Abstract
In eukaryotes, polyadenylation of pre-mRNA plays an essential role in the initiation step of protein synthesis, as well as in the export and stability of mRNAs. Poly(A) polymerase, the enzyme at the heart of the polyadenylation machinery, is a template-independent RNA polymerase which specifically incorporates ATP at the 3′ end of mRNA. We have solved the crystal structure of bovine poly(A) polymerase bound to an ATP analog at 2.5 Å resolution. The structure revealed expected and unexpected similarities to other proteins. As expected, the catalytic domain of poly(A) polymerase shares substantial structural homology with other nucleotidyl transferases such as DNA polymerase β and kanamycin transferase. The C-terminal domain unexpectedly folds into a compact domain reminiscent of the RNA-recognition motif fold. The three invariant aspartates of the catalytic triad ligate two of the three active site metals. One of these metals also contacts the adenine ring. Furthermore, conserved, catalytically important residues contact the nucleotide. These contacts, taken together with metal coordination of the adenine base, provide a structural basis for ATP selection by poly(A) polymerase.
Keywords: crystal structure/mRNA 3′ end processing/phosphoryl transfer/poly(A) polymerase/terminal nucleotidyl transferase
Introduction
Following synthesis by RNA polymerase II, eukaryotic mRNA precursors (pre-mRNA) undergo a series of modifications, including capping at the 5′ end, splicing and addition of a 3′ poly(A) tail. The poly(A) tail has been shown to affect virtually all aspects of mRNA metabolism, including translation initiation and mRNA stabilization (reviewed in Beelman and Parker, 1995; Sachs et al., 1997; Wahle and Rüegsegger, 1999). Polyadenylation is a two-step event: endonucleolytic cleavage of the pre-mRNA is followed by poly(A) addition to the 3′ end of mRNAs. The cleavage and polyadenylation reactions involve a multicomponent machinery of remarkable complexity (reviewed in Colgan and Manley, 1997; Wahle and Rüegsegger, 1999; Zhao et al., 1999). In mammals, the synthesis of the poly(A) tail is catalyzed by poly(A) polymerase (PAP) with the help of the cleavage and polyadenylation specificity factor (CPSF; Christofori and Keller, 1988; Takagaki et al., 1988; Gilmartin and Nevins, 1989; Wahle, 1991b). CPSF binds to the canonical polyadenylation signal AAUAAA, which is located in the pre-mRNA 10–30 nucleotides upstream of the polyadenylation site (Keller et al., 1991). PAP alone binds RNA very weakly and non-specifically and shows a low, distributive polyadenylation activity. This is in contrast to what happens after the polymerase is recruited to AAUAAA-containing RNAs by CPSF; the protein–protein interactions with CPSF stimulate polyadenylation and greatly stabilize the ternary complex composed of CPSF, PAP and the RNA primer (Bienroth et al., 1993; Murthy and Manley, 1995). In the presence of the nuclear poly(A)-binding protein II (PABP2), another factor of the polyadenylation machinery, poly(A) synthesis becomes fast and processive. PABP2 has also been shown to control the final length of poly(A) tails, which on average are 250 nucleotides long in vivo (Wahle, 1991a; Wahle et al., 1993).
Most PAPs are single polypeptide enzymes, and cDNA clones encoding them have been isolated from many different organisms (Martin et al., 1999). PAPs have a modular organization with a catalytic domain near the N-terminus and an RNA-binding region that overlaps with a nuclear localization signal (NLS) near the C-terminus (Zhelkovsky et al., 1995; Martin and Keller, 1996; Martin et al., 1999). C-terminal extensions of ∼20 kDa are only found in vertebrates and are dispensable for catalytic activity in vitro. The extended C-terminal domain of vertebrate PAPs is rich in serines and threonines, and enzyme activity can be downregulated by phosphorylation at multiple sites (reviewed in Colgan and Manley, 1997; Wahle and Rüegsegger, 1999). The extreme C-terminus of PAP is also the target for another type of regulation. The U1A protein, a component of the U1 snRNP which functions in 5′ splice site recognition, is known to inhibit polyadenylation of its own mRNA by binding to PAP (Gunderson et al., 1994; Vagner et al., 2000). The C-terminus of PAP is also involved in protein–protein interactions with the splicing factor U2AF65 (Vagner et al., 2000) and the snRNP protein U1-70K (Gunderson et al., 1998).
PAPs belong to a large superfamily of nucleotidyl transferases containing two signature features: a conserved catalytic domain adorned with three invariant carboxylates that are crucial for activity, and a helical turn motif involved in nucleotide binding (Holm and Sander, 1995; Martin and Keller, 1996; Aravind and Koonin, 1999; Martin et al., 1999). Mammalian DNA polymerase β (pol β) and kanamycin nucleotidyl transferase (KanNt) are also members of this superfamily of nucleotidyl transferases. The structures of pol β (Davies et al., 1994; Pelletier et al., 1994; Sawaya et al., 1997) and KanNt (Sakon et al., 1993; Pedersen et al., 1995) are known. Both crystal structures have shown that conserved residues in the helical turn motif are involved in recognizing the triphosphate moiety of the nucleotide. Pol β is a template-dependent DNA polymerase involved in the base excision repair pathway (Pelletier et al., 1994; Sobol et al., 1996; Sawaya et al., 1997). Until now, a structure of a template-independent polymerase such as PAP was lacking.
Here we report the structure of a C-terminal deletion mutant of bovine PAP bound to an analog of ATP, cordycepin 5′-triphosphate (3′-dATP), at 2.5 Å resolution. The structural results presented here reveal a surprising domain composition and architecture, and provide insight into the ways in which PAP specifically binds to and incorporates ATP.
Results and discussion
Structure determination
Crystallization trials with full-length bovine and Caenorhabditis elegans PAP failed to produce crystals. A C-terminal deletion mutant of the bovine enzyme (bPAP-513) lacking the last 226 residues readily yielded high-quality crystals. Deletion of the C-terminal domain has a modest effect on activity, which decreases to 81% that of wild-type. Most importantly, this deletion mutant still binds to PABP2, and is stimulated by CPSF (Martin and Keller, 1996). In complex with 3′-dATP, the recombinant selenomethionyl-PAP yielded crystals suitable for structure determination by multiwavelength anomalous diffraction (MAD; Table I). The crystal structure was determined by a combination of MAD and multiple isomorphous replacement (MIR), followed by density modification. The model was refined to a crystallographic R-factor of 22.5% (Rfree 26.1%; Table II). All non-glycine residues lie inside the allowed regions of the Ramachandran plot.
