Abstract
The diagnosis of filarial infections among individuals residing in areas where the disease is not endemic requires both strong clinical suspicion and expert training in infrequently practiced parasitological methods. Recently developed filarial molecular diagnostic assays are highly sensitive and specific but have limited availability and have not been closely evaluated for clinical use outside populations residing in areas of endemicity. In this study, we assessed the performance of a panel of real-time PCR assays for the four most common human filarial pathogens among blood and tissue samples collected from a cohort of patients undergoing evaluation for suspected filarial infections. Compared to blood filtration, real-time PCR was equally sensitive for the detection of microfilaremia due to Wuchereria bancrofti (2 of 46 samples positive by both blood filtration and PCR with no discordant results) and Loa loa (24 of 208 samples positive by both blood filtration and PCR, 4 samples positive by PCR only, and 3 samples positive by blood filtration only). Real-time PCR of skin snip samples was significantly more sensitive than microscopic examination for the detection of Onchocerca volvulus microfiladermia (2 of 218 samples positive by both microscopy and PCR and 12 samples positive by PCR only). The molecular assays required smaller amounts of blood and tissue than conventional methods and could be performed by laboratory personnel without specialized parasitology training. Taken together, these data demonstrate the utility of the molecular diagnosis of filarial infections in mobile populations.
Infections due to filarial nematodes are among the most prevalent parasitic diseases throughout the world. Although the transmission of these organisms is geographically restricted to areas in developing countries where the disease is endemic, modern human travel patterns have resulted in the migration of infected individuals to regions where filarial infections have been eradicated or have never been present, including resource-rich countries such as the United States. Despite being relatively infrequent, filarial infections are sporadically diagnosed in refugees and other immigrants from areas of endemicity, in long-term residents of regions where filarial regions are endemic (members of the armed services, students, missionaries, aid workers, and volunteers), and, rarely, among short-term travelers.
Four filarial pathogens account for the vast majority of human disease. Wuchereria bancrofti (transmitted in sub-Saharan Africa, Southeast Asia, the Caribbean, South America, and the Western Pacific) and Brugia malayi (transmitted in Southern and Southeast Asia, Indonesia, and the Philippines) are both agents of lymphatic filariasis and together infect upwards of 120 million people. Onchocerca volvulus causes onchocerciasis, or river blindness, in 20 to 40 million people, mainly in sub-Saharan Africa but also to a lesser extent in Latin America and the Arabian Peninsula. Loa loa, the African eyeworm, infects between 3 and 13 million people in Central and Western Africa.
The diagnosis of filarial infections in mobile populations can be challenging for several reasons. These infections are often subclinical, manifest with nonspecific signs and symptoms (such as itching or subcutaneous edema), or present clinically after a prepatent period of months to years following exposure. Because of the relative inexperience of many physicians in countries where these pathogens are not endemic, the recognition of these infections requires a high degree of clinical suspicion. Patients often come to attention due to incidental laboratory findings such as unexplained eosinophilia. Clinical diagnosis is sometimes possible based on compatible exposure history. More commonly, laboratory confirmation requires expert training in methods that may not be practiced routinely at most medical centers.
Conventional parasitologic assays involve the isolation of microfilariae from patient blood or tissue samples followed by staining and identification of organisms by microscopic examination for key morphological features. Serological assays (a limited number of which are available commercially) are an alternative but suffer from poor specificity and an inability to distinguish between active and prior infection (1, 20, 27). An immunochromatographic card-type assay (Filariasis Now) detects circulating antigen of W. bancrofti but is not useful for any of the other pathogens (28) and is not available commercially in Europe or North America. In recent years both conventional and real-time PCR assays have been developed for all four of the major filarial pathogens (7, 8, 11, 14, 21, 22, 24-26). While these assays have shown great promise with regard to high-level sensitivity and specificity, none is currently commercially available, and none, to our knowledge, has been in use by clinical pathology laboratories.
Selected filarial molecular diagnostic tools have been studied with patient populations in areas where filarial infections are endemic (2, 4, 6, 18, 19), in the context of specific research projects. However, the performance of these assays has not been well described among internationally mobile populations residing in resource-rich countries where the disease is not endemic. The Clinical Parasitology Unit at the National Institute of Allergy and Infectious Diseases serves to evaluate and treat patients with suspected parasitic diseases on a referral basis. The patients are primarily immigrants, returned expatriates, or travelers referred from throughout the United States and, on occasion, internationally. Since 1999, we have incorporated a panel of real-time PCR assays adapted from previously described conventional PCR targets (10, 13, 16, 31, 32) as part of routine clinical care. In this study, we have assessed the performance of this molecular diagnostic panel in comparison to conventional parasitology methods and report its utility over the past decade among patients referred to the NIH for an evaluation of suspected filarial infection.
