/ml) assays were slightly higher than those obtained for either EQ (mean, 3.8 ± 1.0 log10 copies/ml) or bDNA (mean, 3.8 ± 0.9 copies/ml).
Viral load agreement.
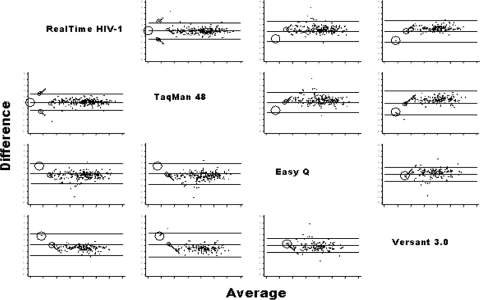
All of the assays showed good agreement across their dynamic ranges compared to the Health Canada HIV-VL standard panel, but the tightest correlation occurred with the m2000rt assay (r = 0.95) (data not shown). A matrix comparison of Bland-Altman pairwise agreement plots showing the quantification differences between each assay is provided in Fig. 2. Although the overall bias was small in these pairwise comparisons, the limits of agreement were wide, as shown in Table 1.
FIG. 2.
Matrix of Bland-Altman plots comparing the pairwise differences between different HIV-1 viral load assays.
TABLE 1.
Mean differences and limits of agreement among the four HIV viral load tests
| Tests | Mean difference (bias) | CI | Limits of agreement |
|---|---|---|---|
| m2000rt vs CAP-CTM | −0.035 | −0.111 to 0.041 | −1.492, 1.422 |
| m2000rt vs EQ | −0.101 | −0.194 to 0.008 | −1.836, 1.634 |
| CAP-CTM vs EQ | −0.036 | −0.132 to 0.061 | −1.814, 1.634 |
| m2000rt vs bDNA | −0.151 | −0.262 to −0.041 | −2.019, 1.716 |
| CAP-CTM vs bDNA | −0.129 | −0.255 to −0.003 | −2.207, 1.949 |
| EQ vs bDNA | −0.012 | −0.087 to 0.063 | −1.218, 1.195 |
Discrepancies in assays were due both to false negatives as well to significant differences in underquantification of viral load by some assays and also missing values, and these varied by subtype (Table 2). Results were obtained for all 371 samples with m2000rt, but for only 357 (97%) for CAP-CTM, 338 (92%) for EQ, and 276 (75%) for bDNA due to errors/equipment failures. A total of 98 (26%) samples showed discordant results; 6 (6%) had missing values, 38 (39%) samples were false negative by one or more of the assays, and another 50 (51%) samples were underquantified by ≥1.0 log10 copies/ml. Both the m2000rt and CAP-CTM assays showed excellent agreement between the overall number of samples that demonstrated undetectable viral load levels (i.e., TND or <LDL) (Table 2). Overall, both the m2000rt and CAP-CTM assays also had low rates of false-negative results and underquantified a small number of samples by ≥1.0 log10 copies/ml compared to the other two assays. EQ demonstrated the most discrepancies for the qRT-PCR assays, both in the number of false-negative results and in underquantification of samples. Although the bDNA assay had a low number of false negatives, a far higher number of samples were underquantified by this qPCR assay compared to either the m2000rt or CAP-CTM assays. Five samples were falsely high, including the CAP-CTM (one sample), EQ (two samples), and bDNA (two samples) assays.
TABLE 2.