Table I. X-ray crystallography data collection statistics.
| Beamline/detector |
|||||
|---|---|---|---|---|---|
| X12C/Brandeis B1 | X12C/Brandeis B1 | X12C/Brandeis B1 | X8C/ADSC Q4 | UVM/Mar345 | |
| Wavelength (Å) | 0.9789 | 0.9785 | 0.9500 | 1.0720 | 1.5418 |
| Resolution (Å) | 2.63 | 2.63 | 2.56 | 2.50 | 2.60 |
| Unique reflections | 20 256 | 19 960 | 21 802 | 20 878 | 20 530 |
| Average redundancy | 3.5 | 3.5 | 3.5 | 4.2 | 5.5 |
| Completeness (%) | 94.0 (84.6) | 96.6(85.0) | 97.9 (94.3) | 90.8 (81.1) | 98.0 (94.6) |
| Rmerge | 0.061 (0.175) | 0.062 (0.170) | 0.062 (0.200) | 0.085 (0.267) | 0.085 (0.326) |
| I/σ | 16.6 (3.9) | 18.9 (4.4) | 14.8 (3.6) | 16.7 (6.9) | 17.7 (4.3) |
| Selenium sites | 6 | ||||
| RCullis iso/ano | –/0.93 | 0.66/0.91 | 0.87/0.94 | 0.77/0.83 | 0.67/0.98 |
| Phasing power iso/ano | –/1.11 | 1.95/1.20 | 0.19/0.98 | 1.37/1.04 | 1.59/0.53 |
Rmerge = Σ|Ih – <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalents.
The numbers in parentheses refer to the highest resolution bin.
RCullis = mean lack of closure error divided by the mean isomorphous or anomalous differences.
Phasing power = mean value of heavy atom structure factor amplitudes divided by the lack of closure error. The numbers given for RCullis and phasing power were calculated by SHARP (de La Fortelle and Bricogne, 1997) and are only for acentric reflections.
Table II. Refinement statistics.
| Resolution (Å) | 20.0–2.5 | |||
| Rfree | 26.1 | |||
| Rwork | 22.0 | |||
| No. of protein atoms | 3734 | |||
| No. of substrate atoms | 30 | |||
| No. of solvent molecules | 171 | |||
| No. of metal atoms | 3 | |||
| R.m.s.d. bonds (Å) | 0.0067 | |||
| R.m.s.d. angles (°) | 1.34 | |||
| R.m.s.d. dihedral (°) | 22.9 | |||
| R.m.s.d. improper (°) | 0.94 | |||
| <B> (Å2) | protein: 28.1 | substrate: 39.2 | metals: 38.5 | solvent: 29.1 |
| Ramachandran plot (%) | favorable: 88.8 | additional: 10.5 | generous: 0.7 | disallowed: 0 |
Rwork and Rfree = Σ|Fo| – |Fc|/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes. Rfree was calculated with 7% of the reflections not used in refinement.
A tripartite domain architecture for PAP
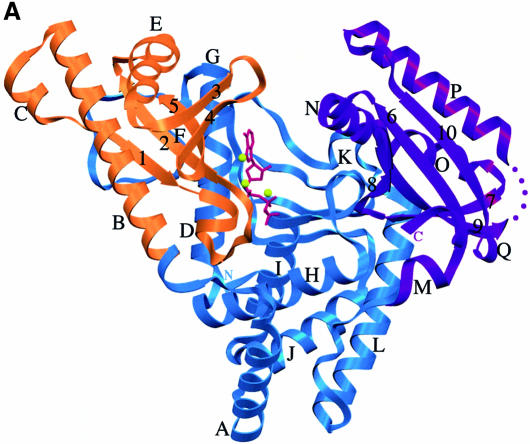
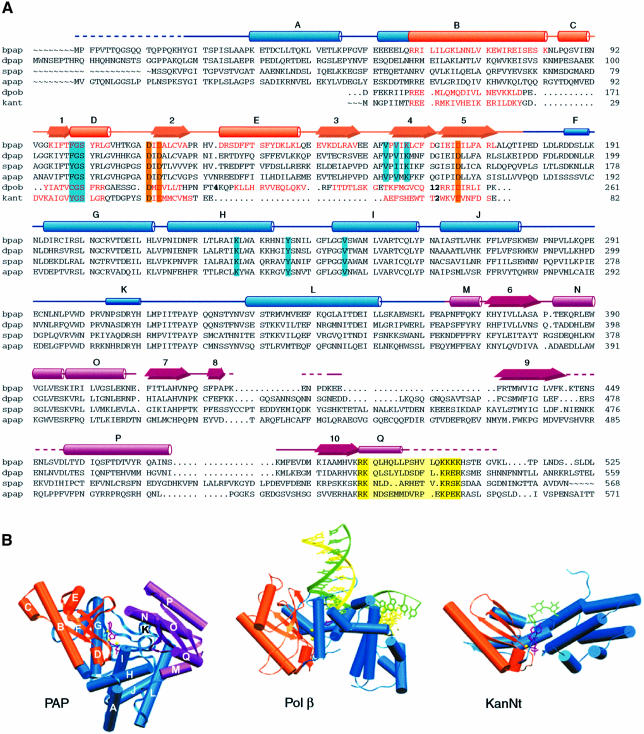
The overall structure of PAP is shown in Figures 1 and 2. PAP is a U-shaped molecule of approximate dimensions 48 × 55 × 75 Å, defining a large cleft measuring ∼20 × 25 Å. The polymerase is composed of three domains: the N-terminal domain is, as expected, homologous to the catalytic domain of nucleotidyl transferases, and comprises a five-stranded mixed β-sheet and two α-helices (Figure 2; Holm and Sander, 1995; Martin and Keller, 1996; Martin et al., 1999). The ATP analog and metals bind to the catalytic domain (Figure 1; see below). A search for structural homologs with the Dali server (Holm and Sander, 1998) confirmed the homology between the N-terminal domain of PAP and the catalytic domain of pol β (Z score = 8.3; Sawaya et al., 1997). This search also unveiled surprising structural homologies for the remaining two domains of PAP. The connection from the catalytic domain to the central domain is through a central β-strand, an architecture reminiscent of pol β. The central domain, however, does not hold any homology to the pol β fingers. Interestingly, this domain instead shares structural similarity with the allosteric activity domain of ribonucleotide reductase R1, which comprises a four-helix bundle and a three-stranded mixed β-sheet (Z score = 3.1; Uhlin and Eklund, 1994). The two four-helix bundles can be superimposed with a root mean square (r.m.s.) deviation of 3.1 Å for 75 corresponding Cα atoms. Even though the two enzymes bind ATP, the ATP-recognition motifs are different, i.e. the superposition of the four-helix bundles does not overlay the two ATP molecules.
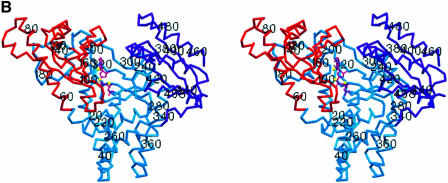
Fig. 1. (A) Ribbon diagram of the bovine PAP complex with 3′-dATP. The catalytic domain (residues 60–173) is shown in orange, the central domain in blue (residues 20–59 and 174–352) and the C-terminal RNA-binding domain in purple (residues 353–498). The 3′-dATP molecule bound in the active site is shown in magenta, and the three metals ions are shown as yellow spheres. Helices are designated with letters, β-strands with numbers. (B) Stereo view of the PAP complex in the same orientation as in (A). Every 20 residues are labeled. Secondary structure elements were determined by Procheck (Laskowski et al., 1993). Figures 1, 3, 4 and 5 were generated with SETOR (Evans, 1993).