MATERIALS AND METHODS
Patients and specimen collection.
Assay data were collected prospectively from all patients referred to the Clinical Parasitology Unit of the Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, between April 1999 and December 2009. All patients were evaluated under protocols approved by the NIAID Institutional Review Board and registered (protocols NCT00001230 and NCT00001645). Written informed consent was obtained from all subjects. Patients were either immigrants from regions of the world where filarial disease is endemic or travelers to these same regions.
Diagnostic assays were selected based on compatible clinical symptoms as well as geographic exposure history in relation to the known distributions of the major human filarial pathogens. Laboratory evaluation for all patients included basic screening studies (complete blood count, complete metabolic profile, urinalysis, and stool examination for ova and parasites) as well as serology for antifilarial IgG and IgG4 (12). For patients with suspected infection with blood-borne filariae, whole-blood samples were obtained by venipuncture either at midday (L. loa) or at midnight (W. bancrofti and B. malayi). For patients with suspected onchocerciasis, six skin snip samples were typically obtained (1 mm3 each from shoulder, iliac crest, and thigh) by using a Walzer-type corneoscleral biopsy instrument. Two of the six snips were submitted for DNA extraction and PCR analysis, while the remaining four snips were examined by conventional methods. Certain patients were evaluated longitudinally, with multiple blood or tissue samples collected over time following treatment, and some patients had duplicate blood specimens submitted on the same day or on consecutive days.
For the purposes of sensitivity and specificity calculations only, the “gold standard” for loiasis required either the demonstration of microfilariae in the blood, having an adult worm extracted, or having positive antifilarial antibodies plus Calabar swellings and response to definitive diethylcarbamazine therapy. For lymphatic filariasis the gold standard required a sample being circulating filarial antigen positive and having positive antifilarial antibodies. For onchocerciasis, the gold standard required either the demonstration of microfilariae in skin snip samples or the presence of antifilarial antibodies and onchocerca-specific antibodies (3), a compatible clinical picture, and a definitive response to ivermectin therapy.
Parasitologic assays.
Blood filtration and microscopic evaluation for microfilariae were performed by using 1 ml of anticoagulated blood as described previously (15). Skin snip samples were immersed individually into normal saline-filled wells of a plastic flat-bottom 96-well plate and examined after 24 h of incubation at 37°C for the presence of O. volvulus microfilariae.
DNA extraction.
Skin snip samples were digested in 200 μl 0.1 M EDTA disodium salt solution (Na2EDTA) (Sigma-Aldrich), 2 μl proteinase K (20 mg/ml) (Invitrogen), and 2 μl 10% sodium dodecyl sulfate (SDS) solution (Sigma-Aldrich) and incubated at 56°C in a heat block for 1 h. Following incubation, 4 μl 1.0 M dithiothreitol (DTT) solution (Sigma-Aldrich) was added, and the samples were vortexed briefly and then incubated at 95°C in a heat block for 1 h. The digested suspension was added to 0.9 ml NucliSens lysis buffer, and DNA was extracted by using the NucliSens nucleic acid isolation kit as recommended by the manufacturer (bioMérieux).
Between 1999 and November 2001, 200 μl of whole blood (EDTA) was aliquoted into 0.9-ml NucliSens lysis buffer, and DNA was extracted by using the NucliSens nucleic acid isolation kit. In an attempt to potentially increase the PCR sensitivity, the volume of blood extracted by this method was increased to 1.0 ml (added to 9.0 ml lysis buffer) beginning in December 2001. To verify the successful recovery of DNA and removal of PCR inhibitors, the lysis buffer containing the digested tissue or whole-blood specimens were spiked with an internal control (pBR322 plasmid DNA) before nucleic acid isolation. All samples were eluted with 50 μl of elution buffer.
PCR assays.
Two different types of assays were performed. Prior to May 2005, all assays were performed as previously described (16), except that probes were labeled with europium instead of fluorescein and detection of the amplification products was performed by using the Delfia plate hybridization assay (Perkin-Elmer Wallac, Inc.), with the resulting time-resolved fluorescence signals being measured on a time-resolved fluorometer. The primers and probes were then modified or redesigned (Table 1) in order to convert each assay to a real-time PCR format.