Performance of HIV-1 viral load assays according to HIV subtype
| Parameter | No. (%) of patients with indicated result based on: |
|||
|---|---|---|---|---|
| m2000rt | TaqMan 48 | NucliSens Easy Q v1.2 | Versant 3.0 | |
| No. of overall results obtained (n = 369 patients) | 371 (100) | 357 (97) | 338 (92) | 276 (75) |
| TND or no amplificationa | 74 (20) | 69 (19) | 23 (7) | NA |
| Subtype B | 12 (16) | 10 (14) | 13 (57) | |
| Non-subtype B | 62 (84) | 59 (86) | 10 (43) | |
| <LDL log10 copies/ml | 28 (7.5) | 24 (7) | 74 (22) | 109 (39) |
| Subtype B | 7 (25) | 7 (29) | 16 (22) | 20 (18) |
| Non-subtype B | 21 (75) | 17 (71) | 58 (88) | 89 (82) |
| False-negative resultb (n = 38 samples) | 8 (2) | 9 (2.5) | 20 (6) | 5 (2) |
| Subtype B | 4 (50) | 6 (67) | 7 (35) | 0 |
| Non-subtype B | 4 (50) | 3 (33) | 13 (65) | 5 (100) |
| Underquantified result ≥1.0 log10 copies/mlc (n = 50 samples) | 5 (1.3) | 7 (2) | 15 (4.4) | 26 (9.4) |
| Subtype B | 2 (25) | 3 (43) | 4 (27) | 5 (19) |
| Non-subtype B | 3 (75) | 4 (57) | 11 (73) | 21 (81) |
| Underquantified result ≥0.5 and <1.0 log10 copies/mlc (n = 95 samples) | 23 (9) | 20 (6) | 61 (18) | 68 (25) |
| Subtype B | 11 (48) | 13 (65) | 26 (43) | 26 (38) |
| Non-subtype B | 12 (52) | 7 (35) | 35 (57) | 42 (62) |
| Falsely high (VL ≥1.0 log10 copies/ml [n = 4]) | None | 1 CRF02_AG | 1 C | 2 (1) (1 B and 1 AE) |
| Total discordant resultsd | 13 (3.5) | 17 (5) | 36 (11) | 33 (12) |
All of the real-time assay results are reported as target not detected (TND) or no amplification (EQ) result if no HIV-1 RNA was detected. NA, not applicable.
Defined as a TND result for a real-time assay where all of the other assays detected HIV-1 RNA above the LDL or where an assay gave a result <LDL while all of the other assays gave an HIV-1 VL of > 2.0 log10 copies/ml higher. Three samples were false negative by more than one assay.
A total of 144 samples were underquantified by one or more assays as follows (number of samples underquantified in parentheses): m2000rt (8), CAP-CTM (8), EQ (40), bDNA (36), m2000rt + EQ (2), m2000rt + CAP-CTM (2), m2000rt + bDNA (10), CAP-CTM + EQ (7), CAP-CTM + bDNA (2), EQ + bDNA (22), CAP-CTM + EQ + bDNA (3), and m2000rt + EQ + bDNA (4).
Results include false negatives, falsely high results, and results underquantified by ≥1.0 log10 copies/ml. The total number of results for each assay was used to calculate the percentages.
Individual comparisons of the number of false-negative and underquantified results between each assay showed the following. (i) For m2000rt versus CAP-CTM, eight (2.2%) samples were not detected by m2000rt while 9 (2.5%) other samples were not detected by CAP-CTM. m2000rt underquantified 23 (22%) samples by ≥0.5 log10 copies/ml, while CAP-CTM underquantified 17 (8%) other samples; 5/23 m2000rt and 3/17 (18%) CAP-CTM results were discrepant by ≥1.0 log10 copies/ml. The target was not detected by one assay, while viral RNA was detected at the limit of detection for the other assay in 29 samples (m2000rt [15] versus CAP-CTM [14]).
(ii) m2000rt versus EQ.
EQ gave no amplification for 14 samples that by m2000rt contained ≥2.0 log10 copies/ml. EQ also underquantified 72 samples by ≥0.5 log10 copies/ml, while m2000rt underquantified 14 other samples; 18/72 (25%) EQ and 2/14 (14.3%) m2000rt results were discrepant by ≥1.0 log10 copies/ml. m2000rt did not detect the target when viral RNA was detected at the limit of detection by EQ in 72 samples.
(iii) m2000rt versus bDNA.