Fig. 2. Sequence and structural alignment of poly(A) polymerases and related nucleotidyl transferases. (A) Alignment of homologous regions of PAPs of bovine origin (bpap; SWISS-PROT: P25500), from Drosophila melanogaster (dpap; DDBJ/EMBL/GenBank: AAF36978), Saccharomyces cerevisiae (spap; SWISS-PROT: P29468) and Arabidopsis thaliana (apap; DDBJ/EMBL/GenBank: CAB80002). The sequences of rat DNA polymerase β (polb; SWISS-PROT: P06766) and kanamycin nucleotidyl transferase (kant; SWISS-PROT: P05057) are also included in the alignment (numbers in bold indicate number of residues not displayed). Structural elements homologous among PAP, pol β and KanNt are shown in orange. Important conserved residues are highlighted in orange for catalytic carboxylates, cyan for residues interacting with the nucleotide, and yellow for the predicted nuclear localization signal. The secondary structure elements are indicated above the bpap sequence, as tubes (α-helices), narrow tubes (3/10 helices), arrows (β-strands) or lines (coil). The three domains in PAP are color-coded as in Figure 1. (B) The conserved catalytic domain (orange) is shown in the same orientation in the crystal structures of PAP, pol β (Protein Databank accession No. 1BPY) and kanamycin nucleotidyl transferase (Protein Databank accession No. 1KNY). For clarity, only one monomer of kanamycin transferase is shown. In the PAP model, helices are labeled according to (A).
The PAP C-terminus folds into a compact globular domain, consisting of a four-stranded antiparallel β-sheet, flanked on one side by two helices. This domain is topologically similar to the RNA-binding domains of several RNA-binding proteins such as ribosomal protein S6 (Z score = 4.4; Lindahl et al., 1994), phenylalanyl-tRNA synthetase (Z score = 3.8; Goldgur et al., 1997) and sex-lethal protein (Z score = 2.9; Handa et al., 1999). Even though they do not possess the conserved ribonucleoprotein (RNP) sequences characteristic of members of the RNA-recognition motif (RRM) protein family, PAP and ribosomal protein S6 share topological similarity with this family of proteins which comprises >200 members, with representatives found from bacteria to human (reviewed in Burd and Dreyfuss, 1994; Varani and Nagai, 1998). Crystallographic studies of RRM proteins bound to RNA have documented that RNA binding is predominantly via the β-sheet surfaces (Oubridge et al., 1994; Price et al., 1998; Deo et al., 1999; Handa et al., 1999). The C-terminal domain is connected to the central domain via a hinge-like region. The temperature factor is higher in the C-terminal domain (average temperature factor = 37.5 Å2 versus 25.5 Å2 for the rest of the polymerase), which may indicate increased mobility. A cDNA clone isolated from a HeLa cDNA library codes for a 43 kDa PAP that was found to be inactive in polyadenylation assays (Wahle et al., 1991). Our crystal structure shows that this HeLa PAP is inactive because it is lacking the entire RNA-binding domain.
The structures of other polymerases such as pol I family DNA polymerases (Beese et al., 1993; Doublié et al., 1998; Kiefer et al., 1998; Li et al., 1998), RB69 gp43 (Wang et al., 1997), pol β (Davies et al., 1994; Pelletier et al., 1994; Sawaya et al., 1997), HIV reverse transcriptase (Huang et al., 1998) and T7 RNA polymerase (Sousa et al., 1993; Jeruzalmi and Steitz, 1998) all share a common architecture, which has been described as a half opened right hand, composed of fingers, palm and thumb subdomains (Ollis et al., 1985; Steitz, 1993). Unlike all polymerases solved to date, PAP is a template-independent RNA polymerase, and its structure is a departure from the common polymerase fold. Interestingly, the PAP active site is located on one side of the U, and not at the confluence between the fingers and thumb (Figure 1). The hand metaphor therefore does not hold for PAP. For this reason, we will refer to the three separate domains, from the N- to C-terminus, as the catalytic, central and RNA-binding domains.
Role of conserved residues and metals
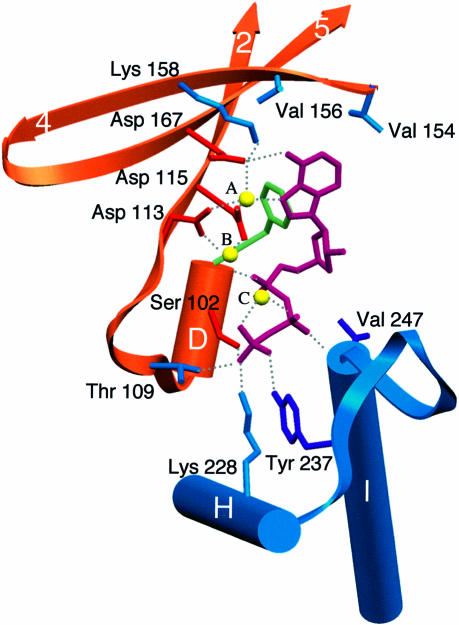
Nucleotidyl transferases catalyze phosphoryl transfer through the nucleophilic attack by the 3′-hydroxyl of the primer (4′-OH for KanNt) on the α-phosphate of the incipient nucleotide, resulting in nucleoside monophosphate incorporation and release of pyrophosphate. Biochemical and structural studies have identified several conserved residues and sequence motifs that play a critical role in this reaction (Figure 2). Prominent among these are the three conserved active site aspartates (Asp113, Asp115 and Asp167 in bovine PAP) and the helical turn motif that is formed by the first four residues of the nucleotidyl transferase superfamily sequence motif (G[G/S]x9–13Dx [D/E]). Mutating any of the three carboxylates into alanine strongly impairs phosphoryl transfer in PAP (Martin and Keller, 1996) and pol β (Davies et al., 1994; Pelletier et al., 1994; Sawaya et al., 1997). In the PAP structure, the three aspartates ligate two metals in the active site in a configuration that is reminiscent of pol β (Figure 3). Ser102, a conserved residue in the helical turn motif, contacts the γ-phosphate in PAP, pol β and KanNt. This residue also provides a hydrogen bond via a main chain interaction with a non-esterified oxygen of the α- or β-phosphate. Pyrophosphate inhibition studies of PAP mutants showed that S102A is more sensitive to pyrophosphate addition than the wild-type enzyme (Martin et al., 1999), confirming the role of this conserved residue in contacting the β- and/or γ-phosphates. The ribose and base moieties are within van der Waals contact of Phe100. This residue is the structural homolog of Tyr37 in KanNt, where it rests against the sugar moiety (Pedersen et al., 1995). Steady-state kinetic studies showed that mutating Phe100 into an aspartate has a devastating effect on kcat (Martin et al., 1999).
Fig. 3. PAP active site showing the major interacting residues. The secondary structure elements are color-coded as in Figure 1. Residues are shown in blue, except when they share structural homology with other proteins: orange (pol β and KanNt), green (KanNt) and purple (T7 DNA polymerase). Hydrogen bonds are shown as dashed gray lines.