TABLE 1.
Primer and probe sequences, product size, and target DNA for each PCR assay
| Assay target and primer or probe | Sequencea | Product size (bp) | Target |
|---|---|---|---|
| Brugia malayi | 324 | HhaI repeat | |
| Bmal.FOR primer | 5′-GCGCATAAATTCATCAGC-3′ | ||
| Bmal.REV primer | 5′-CAAAACTTAATTACAAAAGCGT-3′ | ||
| Bmal.FRET.up probe | 5′-TTGAACCTGATTGACTATGTTACGT-Fluor.-3′ | ||
| Bmal.FRET.dn probe | 5′-Red640-ATTGTACCAGTGCTGGTCT-Phos.-3′ | ||
| Loa loa | 503 | Loa interspersed repeat | |
| Lloa.FOR primer | 5′-TCCTCGTTTTAGTGCT-3′ | ||
| Lloa.REV primer | 5′-GTGCTTCTTGTTATAAATGC-3′ | ||
| Lloa.FRET.up probe | 5′-GCTTAGTTTTTTTTTGAACACTGTT-Fluor.-3′ | ||
| Lloa.FRET.dn probe | 5′-Red640-ATAACCATATAAGTATCATAAATGTAAACATGT-Phos.-3′ | ||
| Wuchereria bancrofti | 134 | SspI repeat | |
| Wban.FOR primer | 5′-CGTGATGGCATCAAAG-3′ | ||
| Wban.REV primer | 5′-AAATAAGGTTATACCAAGCA-3′ | ||
| Wban.FRET.up probe | 5′-GAATTGTTTTTTTAATATTTTCAAGTATGAAT-Fluor.-3′ | ||
| Wban.FRET.dn probe | 5′-Red640-GAATTTTTAGCAATTTTTTTGTTTATATTTTTA-Phos.-3′ | ||
| Onchocerca volvulus | 154 | O-150 repeat | |
| Ovol.FOR primer | 5′-TYTTCCGRCGAANARCGCATTTTG-3′ | ||
| Ovol.REV primer | 5′-TCGCNRTRTAAATNTGNAAATTCACC-3′ |
Fluor., fluorescein label; Phos., phosphate cap.
Since May 2005, the B. malayi, L. loa, and W. bancrofti assays were performed by using a LightCycler (LC) 1.2 instrument (Roche Diagnostics) with a 20-μl reaction mixture consisting of 1× LC FastStart DNA Master HybProbe mixture containing FastStart Taq polymerase, reaction buffer deoxynucleoside triphosphate (dNTP) mix (with dUTP substituted for dTTP), 1.0 mM MgCl2 (Roche), an additional 3.0 mM MgCl2, 1.0 μM each primer, 0.2 μM each fluorescence resonance energy transfer (FRET) probe, 1 U uracil-DNA-glycosylase (UNG), and 10 μl of extracted DNA. The reaction mixture was preincubated for 10 min at 30°C to activate UNG, and DNA was denatured and UNG was inactivated at 95°C for 10 min. The template amplification consisted of 45 cycles of 5 s at 95°C, 10 s at 55°C, and 20 s at 72°C. The O. volvulus real-time PCR was performed in a 25-μl reaction mixture consisting of 1× QuantiTect SYBR green PCR master mix (Qiagen) containing HotStarTaq DNA polymerase, reaction buffer, 2.5 mM MgCl2, dNTP mix (containing a dTTP-dUTP mixture), SYBR green I and ROX (6-carboxy-X-rhodamine) fluorescent dyes, 0.5 μM each primer, 0.5 U UNG, and 5 μl of extracted DNA. Amplification was performed with a Rotorgene-3000 instrument (Qiagen) with cycling parameters of 10 min at 30°C, 10 min at 95°C, and 45 cycles of 15 s at 95°C, 30 s at 57°C, and 30 s at 72°C. A melt curve analysis was then performed by reducing the temperature to 60°C for 45 s and then raising the temperature 1°C every 5 s up to 99°C. To be considered positive, the melt peak temperature from the patient specimen must match the positive control within the specified range (72°C ± 2°C). The internal control in extracted samples was detected by amplification in a separate qualitative LC real-time PCR as described previously (5).
The analytical sensitivity analysis of the assays used in this study demonstrated lower limits of detection equal to 10 fg/μl for B. malayi, 2.5 fg/μl for L. loa, 100 fg/μl for W. bancrofti, and 400 fg/μl for O. volvulus.