Both the m2000rt and bDNA assays gave false negatives, including three (1.1%) samples that were not detected by bDNA and one other sample that was not detected by m2000rt. bDNA underquantified 73 samples by ≥0.5 log10 copies/ml; 12/73 (16.5%) bDNA results were discrepant by ≥1.0 log10 copies/ml. m2000rt did not detect the target when viral RNA was detected at the limit of detection by bDNA in 63 samples.
(iv) CAP-CTM versus EQ.
CAP-CTM results were false negative in two samples, while EQ gave no amplification for 11 samples that by CAP-CTM were found at ≥2.0 log10 copies/ml. EQ also underquantified 77 samples by ≥0.5 log10 copies/ml, while CAP-CTM underquantified 12 other samples; 22/77 (28.5%) EQ and 2/12 (16.7%) CAP-CTM viral load measurements were discrepant by ≥1.0 log10 copies/ml. CAP-CTM did not detect the target when viral RNA was detected at the limit of detection by EQ in 65 samples.
(v) CAP-CTM versus bDNA.
Five samples were not detected by CAP-CTM, while four samples were not detected by bDNA. CAP-CTM underquantified six samples by ≥0.5 log10 copies/ml, while bDNA underquantified 79 other samples; 20/79 (25.3%) bDNA measurements but none of the CAP-CTM viral load measurements was discrepant by ≥1.0 log10 copies/ml. CAP-CTM did not detect the target when viral RNA was detected at the limit of detection by bDNA in 64 samples.
(vi) EQ versus bDNA.
Three samples were not detected by EQ, while two samples were not detected by bDNA. EQ underquantified 19 samples by ≥0.5 log10 copies/ml, while 43 other samples were underquantified by bDNA; 4/19 (21%) EQ and 5/43 (11.6%) bDNA results were discrepant by ≥1.0 log10 copies/ml. Both assays detected viral RNA at the limit of detection in 90 samples.
Assay performance according to HIV-1 subtype.
The overall performance of each assay according to HIV subtype is shown in Table 2. m2000rt had a similar number of false-negative results as CAP-CTM, but for different samples. A total of 17 samples were false negative by either the m2000rt (8 [2%] samples; B [4 samples], C [1 sample], CRF_01AE [1 sample], and non-B [2 samples] subtypes) or CAP-CTM (9 [2.5%] other samples; B [6 samples] and non-B subtypes [3 samples]). Both of these assays had higher rates of target nondetection for subtype B patients (10 [59%]) compared to non-B subtype patients 7 [41%]). A similar rate of underquantification was also found for these qRT-PCR assays, but for different samples. m2000rt underquantified five samples (B [two samples], C [one sample], CRF02_AG [one sample], and non-B [one sample] subtypes), while the CAP-CTM underquantified seven other samples (B [one sample], C [one sample], G [one sample], AE [one sample], F1 [one sample], and non-B [two samples] subtypes) by ≥ 1.0 log10 copies/ml. Both of these assays had a higher rate of viral load discrepancy by ≥0.5 < 1.0 log10 copies/ml for subtype B (24/43; 56%) than for non-subtype B infections (19/43; 44%). A total of 43 samples were underquantified by either the m2000rt (23 [9%]; B [11], C [11], and non-B [1]) or CAP-CTM (20 [6%]; B [13], C [1], G [1], AG [2], and non-B [3] subtypes). CAP-CTM was falsely high for one CRF02_AG sample.