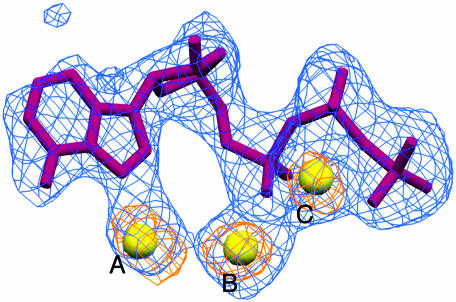
Crystallographic analyses of DNA polymerases showed that there are two metals in the polymerase active site that participate in catalysis (Sawaya et al., 1997; Huang et al., 1998; Li et al., 1998; Doublié et al., 1999; Steitz, 1999). In order to identify the position of the metals in the PAP active site, we calculated an anomalous difference Fourier map (Figure 4), which revealed that there are three metals in the PAP active site, instead of the two usually seen in DNA polymerases. PAP was co-crystallized with 3′-dATP in the presence of MnCl2, so the metals are likely to be Mn2+. The manganese ions are located 3.6–3.7 Å apart and make key interactions that help position the nucleotide in the active site. The octahedral coordination shell of metal A includes all three aspartic acid side chains, the N7 atom of adenine, a water molecule and a non-bridging oxygen of the α-phosphate of a second nucleotide. Only the triphosphate tail of the second nucleotide shows identifiable density. The strength of the electron density and its shape strongly suggest that this is a second triphosphate, and not ordered waters or other small molecules. The average temperature factor for the triphosphate is 50% higher than that calculated for the phosphates of the first 3′-dATP, indicating increased motion in the second molecule. Moreover, there is no density for the ribose moiety, and poorly defined density for the base, which lies within van der Waals distance of the adenine base of the first 3′-dATP. This observation may indicate that the second nucleotide is stacking against the first. The significance of this finding is uncertain at this time. Metal B has only four clear ligands: the side chains of Asp113 and Asp115, the α-phosphate of 3′-dATP and a water molecule. Both triphosphate tails wrap around metal C. A non-bridging oxygen from each of the phosphates contributes to the octahedral coordination of this metal.
Fig. 4. The refined model of 3′-dATP is shown in magenta, and the three metal ions are shown as yellow spheres. The superimposed simulated annealing omit map (blue) and anomalous difference Fourier map (orange) are contoured at 3.0σ and 4.5σ above the mean, respectively. The molecule of 3′-dATP and the three metal ions were omitted in the calculation of the omit map. The peak heights for metals A, B and C in the anomalous difference map are: 8.3, 9.7 and 8.5σ, respectively.
All of the non-bridging phosphate oxygens not contacted by metals interact with one or more conserved residues. In pol β, the phosphoryl oxygens are held in place by side chains coming exclusively from the helical loop motif of the palm domain (Pelletier et al., 1994; Sawaya et al., 1997). In the PAP structure, in addition to the helical turn motif, residues from helices H and I contact the phosphates in a situation that is analogous to the conserved motif B of pol I family polymerases (Doublié et al., 1998; Li et al., 1998; Martin et al., 1999). Conserved residues from helices H and I contact the β- and γ-phosphates, while residues from helix D of the helical turn motif interact with the α- and γ-phosphates. The ε-amino group of Lys228 and the hydroxyl group of Tyr237 donate a hydrogen bond to the γ-phosphate. Mutating Tyr237 into alanine results in an increased KM for ATP, while the same mutation for Lys228 has an effect on both KM and kcat (Martin et al., 1999). The β-phosphate rests against a glycine (Gly246) that is strictly conserved among eukaryotic PAPs, in such a manner that a larger side chain would alter the position of the triphosphate tail. Val247 makes a main chain interaction with the non-esterified oxygen of the β-phosphate. The valine side chain is within van der Waals distance of the ribose moiety.
Selectivity for ATP
Despite the fact that PAP does not use a nucleic acid template, incorporation of nucleotides in the growing poly(A) tail is highly specific for ATP, and shows a low error frequency under physiological conditions (Edmonds, 1982). This poses an interesting problem: how do template-independent polymerases select for ATP, in the case of PAP, or two Cs and an A, in the case of CCA-adding enzymes (Yue et al., 1998)? In template-dependent DNA polymerases, the shape of the active site dictates which nucleotide will be incorporated. After binding the correct nucleotide, the fingers domain rotates towards the palm, assembling the polymerase active site for nucleotide incorporation. The tight fit of the nascent base pair in the polymerase active site precludes a mismatch between the template base and the incipient nucleotide. Both the polymerase and template therefore act in concert to assemble an active site that will select for Watson–Crick base pairs (Pelletier et al., 1994; Sawaya et al., 1997; Doublié et al., 1998; Li et al., 1998).
In the case of PAP, the nucleotide selection proceeds in the absence of a template. The other two members of the nucleotidyl transferase family for which the structure is known do not show a strong preference for one nucleotide over another. As with any DNA polymerase, pol β can bind and incorporate any of the four dNTPs in a template-dependent manner. KanNt is known to utilize ATP, GTP or UTP as nucleoside monophosphate donors (Pedersen et al., 1995). Inspection of the crystal structures of pol β and KanNt showed, as expected, that the nucleotide base is not contacted specifically by the enzyme (Sawaya et al., 1994, 1997; Pedersen et al., 1995). Of the three nucleotidyl transferases, PAP is the only enzyme that displays a strong preference for ATP (Wahle, 1991b). In our crystal structure, the N6 amino group of the adenine base donates a hydrogen bond to Asp167, the third aspartate of the catalytic triad, and metal A contacts N7, an atom that is a good ligand for alkali and transition metals (Figure 3). In pol β, the base moiety of the incipient nucleotide is in a different conformation, precluding its interaction with the homologous Asp256. This aspartate is shown to contact the activating metal in the pol β ternary complex (Sawaya et al., 1997), while in pol I family polymerases, the analogous carboxylate (Glu655) does not seem to participate in metal binding (Doublié et al., 1999).
Recognition via the N7 position of adenine is not unprecedented. The crystal structure of an archaeal aspartyl-tRNA synthetase revealed the presence of three metals in the active site, one of them coordinating the N7 of the ATP ligand (Schmitt et al., 1998). The possibility that Asp167 and metal A may play a part in selecting for ATP is in agreement with studies of another member of the nucleotidyl transferase family, the CCA-adding enzyme from Sulfolobus shibatae, where mutating the aspartate corresponding to Asp167 markedly reduced ATP-adding activity, but did not affect CTP-adding activity (Yue et al., 1998). The adenine moiety is stabilized further in the active site of PAP via hydrophobic interactions with two conserved valines, Val154 and Val156. Mutating these two residues into alanine substantially decreases kcat (unpublished results).
These hydrogen bond and van der Waals contacts to the adenine base provide an answer to the question of how PAP distinguishes adenine from the other three RNA bases. Purines can be differentiated from pyrimidines on the basis of their size (bicyclic versus monocyclic) and of their substituents. Pyrimidines lack the hydrogen bond acceptor found in the N7 of purines, and therefore lack the interaction with metal A. The discrimination against the guanine base is achieved through interactions with C6 and C2. The C6 carbonyl of guanine would interact unfavorably with Asp167. Furthermore, Val154 makes a van der Waals contact with C2 (CG2–C2 = 3.6 Å), which would preclude binding of any base with a substituent at that position, thus providing a structural basis for discrimination against GTP incorporation.