RESULTS
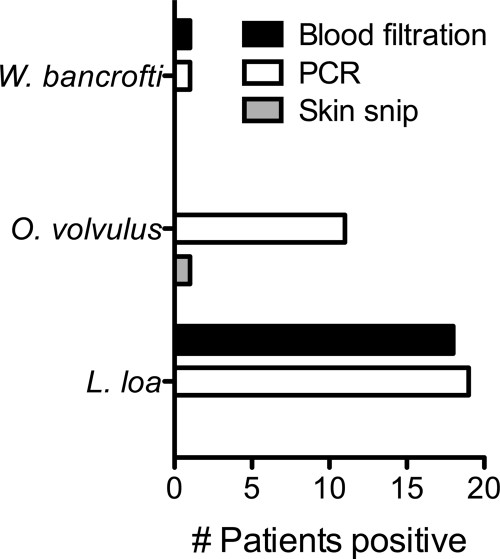
Two hundred patients were evaluated at our clinic for suspected filarial infections between April 1999 and December 2009. Parasitologic and molecular diagnostic testing for these patients is summarized in Table 2. In total, 392 specimens were collected (256 blood samples and 136 skin snip sets), and 887 diagnostic assays were performed on these specimens (356 conventional parasitologic studies and 531 PCR assays). All skin snip microscopic studies were paired with at least one (and usually two) PCR assay performed on the same tissue, while the pairing of blood filtration studies with PCR assays performed on the same blood samples was variable. In total, 19 patients were diagnosed with L. loa infection, 11 were diagnosed with onchocerciasis, 1 was diagnosed with lymphatic filariasis due to W. bancrofti, and none was diagnosed with infection due to B. malayi (Fig. 1).
TABLE 2.
Summary data for parasitologic and molecular testing for suspected filarial infections
| Test | No. of patients | No. of assaysa |
|---|---|---|
| Evaluation for suspected filarial infection | 200 | 887 |
| Parsitologic assays | ||
| Blood filtration | 139 | 220 |
| Skin snips | 129 | 136 |
| Antifilarial IgG/IgG4 | 200 | NA |
| Molecular assays | ||
| PCR for L. loa | 151 | 237 |
| PCR for W. bancrofti | 47 | 60 |
| PCR for B. malayi | 14 | 16 |
| PCR for O. volvulus | 129 | 218 |
NA, not applicable.
FIG. 1.
Diagnosis of filarial infections by PCR compared to blood filtration and skin snip microscopy.
The performance of PCR assays on blood samples in comparison to blood filtration is summarized in Table 3. Of 208 specimens tested for L. loa microfilaremia, 24 were positive by both PCR and blood filtration, while 177 were negative by both methods. L. loa microfilaremia levels among positive blood filtration assays ranged from 1 organism/ml to 7,400 organisms/ml. Discordant results occurred for 7 specimens, with 4 being positive by PCR only and 3 being positive by blood filtration only (1 to 2 organisms/ml). Removing posttreatment samples, there were 17 diagnoses of loiasis made concurrently by PCR and blood filtration, 1 diagnosis by blood filtration alone, and 2 diagnoses by PCR alone.
TABLE 3.
Comparison of results from paired blood filtration and PCR assays on blood samples from patients with suspected filarial infections
| Microscopy result | No. of samples with result |
|||||
|---|---|---|---|---|---|---|
|
L. loa |
W. bancrofti |
B. malayi |
||||
| PCR+ | PCR− | PCR+ | PCR− | PCR+ | PCR− | |
| Blood filtration positive | 24 | 3 | 2 | 0 | 0 | 0 |
| Blood filtration negative | 4 | 177 | 0 | 44 | 0 | 11 |
Comparative data were available for fewer samples when the performance of the W. bancrofti and B. malayi PCR assays were evaluated. Forty-six paired assays for W. bancrofti microfilaremia resulted in 2 specimens that were positive by both PCR and blood filtration (100 to 800 organisms/ml), 44 specimens that were negative by both methods, and no discordant results. B. malayi was not detected in any sample either by PCR or by blood filtration.
Mansonella perstans was detected by blood filtration in 3 samples (7 to 64 organisms/ml), including one patient who had a coinfection with L. loa detected by both PCR and blood filtration (data not shown). No other filarial coinfections were diagnosed. There were no paired blood samples for which discordant pathogens were identified by blood filtration and PCR.