EQ had a much higher rate of false-negative and underquantified results than either of the other qRT-PCR assays (Table 2). A total of 20 (6%) patients had false-negative viral load results according to EQ, including 7 subtype B and 13 non-subtype B infections (C [5], CRF01_AE [1], AG [2], and non-B [5] subtypes). EQ also underquantified 15 samples by ≥1.0 log10 copies/ml, including B (4), AG (1), A1 (1), C (3), D (1), G (2), and non-B (2) subtypes. Another 61 samples were underquantified by ≥0.5 but <1.0 log10 copies/ml, including B (26), C (21), D (3), A1 (3), AG (3), G (1), and non-B (4) subtypes. EQ was falsely high for one subtype C sample.
bDNA had a low rate of false negatives (n = 5), but four of the patients had subtype C infections and only one had a subtype B infection. This assay also underquantified 26 (9.4%) samples by ≥1.0 log10 copies/ml, including B (5), C (14), D (1), A1 (1), AG (1) and non-B (4) subtypes. The 68 samples underquantified by bDNA by ≥0.5 but <1.0 log10 copies/ml included B (26), C (26), D (2) A1 (1), AG (3), F1 (1), K (1), CRF02-AG (1) and non-B (7) subtypes.
DISCUSSION
To the best of our knowledge this study is one of the largest comparative evaluations to date of the four commercially available HIV-1 viral load assays in a population of patients with diverse HIV-1 subtype infections. Although no patients with HIV-1 group O, N, or P infections were included, we did include the most common group M subtypes as well as CRFs reported globally. Genetic variation in HIV-1 subtypes or extreme divergence within an HIV subtype may significantly affect the ability to detect and quantify the viral RNA in clinical specimens. Either nondetection or underquantification of HIV-1 plasma viremia, particularly at the lower dynamic range of viral load assays, has the potential to cause serious clinical errors in making accurate decisions about the use or changes in antiretroviral therapy.
Our study shows that none of the assays detected or accurately quantified all of the samples within the accepted clinical range required. Although m2000rt (eight samples) and CAP-CTM (nine samples) had low rates of false negatives, the nondetected samples were different for each assay, likely reflections of primer and probe mismatches in the pol-int and gag target regions, respectively. However, both of these qRT-PCR assays had a much lower rate of false negatives or underquantification of samples by ≥1.0 log10 copies/ml than did EQ. Although bDNA had a low number of false negatives, this assay had a higher rate of underquantification than the three qRT-PCR assays. Some interesting discrepancies were found in the abilities of these assays to detect and quantify HIV subtype B and non-B subtype infections. Most false-negative results by either m2000rt or CAP-CTM occurred for patients with subtype B infections (10/17; 59%), while most of the underquantification with either of these assays occurred for non-B subtypes (8/12; 67%). However, the EQ and bDNA assays mainly gave false-negative results (18/25; 72%) or underquantified the results (32/41; 78%) by ≥1.0 log10 copies/ml for patients with non-B subtype infections. However, some subtype B strains were also not detected by EQ or were underquantified by both assays. Although a wide range of non-B subtypes were not detected or were underquantified by the EQ and bDNA tests, EQ had difficulty with subtype C, A1, AG, G, and CRF02-AG templates, while bDNA had difficulty with subtype C templates. This finding confirms the previously reported difficulties these platforms have with particular HIV-1 viral templates (18, 22, 34). Although the EQ assay also uses qRT-PCR detection, the lower correlation with either m2000rt or CAP-CTM results may be due to a combination of instability in the method (i.e., molecular beacon approach) as well as mismatches between the probe and primers and the gag target region, particularly when presented with a diversity of HIV-1 viral strains. Our results confirm the concern that commercial qRT-PCR assays utilizing a molecular beacon (EQ) and to some extent TaqMan qRT-PCR methods may not be as reliable as other molecular strategies for measuring highly polymorphic targets (18, 41).