Comparison with terminal nucleotidyl transferases and mechanism of phosphoryl transfer
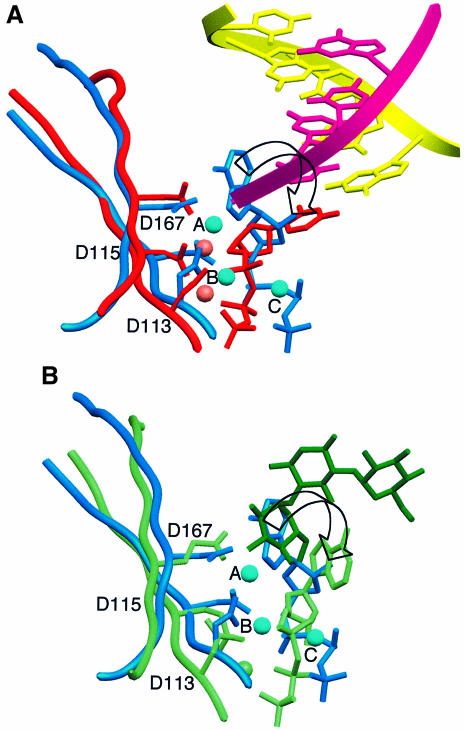
Structural homology of residues ∼60–170 of PAP with the palm domain of pol β and the catalytic domain of KanNt had been predicted based on sequence alignments and mutagenesis studies (Holm and Sander, 1995; Martin and Keller, 1996; Martin et al., 1999). A superposition of the three crystal structures demonstrates that indeed the three proteins superimpose well, despite some structural differences in connecting loops (the r.m.s. deviation between PAP and pol β is 1.85 Å for 76 corresponding Cα atoms, and that between PAP and KanNt is 2.70 Å for 61 pairs of Cα atoms). This superposition overlays the three strictly conserved active site carboxylates, as well as the triphosphate tails and ribose moieties of the three nucleotides (Figure 5). When pol β and PAP are overlaid on the basis of the three conserved active site aspartates (Asp113, Asp115 and Asp167 in PAP; Asp190, Asp192 and Asp256 in pol β), metal A of PAP is within 1.2 Å of the activating metal in pol β (Mg340 in pol β) and metal B is within 0.5 Å of the nucleotide-binding metal (Mg339; Sawaya et al., 1997; Protein Databank accession No. 1BPY). The orientation of the adenine base, however, is markedly different in PAP. The base is actually located where the attacking moiety sits in pol β (3′-OH of the ribose) and KanNt (4′-OH of kanamycin), and would need to rotate ∼90° to overlay on top of the corresponding base in pol β and KanNt (Figure 5).
Fig. 5. Superposition of the PAP active site with those of pol β and kanamycin transferase. The orientation of the molecules is the same as in Figure 2. For clarity, only residues of β-strands 2 and 5 are shown, and the triphosphate tail of the second nucleotide is omitted. (A) Superposition of PAP (blue) and pol β (red; Protein Databank accession No. 1BPY). The primer and template strands in pol β are shown in magenta and yellow, respectively. For clarity, only five base pairs are shown. The 3′ end of the primer (magenta) in pol β overlays the adenine base of 3′-dATP (blue) in PAP. (B) Superposition of PAP and kanamycin transferase (green; Protein Databank accession No. 1KNY). The attacking group of kanamycin (shown in dark green) overlays the adenine base of 3′-dATP (blue) in PAP. An arrow indicates the rotation that would bring the adenine base in PAP on top of the corresponding base in pol β or KanNt.
How might PAP catalyze phosphoryl transfer? Although polymerization by PAP is a template-independent reaction, its mechanism is a typical polymerase reaction in all other respects: AMP incorporation is primer dependent, the products are poly(A) RNA and pyrophosphate and, finally, the incorporation of AMP results in the inversion of configuration at the α-phosphate (Wittmann and Wahle, 1997). This indicates that the reaction most probably involves an in-line attack of the 3′-OH of the primer on the incoming ATP, without a covalent intermediate. This is known to be the case for DNA polymerases (Sawaya et al., 1997; Doublié et al., 1998; Steitz, 1999). In our crystal structure, the nucleotide configuration in the PAP active site is not compatible with phosphoryl transfer. The α-phosphate of 3′-dATP is in a configuration that would not allow attack from the 3′-OH of an RNA primer. Instead, Asp115 is aligned with the α- and β-phosphates, and seems poised for attack on the α-phosphate. Attack from the aspartate carboxylate, however, is unlikely since no covalent intermediate has ever been isolated for mammalian PAPs. Our crystal structure may therefore represent the first stage of a two-step reaction, where the nucleotide is first selected, followed by the chemical step and incorporation into the poly(A) tail (see below).
PAP was co-crystallized with 3′-dATP and Mn2+, in the absence of RNA. We expect that in the presence of both RNA and ATP substrates, the polymerase undergoes conformational changes that would bring the ATP molecule into a position compatible with phosphoryl transfer. The crystal structures of binary and ternary complexes of DNA polymerases have clearly illustrated that a conformational change must occur after binding of the correct nucleotide and before incorporation of a nucleoside monophosphate into the growing chain (Sawaya et al., 1997; Doublié et al., 1998; Kiefer et al., 1998; Li et al., 1998). The closure of the fingers towards the polymerase active site precisely aligns the 3′ end of the primer with the α-phosphate of the incipient nucleotide, thereby facilitating phosphoryl transfer. When T7 DNA polymerase and PAP are superimposed based on the three active site carboxylates (Asp475, Asp654 and Glu655 for T7 DNA polymerase, and Asp113, Asp115 and Asp167 for PAP), the O helix of T7 DNA polymerase, which provides several crucial side chain interactions with the incoming nucleotide, overlays on top of helices H and I in PAP, such that Arg518 of T7 DNA polymerase overlaps with Tyr237 of PAP, and Tyr526 overlaps with Val247. In T7 DNA polymerase, the guanidinium group of Arg518 contacts the γ-phosphate, while Tyr526 stacks against the ribose and base moieties of the incipient nucleotide. Tyr237 and Val247 have similar functions in PAP.
In crystallographic studies of complexes with DNA primer/template and incoming nucleotide, the O helix of pol I family polymerases was shown to rotate by ∼40° to bring conserved, catalytically important residues into contact with the incoming nucleotide (Doublié et al., 1998; Li et al., 1998). We expect more subtle conformational changes to take place in PAP, as only the base and ribose moities need to rotate to bring the α-phosphate into a position where it could be attacked by the 3′ end of the RNA primer. Interestingly, the ATP molecule in the PAP active site is contacted by residues from both the catalytic and central domains (Figure 3). Subtle rearrangements between the two domains could help position the ATP molecule in a configuration compatible with phosphoryl transfer.