Laboratory evaluation of patients with suspected onchocerciasis, summarized in Table 4, included 218 skin snip assays with paired microscopic and PCR assay results. There were 14 positive PCR assays from 11 patients, only 2 of which (both from a single patient's skin snip set) were also positive by conventional parasitology. No skin snip samples were positive by microscopic evaluation but negative by PCR.
TABLE 4.
Results of PCR assays on skin snip samples from patients with suspected onchocerciasis
| Microscopy result | No. of samples with result |
|
|---|---|---|
| PCR+ | PCR− | |
| Skin snip positive | 2 | 0 |
| Skin snip negative | 12 | 204 |
The clinical performance statistics for L. loa and O. volvulus PCR assays that were performed during initial patient evaluations showed that the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were all 100% for the L. loa and O. volvulus assays based on the outlined gold standard criteria. The inclusion of follow-up (posttreatment) assays in the statistical analysis decreased the sensitivity and NPV of the L. loa PCR assay to 90.3% (28/31 samples) and 98.6% (206/209), respectively, with no changes in the other L. loa or O. volvulus statistics. Performance statistics were not calculated for the B. malayi and W. bancrofti assays due to an insufficient number of infections.
DISCUSSION
An accurate diagnosis of filarial infections in mobile populations can be challenging. Infected individuals may present with nonspecific symptoms or laboratory findings, and proper evaluation requires both a strong degree of clinical suspicion as well as specialized knowledge of filarial epidemiology and pathogenesis and expertise in the morphological classification of filarial parasites by microscopic examination. Further difficulties arise from the extremely limited commercial availability of filarial diagnostic assays that can distinguish not only between specific pathogens but also between active current infections and those that occurred in the past.
In this study, we assessed the performance and feasibility in a clinical diagnostic setting of a panel of real-time PCR assays designed to detect species-specific genomic DNA target sequences of the four most prevalent pathogenic filariae of humans: W. bancrofti, B. malayi, O. volvulus, and L. loa. Compared to conventional parasitology methods, our PCR assays were overall equal to or significantly more sensitive among blood and skin snip samples collected from a cohort of 200 patients undergoing evaluations for suspected filarial infections.
Differences in assay performance were most striking for the detection of O. volvulus in skin snip samples, with diagnosis by a positive PCR assay for 11 patients compared to diagnosis by microscopy for only one patient. One possible interpretation of this discordance is that only one patient was truly infected with O. volvulus, with the remaining diagnoses representing false-positive PCR assays. This situation is unlikely, however, since DNA extraction and assay setup were conducted under rigorous conditions to protect against cross-sample contamination, and all PCR runs included internal negative controls with verified negative assay results. Moreover, each of the positive patients was treated definitively with ivermectin, and each patient had a clinical response. Some patients also had a Mazzotti-type reaction following ivermectin treatment, which indicates a high likelihood of O. volvulus infection. Each of the PCR-positive patients was also found retrospectively to be positive by highly O. volvulus-specific serological assays (3, 28; data not shown).
More likely, discordances represent situations in which very low numbers of O. volvulus microfilariae are present in skin snip samples such that microscopy is truly insensitive compared to PCR (23). It is possible that PCR allows the detection of O. volvulus DNA found within the entirety of the tissue, while microscopy detects only organisms that are capable of extruding themselves from submerged tissue samples. The increased sensitivity of our O. volvulus PCR assay is notable in that for each patient evaluation, 4 to 6 skin snip samples were typically examined microscopically, while only 2 samples were processed for DNA extraction and PCR. Our PCR assay therefore achieved a higher rate of detection despite having to overcome a potential loss of sensitivity due to sampling error. Furthermore, there were no patients diagnosed with O. volvulus infection by conventional microscopy but negative by PCR assay.
The performance of our PCR assay for L. loa infection was similar to that of blood filtration. Three paired assays were positive by blood filtration but negative by PCR, all of which were posttreatment samples collected from patients whose pretreatment evaluation included positive blood filtration and PCR assays on paired samples. Quantification of microfilaremia by blood filtration in each of these instances was only 1 or 2 organisms/ml, and there were several positive PCR assays for which the paired blood filtration identified microfilaremia at only 1 to 2 organisms/ml. Therefore, the false-negative PCR assays were most likely due to sampling error in the context of very low-level microfilaremia rather than any inherent inability of the assay to detect small quantities of parasite DNA. In further support of this interpretation, there were four paired blood samples (from four patients) for which PCR was positive but blood filtration was negative. Two of the patients had recently undergone medical therapy for loiasis, with pretreatment blood samples being positive by both PCR and blood filtration, while the other two patients likely had low-level microfilaremia upon initial evaluation. These four cases illustrate the utility of our L. loa PCR assay for the detection of low-level infection both at the time of diagnosis and during the course of monitoring the response to therapy.