Previous studies of the newer qRT-PCR assays have shown similar but more limited results when testing patients with diverse types of HIV-1 infection, but none has studied the performance of all of the currently available commercial assays. Most recently, Holguin et al. (19) compared the performance of Versant v3.0 (bDNA), CAP-CTM, and EQ in testing 83 plasma specimens from patients infected with HIV-1 non-B subtypes and recombinants as defined by phylogenetic analysis of the pol gene. Only 32 (58.2%) samples from naïve patients were quantified by the three methods; EQ gave the highest HIV RNA values (mean, 4.87 log10 copies/ml), and bDNA gave the lowest values (mean, 4.16 log10 copies/ml). Viremia differences of ≥1.0 log10 copies/ml were found in 8 of 55 (14.5%) specimens, occurring in 10.9, 7.3, and 5.4%, respectively, of the specimens in comparisons of bDNA versus EQ, bDNA versus CAP-CTM, and CAP-CTM versus EQ. Differences greater than 0.5 logs, considered significant for clinicians, occurred in 45.4, 27.3, and 29% when the same assays were compared. Some HIV-1 strains, including subtype G and CFR02_AG, showed more distinct discrepancies in comparing the results from the different assay platforms. In another recent study, Gueudin et al. (15) compared the performance of the Abbott m2000rt and Roche CAP-CTM assay platforms for their capacities to quantify various HIV-1 subtypes. The systems were tested on culture supernatants belonging to HIV-1 group M and group O and HIV-2 as well as on patient samples infected with HIV-1 group M and other strains. The m2000rt system quantified all 29 HIV-1 group M supernatants and 7/8 group O viruses, while the CAP-CTM system didn't detect 1 CRF02 strain. Both of these assays did not detect any of the HIV-2 strains. Four samples were underquantified by CAP-CTM by >1 log10 copies/ml. Another recent study by Rouet et al. (46) compared a generic HIV viral load assay with EQ and the Amplicor HIV-1 Monitor v1.5 assay for quantification of non-B subtypes in a resource-limited setting (i.e., the Kesho Bora preparatory study). Although the generic assay detected all of the non-B subtypes, nine samples were not detected by either the EQ (n = 2) or Monitor (n = 7) assays.
Clinical laboratories should implement one of the newer qRT-PCR methods (i.e., m2000rt or CAP-CTM) for optimal performance of HIV viral load testing of patients with subtype B as well as patients with non-B subtype infections. According to the June 2010 College of American Pathologists (CAP) HIV viral load (HIV-B) proficiency testing survey, there are still 107/395 (27.1%) participant laboratories that continue to use either the Roche Monitor 1.5 standard/ultrasensitive test (Amplicor, COBAS, or COBAS AmpliPrep) or bDNA (Versant v3.0 340 or 440) (9). Our study shows that both the EQ and bDNA assays are considerably less reliable for accurate viral load measurements across HIV subtypes. However, even when HIV-1 viral load testing is routinely performed using one of the new FDA-cleared HIV-1 viral load tests (i.e., Abbott's m2000 or the Roche CAP-CTM assay), discrepant results may not be apparent unless the test result is correlated to the patient's HIV status and treatment history. If HIV-1 viral load testing is centralized in large clinical laboratories that do not have a close clinical collaboration with the treating physician or clinic, such discordance may be missed. HIV viral load testing laboratories frequently do not know the HIV-1 subtype of the patient being tested. Discrepant HIV-1 results are therefore most often not identified by the laboratory but by the physician, who may or may not notify the laboratory of their concerns (they simply repeat the test). In our experience, most discrepant HIV-1 results (i.e., subtype B or non-B subtype infections) could not be easily identified by the laboratory for routine send-out to a reference laboratory for further testing.
Further evaluation should be done on an even higher number of clinical samples from patients with long-standing subtype B or known non-B subtype infection, using all commercially licensed HIV-1 viral load assays. As HIV strains continue to diverge within a given geographic area as well as globally, it will become increasingly important for clinical laboratories to establish the viral genotype causing infection in their test population. Patients with documented non-B subtype infection as well as those with extreme divergence within subtype B may require HIV viral load testing on a different platform if their results seem discordant with either their clinical presentation or status. Discordant samples should be retested using a second HIV-1 viral load assay that targets an alternate gene region (i.e., gag if a pol-based assay was initially used, and vice versa). Molecular sequencing studies should also characterize mismatches between an assay's probes and primers and the HIV subtype or strain target template. Periodic evaluation of assays may be necessary to assess their performance in detecting and accurately quantifying divergent HIV subtypes, in order to provide optimal performance for patients in both developed and resource-poor countries.