Putative RNA path
In the vast majority of DNA- and RNA-binding proteins, the regions containing an NLS overlap with polynucleotide-binding domains. UV cross-linking experiments with PAP protein fragments confirmed the presence of an RNA-binding region between amino acids 488 and 508, which encompasses NLS-1 (Martin and Keller, 1996). In our crystal structure, NLS-1 forms a 3/10 helix that is located at the C-terminus (helix Q). Mammalian PAPs possess a second NLS near residue 650, which was found to cross-link weakly with RNA. These residues are not present in our structure because they were deleted in the design of this PAP C-terminal deletion mutant.
In addition to the 3/10 helix of NLS-1, two central β-strands of the PAP C-terminal domain may be involved in binding RNA. As mentioned earlier, the PAP C-terminal domain is topologically similar to proteins of the RRM family. Structures of some of these proteins have been solved in complex with RNA: sex-lethal protein (Handa et al., 1999), U1A (Oubridge et al., 1994) and poly(A)-binding protein (Deo et al., 1999). These structures revealed that the central β-strands of RRMs provide an interaction surface with RNA. NLS-1 and the central two β-strands of the C-terminal domain therefore present potential interaction surfaces with poly(A) RNA.
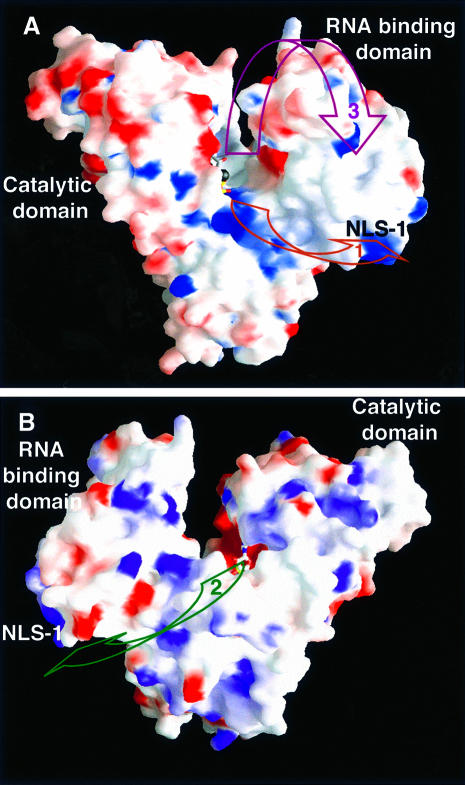
The U-shaped PAP molecule exhibits grooves that could accommodate single-stranded RNA, from the active site to the C-terminal domain. The first groove originates at the polymerase active site, follows the top of the central domain and finally reaches NLS-1 (Figure 6). This path is lined with conserved basic residues (Lys228, Lys232, Arg233 and Lys370), which could interact with the RNA phosphate backbone (Figure 6; path No. 1). It should be noted, however, that the domains containing these conserved basic residues did not cross-link with RNA (Martin and Keller, 1996).
Fig. 6. View of the PAP model showing the surface of the enzyme colored according to its electrostatic potential (blue, positively charged; red, negatively charged). (A) The PAP molecule is shown in the same orientation as in Figure 1. The putative RNA paths are shown with maroon and purple arrows. (B) The PAP molecule was rotated 180° around a vertical axis relative to (A). The putative RNA path is shown with a green arrow. This Figure was generated with the program GRASP (Nicholls et al., 1991).
The second path also originates at the active site, but it reaches the C-terminal domain following a direction that is opposite to the path described above (Figure 6; path No. 2). Finally, a third path goes to the top of the RNA-binding domain, and follows the two central β-strands, before reaching NLS-1 (Figure 6; path No. 3). This path would require the RNA-binding domain to move closer towards the PAP active site. Paths 2 and 3 are consistent with a superposition of the active sites of PAP, pol β and KanNt. The three enzymes were superimposed in such a manner that all three active sites were oriented similarly (Figures 2 and 5). In this superposition, the attacking moiety comes from the top of the nucleotide for both pol β (3′-OH of the primer ribose) and KanNt (4′-OH of kanamycin; Figure 5). We propose that the 3′ end of the RNA primer may be positioned similarly in the PAP active site. Furthermore, the putative position of the RNA primer in the third path is consistent with the position of the DNA primer in the ternary complex of pol β (Pelletier et al., 1994; Sawaya et al., 1997), as well as that of the RNA vis-a-vis ribosomal protein S6 in the recently published structure of the S15,S6,S18–rRNA complex (Agalarov et al., 2000).
CPSF is known to bind the ubiquitous recognition sequence AAUAAA upstream of the poly(A) site in the pre-mRNA (Keller et al., 1991). The interaction of PAP and CPSF has been studied by co-immunoprecipitation, by specific polyadenylation assays and by gel retardation experiments. The 160 kDa subunit of CPSF was shown to bind specifically to full-length PAP but not to a naturally occurring inactive splice variant of PAP that lacks the entire RNA-binding domain (Murthy and Manley, 1995). Specific polyadenylation assays and gel retardation with RNA, CPSF and a series of C-terminal PAP deletion mutants have shown that the last 219 residues of PAP are not required for specific polyadenylation activity nor for the formation of a CPSF–PAP–RNA complex (G.Martin and W.Keller, unpublished results). Taken together, these results suggest that CPSF interacts with amino acids 375–520 in PAP, a region that represents the RNA-binding domain. It is therefore tempting to speculate that following the endonucleolytic cleavage step, CPSF hands over the RNA primer to the RNA-binding domain of PAP. These data on the PAP–CPSF interactions do not, however, allow us to select one RNA path unequivocally over the others. The unambiguous assignment of the RNA path must therefore await the crystal structure determination of the complex of PAP with poly(A) RNA.
Conclusion
This study of bovine PAP complexed to 3′-dATP provides the first structure of any component of the mammalian polyadenylation machinery and the first example of a template-independent polymerase. In the active site, conserved, catalytically important amino acids contact the nucleotide. The three aspartates that are conserved in this family of terminal nucleotidyl transferases, and amongst other families of DNA and RNA polymerases, ligate two metals. One of these metals contacts the N7 position of the adenine ring, and could play a role in the selective incorporation of adenine. A third metal ligates three non-esterified oxygens of the triphosphate tail. This crystal structure will guide additional studies of PAP complexed to poly(A) RNA and proteins of the polyadenylation machinery that are known to interact with the polymerase, such as CPSF and PABP2.
Materials and methods
Crystallization and data collection
Recombinant his-tagged bovine PAP (residues 1–513; bPAP-513) was expressed in Escherichia coli essentially as described (Martin and Keller, 1996). During lysis and affinity purification on Ni2+ NTA–agarose, buffers contained 10 mM β-mercaptoethanol. Thereafter, the protein was kept in buffers containing 4 mM dithiothreitol (DTT). After elution from Ni2+ NTA, PAP was purified further on three additional columns: Hitrap heparin, Hitrap Blue and Hitrap Q on an FPLC system (all from Pharmacia). After binding the protein to the Hitrap heparin, the column was washed with 10 column volumes of buffer containing 5 mM EDTA in order to remove any residual Ni2+. The protein was concentrated to 10–20 mg/ml in Centricon-30 filter cartridges, flash frozen in N2 and stored at –80°C. The selenomethionyl variant was produced by the method of methionine pathway inhibition (Doublié, 1997). The selenomethionyl protein was purified using essentially the same protocol as for the wild-type PAP. Crystals of PAP complexed to 3′-dATP were obtained by using a 24-condition incomplete factorial design (Carter and Carter, 1979). After microseeding, crystals grow overnight at 24°C against a reservoir solution made of 22% (w/v) polyethylene glycol 8000, 100 mM MES pH 6, 120 mM ammonium sulfate, 5 mM CaCl2, 2 mM MnCl2 and 2 mM β-mercaptoethanol. Crystals grow as clusters of long needles, and are of the space group P212121, with one monomer in the asymmetric unit and a solvent content of ∼55%. The cell dimensions are a = 57.55 Å, b = 62.72 Å, c = 179.69 Å. Diffraction data on flash-frozen crystals cryoprotected with 25% (v/v) glycerol were collected at beamline X12C of the National Synchrotron Light Source (NSLS; Upton, NY) on a CCD detector (Brandeis B1). One of the MAD data sets (λ = 1.072 Å) was obtained through a remote-controlled Fedex data collection at beamline X8C (NSLS) on a CCD detector (ADSC Q4). Data acquired using a laboratory X-ray source at the University of Vermont were collected on a Mar345 image plate detector. The monochromator positions at the X12C beamline were based on X-ray absorption fluorescence of the selenomethionine-containing crystal. Two crystals were used for the MAD data collection. Data were integrated with Denzo/Scalepack (Otwinowski and Minor, 1997). Details of the crystallographic data sets used for structure solution and refinement are given in Table I. Unless otherwise indicated, subsequent crystallographic analysis was done using the CCP4 package (CCP4, 1994).
Structure determination and refinement
We were unable to locate the selenium sites with any of the programs routinely used for finding heavy atom positions. We therefore screened for heavy metal derivatives. PAP crystals are very sensitive to heavy metal addition. Numerous heavy atom compounds were screened at different concentrations and varying incubation times. In most cases, derivatization resulted in loss of diffraction or severe non-isomorphism. The sites from non-isomorphous derivatives (mercury chloride, PIP [di-m-iodobis(ethylenediamine) diplatinum nitrate], trimethyl lead acetate and ytterbium chloride) were refined with MLPHARE against solvent-flattened MIR phases (Rould et al., 1992). The refinement converged after three cycles and resulted in substantially improved phases. Heavy atom parameters subsequently were input into SHARP, which was then used for further refinement and to calculate phases (de La Fortelle and Bricogne, 1997). The resulting electron density map was uninterpretable, but the phases were accurate enough to locate six of the nine selenium sites in anomalous difference Fourier maps. The electron density maps later revealed that three of the selenomethionines are disordered (Met1, Met476 and Met486). The selenium sites were refined and phases calculated with SHARP (de La Fortelle and Bricogne, 1997). The data set collected at the inflection point (λ = 0.9789 Å) was chosen as the native. The overall Figure of merit of the SHARP phases increased from 0.35 to 0.87 after solvent flattening, assuming 50% solvent. The resulting map was of excellent quality. Model building and adjustments were done with the program O (Jones et al., 1991), first building into the solvent-flattened MAD map, then into the σA-weighted maps. Final adjustments were done with Vusette ZC (M.A.Rould, unpublished program). Refinement was performed with CNS using standard protocols, including maximum likelihood target (mlhl target using amplitudes and unmodified SHARP phases), bulk solvent correction and anisotropic B-factor correction (Brünger et al., 1998). The model was inspected manually with simulated annealing omit maps and σA-weighted 2Fo – Fc and Fo – Fc maps, and progress in the model refinement was gauged by the decrease in the free R-factor. Refinement statistics can be found in Table II.
The current model includes 463 residues (20–423, 430–444 and 455–498, one molecule of 3′-dATP, one molecule of triphosphate, three metal ions and 171 water molecules). There are two cis-prolines in the complex: Pro282 and Pro321. The following residues were modeled as alanines because their side chain density was either poorly defined or non-existent: Glu292, Asn294, Lys355, Glu409, Gln419, Met476, Met486 and His494. Coordinates of the mammalian PAP complex have been deposited in the RSCB Protein Databank (accession No. 1F5A).
Acknowledgments
Acknowledgements
We thank R.M.Sweet and M.Becker for access and assistance with the use of beamline X12C and data collection at X8C. We are grateful to Drs T.Ellenberger, C.S.Francklyn, M.R.Sawaya and M.Tierney for critically reading the manuscript, and to M.A.Rould for the use of unpublished programs and for insightful comments on the manuscript. We thank D.Blank for invaluable help with Figure 2. S.D. is a Pew Scholar. This work was supported by an award to the University of Vermont under the Howard Hughes Medical Institute Biomedical Research Support Program for Medical Schools. G.M. and W.K. were supported by the University of Basel, the Swiss National Science Foundation, the European Union (via the Bundesamt für Bildung und Wissenschaft, Bern) and the Louis-Jeantet Foundation for Medicine. Diffraction data for this study were collected at the National Synchrotron Light Source, a facility supported by the US Department of Energy, by the National Science Foundation and by a grant from the National Institutes of Health.
Note added in proof
The crystal structure of poly(A) polymerase from yeast has recently been solved [Bard,J., Zhelkovsky,A.M., Helmling,S., Earnest,T.N., Moore,C.L. and Bohm,A. Structure of yeast poly(A) polymerase alone and in complex with 3′-deoxy ATP. Science, in press].