Acknowledgments
This work was funded by a peer-reviewed grant from Calgary Laboratory Services.
EQ assays were run using CLS equipment and reagents. Abbott Molecular Diagnostics, Roche Molecular Diagnostics, and Siemens Medical Solutions, Canada, supported this study by providing company training and certification to T.L. and by supplying the required equipment and reagent kits. Abbott Canada also provided funding to the two clinics (SAC and MHC) to support patient recruitment and the procurement of blood samples.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Akouamba, B. S., et al. 2005. HIV-1 genetic diversity in antenatal cohort, Canada. Emerg. Infect. Dis. 11:1230-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alaeus, A., K. Lidman, A. Sonnerborg, and J. Albert. 1997. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS 11:859-865. [DOI] [PubMed] [Google Scholar]
- 3.Amendola, A., et al. 2002. Under evaluation of HIV-1 plasma viral load by a commercially available assay in a cluster of patients infected with HIV-1 A/G circulating recombinant form (CRF02). Acquir. Immune Defic. Syndr. 31:488-494. [DOI] [PubMed] [Google Scholar]
- 4.Antunes, R., et al. 2003. Evaluation of the clinical sensitivities of three viral load assays with plasma samples from a pediatric population predominantly infected with human immunodeficiency virus type 1-subtype G and BG recombinant forms. J. Clin. Microbiol. 41:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldanti, F., et al. 2010. Early emergence of raltegravir resistance mutations in patients receiving HAART salvage regimens. J. Med. Virol. 82:116-122. [DOI] [PubMed] [Google Scholar]
- 6.Burgisser, P., et al. 2000. Performance of five different assays for the quantification of viral load in persons infected with various subtypes of HIV-1. Swiss HIV Cohort Study. J. Acquir. Immune Defic. Syndr. 23:138-144. [DOI] [PubMed] [Google Scholar]
- 7.Chew, C. B., et al. 1999. Comparison of three commercial assays for the quantification of plasma HIV-1 RNA from individuals with low viral loads. AIDS 13:1977-1978. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, J. R., et al. 2000. Comparative quantification of diverse serotypes of HIV-1 in plasma from a diverse population of patients. J. Med. Virol. 62:445-449. [DOI] [PubMed] [Google Scholar]
- 9.College of American Pathologists. 2010. Participant summary, HIV-B viral load survey, June 2010. College of American Pathologists, Chicago, IL.
- 10.Colson, P., et al. 2007. Impaired quantification of plasma HIV-1 RNA with a commercialized real time PCR assay in a couple of HIV-1-infected individuals. J. Clin. Virol. 39:226-229. [DOI] [PubMed] [Google Scholar]
- 11.Crump, J. A., et al. 2009. Evaluation of the Abbott m2000rt RealTime HIV-1 assay with manual sample preparation compared with the Roche COBAS AmpliPrep/Amplicor HIV-1 Monitor v1.5 using specimens from East Africa. J. Virol. Methods 162:218-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debyser, Z., et al. 1998. Failure to quantify viral load with two of the three commercial methods in a pregnant woman harboring an HIV type 1 subtype G strain. AIDS Res. Hum. Retroviruses 14:453-459. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, B. S., et al. 2004. Measurement of HIV RNA in patients infected by subtype C by assays optimized for subtype B results in an underestimation of the viral load. J. Med. Virol. 73:167-171. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman, B. S., et al. 2006. Comparative performance of the Amplicor HIV-1 Monitor assay versus NucliSens EasyQ in HIV subtype C-infected patients. J. Med. Virol. 78:883-887. [DOI] [PubMed] [Google Scholar]
- 15.Gueudin, M., et al. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500-505. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, R. K., et al. 2009. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. Lancet Infect. Dis. 9:409-417. [DOI] [PubMed] [Google Scholar]
- 17.Hammer, S. M., et al. 2008. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society USA panel. JAMA 300:555-570. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch, M. S., et al. 2008. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society USA panel. Top. HIV Med. 16:266-285. [PubMed] [Google Scholar]
- 19.Holguin, A., M. Lopez, M. Molinero, and V. Soriano. 2009. Performance of three commercial viral load assays: Versant human immunodeficinecy virus type 1 (HIV-1) RNA bDNA v3.0, COBAS AmpliPrep/Cobas TaqMan HIV-1, and NucliSens HIV-1 EasyQ v1.2, testing HIV-1 non-B subtypes and recombinant variants. J. Clin. Microbiol. 46:2918-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenny-Avital, E. R., and S. T. Beatrice. 2001. Erroneously low or undetectable plasma human immunodeficiency virus type 1 (HIV-1) ribonucleic acid load, determined by polymerase chain reaction, in West African and American patients with non-B subtype HIV-1 infection. Clin. Infect. Dis. 32:1227-1230. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. E., et al. 2007. Short communication: identification of a novel HIV type 1 subtype H/J recombinant in Canada with discordant HIV viral load (RNA) values in three different commercial assays. AIDS Res. Hum. Retroviruses 23:1309-1313. [DOI] [PubMed] [Google Scholar]
- 22.Korenromp, E. L., B. G. Williams, G. P. Schmid, and C. Dye. 2009. Clinical prognostic value of RNA viral load and CD4 cell counts during untreated HIV-1 infection: a quantitative review. PLoS One 4:e5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kredo, T., J. S. Van der Walt, N. Siegfried, and K. Cohen. 2009. Therapeutic drug monitoring of antiretrovirals for people with HIV. Cochrane Database Syst. Rev. 3:CD007268. [DOI] [PubMed] [Google Scholar]
- 24.Krentz, H., and M. J. Gill. 2009. The five-year impact of an evolving global epidemic, changing migration patterns, and policy changes in a regional Canadian HIV population. Health Policy 90:296-302. [DOI] [PubMed] [Google Scholar]
- 25.Kumwenda, N. I., et al. 2008. Extended antiretroviral prophylaxis to reduce breast-milk HIV-1 transmission. N. Engl. J. Med. 359:119-129. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S., et al. 2006. Development and evaluation of HIV-1 subtype RNA panels for the standardization of HIV-1 NAT assays. J. Virol. Methods 137:287-291. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz, M., et al. 2009. Sustained antiretroviral effect of raltegravir after 96 weeks of combination therapy in treatment-naive patients with HIV-1 infection. J. Acquir. Immune Defic. Syndr. 52:350-356. [DOI] [PubMed] [Google Scholar]
- 28.Mellors, J. W., et al. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 29.Michael, N. L., et al. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved Amplicor HIV-1 Monitor test with isolates of diverse subtypes. J. Clin. Microbiol. 37:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Bryan, T., T. Jadavji, J. Kim, and M. J. Gill. 2010. A “perfect storm” leading to an avoidable mother to child HIV transmission (MTCT). Can. Med. Assoc. J. doi: 10.1053/CMAJ.091137. [DOI]
- 31.Parekh, B., et al. 1999. Impact of HIV type 1 subtype variation on viral RNA quantitation. AIDS Res. Hum. Retroviruses 15:133-142. [DOI] [PubMed] [Google Scholar]
- 32.Plantier, J. C., et al. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871-872. [DOI] [PubMed] [Google Scholar]