References
- Agalarov S.C., Sridhar Prasad,G., Funke,P.M., Stout,C.D. and Williamson,J.R. (2000) Structure of the S15,S6,S18–rRNA complex: assembly of the 30S ribosome central domain. Science, 288, 107–113. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Koonin,E.V. (1999) DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res., 27, 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A. and Parker,R. (1995) Degradation of mRNA in eukaryotes. Cell, 81, 179–183. [DOI] [PubMed] [Google Scholar]
- Beese L.S., Derbyshire,V. and Steitz,T.A. (1993) Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science, 260, 352–355. [DOI] [PubMed] [Google Scholar]
- Bienroth S., Keller,W. and Wahle,E. (1993) Assembly of a processive messenger RNA polyadenylation complex. EMBO J., 12, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- Carter C.W. Jr and Carter,C.W. (1979) Protein crystallization using incomplete factorial experiments. J. Biol. Chem., 254, 12219–12223. [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Christofori G. and Keller,W. (1988) 3′ Cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor and a snRNP. Cell, 54, 875–889. [DOI] [PubMed] [Google Scholar]
- Colgan D.F. and Manley,J.L. (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- Davies J.F. 2nd, Almassy,R.J., Hostomska,Z., Ferre,R.A. and Hostomsky,Z. (1994) 2.3 Å crystal structure of the catalytic domain of DNA polymerase β. Cell, 76, 1123–1133. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- Doublié S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol., 276, 523–530. [PubMed] [Google Scholar]
- Doublié S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- Doublié S., Sawaya,M.R. and Ellenberger,T. (1999) An open and closed case for all polymerases. Structure Fold. Des., 7, R31–R35. [DOI] [PubMed] [Google Scholar]
- Edmonds M. (1982) Poly(A) adding enzymes. In Boyer,P. (ed.), The Enzymes. Academic Press, Inc., New York, Vol. 15, pp. 217–240. [Google Scholar]
- Evans S.V. (1993) SETOR: hardware lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics, 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Gilmartin G.M. and Nevins,J.R. (1989) An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev., 3, 2180–2190. [DOI] [PubMed] [Google Scholar]
- Goldgur Y., Mosyak,L., Reshetnikova,L., Ankilova,V., Lavrik,O., Khodyreva,S. and Safro,M. (1997) The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure, 5, 59–68. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Beyer,K., Martin,G., Keller,W., Boelens,W.C. and Mattaj,L.W. (1994) The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell, 76, 531–541. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Polycarpou-Schwarz,M. and Mattaj,I.W. (1998) U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell, 1, 255–264. [DOI] [PubMed] [Google Scholar]
- Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1995) DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci., 20, 345–347. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1998) Touring protein fold space with Dali/FSSP. Nucleic Acids Res., 26, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Chopra,R., Verdine,G.L. and Harrison,S.C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science, 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D. and Steitz,T.A. (1998) Structure of T7 RNA polymerase complexed to the transcriptional inhibitor T7 lysozyme. EMBO J., 17, 4101–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Keller W., Bienroth,S., Lang,K.M. and Christofori,G. (1991) Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing signal AAUAAA. EMBO J., 10, 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer J.R., Mao,C., Braman,J.C. and Beese,L.S. (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature, 391, 304–307. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., McArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Li Y., Korolev,S. and Waksman,G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J., 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M. et al. (1994) Crystal structure of the ribosomal protein S6 from Thermus thermophilus. EMBO J., 13, 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. and Keller,W. (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases and to other nucleotidyltransferases. EMBO J., 15, 2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Martin G., Jenö,P. and Keller,W. (1999) Mapping of ATP binding regions in poly(A) polymerases by photoaffinity labeling and by mutational analysis identifies a domain conserved in many nucleotidyltransferases. Protein Sci., 8, 2380–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K.G. and Manley,J.L. (1995) The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from interfacial and thermodynamics properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Ollis D.L., Brick,P., Hamlin,R., Xuong,N.G. and Steitz,T.A. (1985) Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature, 313, 762–766. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Oubridge C., Ito,N., Evans,P.R., Teo,C.H. and Nagai,K. (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Pedersen L.C., Benning,M.M. and Holden,H.M. (1995) Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry, 34, 13305–13311. [DOI] [PubMed] [Google Scholar]
- Pelletier H., Sawaya,M.R., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer and ddCTP. Science, 264, 1891–1903. [PubMed] [Google Scholar]
- Price S.R., Evans,P.R. and Nagai,K. (1998) Crystal structure of the spliceosomal U2B′′–U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature, 394, 645–650. [DOI] [PubMed] [Google Scholar]
- Rould M.A., Perona,J.J. and Steitz,T.A. (1992) Improving multiple isomorphous replacement phasing by heavy-atom refinement using solvent-flattened phases. Acta Crystallogr. A, 48, 751–756. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- Sakon J., Liao,H.H., Kanikula,A.M., Benning,M.M., Rayment,I. and Holden,H.M. (1993) Molecular structure of kanamycin nucleotidyl transferase determined to 3.0-Å resolution. Biochemistry, 32, 11977–11984. [DOI] [PubMed] [Google Scholar]
- Sawaya M.R., Pelletier,H., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science, 264, 1930–1935. [DOI] [PubMed] [Google Scholar]
- Sawaya M.R., Prasad,R., Wilson,S.H., Kraut,J. and Pelletier,H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry, 36, 11205–11215. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Moulinier,L., Fujiwara,S., Imanaka,T., Thierry,J.C. and Moras,D. (1998) Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation. EMBO J., 17, 5227–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobol R.W., Horton,J.K., Kuhn,R., Gu,H., Singhal,R.K., Prasad,R., Rajewsky,K. and Wilson,S.H. (1996) Requirement of mammalian DNA polymerase-β in base-excision repair. Nature, 379, 183–186. [DOI] [PubMed] [Google Scholar]
- Sousa R., Chung,Y.J., Rose,J.P. and Wang,B.C. (1993) Crystal structure of bacteriophage T7 RNA polymerase at 3.3 Å resolution. Nature, 364, 593–599. [DOI] [PubMed] [Google Scholar]
- Steitz T.A. (1993) DNA- and RNA- dependent DNA polymerases. Curr. Opin. Struct. Biol., 3, 31–38. [Google Scholar]
- Steitz T.A. (1999) DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem., 274, 17395–17398. [DOI] [PubMed] [Google Scholar]
- Takagaki Y., Ryner,L.C. and Manley,J.L. (1988) Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell, 52, 731–742. [DOI] [PubMed] [Google Scholar]
- Uhlin U. and Eklund,H. (1994) Structure of ribonucleotide reductase protein R1. Nature, 370, 533–539. [DOI] [PubMed] [Google Scholar]
- Vagner S., Vagner,C. and Mattaj,I.W. (2000) The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev., 14, 403–413. [PMC free article] [PubMed] [Google Scholar]
- Varani G. and Nagai,K. (1998) RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct., 27, 407–445. [DOI] [PubMed] [Google Scholar]
- Wahle E. (1991a) A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell, 66, 759–768. [DOI] [PubMed] [Google Scholar]
- Wahle E. (1991b) Purification and characterization of a mammalian polyadenylate polymerase involved in the 3′ end processing of messenger RNA precursors. J. Biol. Chem., 266, 3131–3139. [PubMed] [Google Scholar]
- Wahle E. and Rüegsegger,U. (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- Wahle E., Martin,G., Schiltz,E. and Keller,W. (1991) Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J., 10, 4251–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Lustig,A., Jenö,P. and Maurer,P. (1993) Mammalian poly(A)-binding protein II. Physical properties and binding to polynucleotides. J. Biol. Chem., 268, 2937–2945. [PubMed] [Google Scholar]
- Wang J., Sattar,A.K., Wang,C.C., Karam,J.D., Konigsberg,W.H. and Steitz,T.A. (1997) Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell, 89, 1087–1099. [DOI] [PubMed] [Google Scholar]
- Wittmann T. and Wahle,E. (1997) Purification and characterization of full-length mammalian poly(A) polymerase. Biochim. Biophys. Acta, 1350, 293–305. [DOI] [PubMed] [Google Scholar]
- Yue D., Weiner,A.M. and Maizels,N. (1998) The CCA-adding enzyme has a single active site. J. Biol. Chem., 273, 29693–29700. [DOI] [PubMed] [Google Scholar]
- Zhao J., Hyman,L. and Moore,C. (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev., 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky A.M., Kessler,M.M. and Moore,C.L. (1995) Structure–function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s). J. Biol. Chem., 270, 26715–26720. [DOI] [PubMed] [Google Scholar]