- 33.Rouet, F., et al. 2010. Comparison of the generic HIV viral load assay with the Amplicor HIV-1 Monitor v. 1.5 and NucliSens HIV-1 EasyQ v1.2 techniques for plasma HIV-1 RNA quantitation of non-B subtypes: the Kesho Bora preparatory study. J. Virol. Methods 163:253-257. [DOI] [PubMed] [Google Scholar]
- 34.Saag, M. S., et al. 1996. HIV viral load markers in clinical practice. Nat. Med. 2:625-629. [DOI] [PubMed] [Google Scholar]
- 35.Schooley, R. T. 1995. Correlation between viral load measurements and outcome in clinical trials of antiviral drugs. AIDS 9(Suppl. 2):S15-S9. [PubMed] [Google Scholar]
- 36.Scott, L. E., et al. 2009. Evaluation of the Abbott m2000 RealTime human immunodeficiency virus type 1 (HIV-1) assay for HIV load monitoring in South Africa compared to the Roche Cobas AmpliPrep-Cobas Amplicor, Roche Cobas AmpliPrep-Cobas TaqMan HIV-1, and BioMerieux NucliSens EasyQ HIV-1 assays. J. Clin. Microbiol. 47:2209-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sivapalasingam, S., et al. 2009. Monitoring virologic responses to antiretroviral therapy in HIV-infected adults in Kenya: evaluation of a low-cost viral load assay. PLoS One 4:e6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson, P., et al. 2006. Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from Brazil. J. Virol. Methods 134:237-243. [DOI] [PubMed] [Google Scholar]
- 39.Swanson, P., et al. 2006. Performance of the automated Abbott RealTime HIV-1 assay on a genetically diverse panel of specimens from London: comparison to Versant HIV-1 RNA 3.0, Amplicor HIV-1 Monitor v1.5, and LCx HIV RNA quantitative assays. J. Virol. Methods 137:184-192. [DOI] [PubMed] [Google Scholar]
- 40.Swanson, P., et al. 2007. Evaluation of performance across the dynamic range of the Abbott RealTime HIV-1 assay as compared to Versant HIV-1 RNA 3.0 and Amplicor HIV-1 Monitor v1.5 using serial dilutions of 39 group M and O viruses. J. Virol. Methods 141:49-57. [DOI] [PubMed] [Google Scholar]
- 41.Tang, N., et al. 2007. A RealTime HIV-1 viral load assay for automated quantitation of HIV-1 RNA in genetically diverse group M subtypes A-H, group O and group N samples. J. Virol. Methods 146:236-245. [DOI] [PubMed] [Google Scholar]
- 42.Taylor, B. S., M. E. Sobieszczyk, F. E. McCutchan, and S. M. Hammer. 2008. The challenge of HIV-1 subtype diversity. N. Engl. J. Med. 358:1590-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Luin, M., et al. 2009. The effect of raltegravir on the glucuronidation of lamotrigine. J. Clin. Pharmacol. 49:1220-1227. [DOI] [PubMed] [Google Scholar]
- 44.von Truchsess, I., B. Harris, H. M. Schatzl, and J. Hackett, Jr. 2006. The first B/G intersubtype recombinant form of human immunodeficiency virus type 1 (HIV-1) identified in Germany was undetected or underquantitated by some commercial viral load assays. J. Med. Virol. 78:311-317. [DOI] [PubMed] [Google Scholar]
- 45.Xu, S., et al. 2008. Comparative evaluation of the COBAS AmpliPrep/COBAS TaqMan HIV type 1 test (CAP/CTM) and Versant HIV type 1 RNA 3.0 assay (bDNA) for quantifying HIV type 1 viral loads in China. AIDS Res. Hum. Retroviruses 24:1365-1373. [DOI] [PubMed] [Google Scholar]
- 46.Yazdanpanah, Y., et al. 2009. High rate of virologic suppression with raltegravir plus etravirine and darunavir/ritonavir among treatment-experienced patients infected with multidrug-resistant HIV: results of the ANRS 139 TRIO trial. Clin. Infect. Dis. 49:1441-1449. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, M., and J. Versalovic. 2002. HIV update. Diagnostic tests and markers of disease progression and response to therapy. Am. J. Clin. Pathol. 118(Suppl.):S26-S32. [DOI] [PubMed] [Google Scholar]