An important limitation of our study is that our clinic evaluated on average only 20 new patients each year, reflecting the scarcity of opportunities to diagnose filarial infections in the United States, even at a national referral center. In particular, there were relatively few evaluations for suspected lymphatic filariasis. Due to the limited number of positive assays for W. bancrofti and B. malayi (either PCR or blood filtration), it is difficult to draw firm conclusions regarding the performance of our PCR diagnostics for these organisms, although these types of assays have been used successfully in research laboratories in countries where filarial disease is endemic.
In addition to observed gains in sensitivity, there are some distinct advantages of PCR in comparison to other available diagnostic methodologies. First, PCR directly detects filarial DNA, in contrast to serological and circulating-antigen assays, which measure indirect indicators of infection (e.g., antibodies) and which may be persistently positive long after all organisms have died (for both antibody and circulating-antigen assays). Second, our PCR assays achieve similar increased sensitivities compared to that of conventional microscopy while requiring smaller amounts of patient material as a starting point (200 μl of blood for PCR versus 1 ml or greater for filtration and 1 to 2 skin snip samples for PCR versus 6 skin snip samples for microscopy). Finally, PCR assays can be run by laboratory personnel with generalized training and do not require a specialized knowledge of parasite morphology and classification.
There are several disadvantages of PCR that must also be recognized. At this time, PCR assays require costly reagents such as kits for DNA extraction from blood or tissue, enzymes and primers that must be stored frozen, and thermal cycler machines with the ability to detect fluorescence emission (for real-time PCR assays). On a per-assay cost (between $10 and $12 [2]), once equipment is in place, PCR is likely to be of equal or lower cost than antibody-based or parasitologic methods because of significantly lower labor costs than those of classical methods.
PCR may soon become more suitable for point-of-care use in resource-poor countries with ongoing advances in the development of hand-held, battery-operated devices using microfluidic methods for all-in-one DNA extraction, amplification, and detection (9, 17, 29, 30). Unlike the situation with blood filtration, our PCR assay results are currently not reported quantitatively. However, the generation of a standard curve using defined numbers of organisms would be a relatively easy adjustment to the real-time PCR format to allow a quantitative assessment of microfilaremia (methods which we have recently developed for L. loa). Another avenue for improvement would be to multiplex the PCR assays using a different fluorescent reporter for each organism. Such an adjustment would allow the detection of microfilarial coinfections by a single assay, negating the advantage that blood filtration provides in this regard. The development and validation of an M. perstans-specific PCR assay would also be necessary for this purpose.
In summary, we have demonstrated the utility and feasibility of a panel of real-time PCR assays for the diagnosis of filarial infections among immigrants and travelers which have been used clinically for more than a decade. Our data demonstrate that the PCR panel is exquisitely species specific and slightly more sensitive than blood filtration for the detection of microfilaremia. Additionally, our real-time PCR assay for O. volvulus is significantly more sensitive than conventional microscopy for the detection of skin microfilariae. Although not quite ready for widespread use in areas of endemicity, the successful performance of these molecular assays is an important step forward in making accurate filarial diagnostic tools more accessible to clinical parasitology programs that serve internationally mobile populations.
Acknowledgments
This research was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 27 October 2010.
REFERENCES
- 1.Akue, J. P., T. G. Egwang, and E. Devaney. 1994. High levels of parasite-specific IgG4 in the absence of microfilaremia in Loa loa infection. Trop. Med. Parasitol. 45:246-248. [PubMed] [Google Scholar]
- 2.Boatin, B. A., L. Toe, E. S. Alley, N. J. Nagelkerke, G. Borsboom, and J. D. Habbema. 2002. Detection of Onchocerca volvulus infection in low prevalence areas: a comparison of three diagnostic methods. Parasitology 125:545-552. [PubMed] [Google Scholar]
- 3.Burbelo, P. D., H. P. Leahy, M. J. Iadarola, and T. B. Nutman. 2009. A four-antigen mixture for rapid assessment of Onchocerca volvulus infection. PLoS Negl Trop. Dis. 3:e438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chansiri, K., and S. Phantana. 2002. A polymerase chain reaction assay for the survey of bancroftian filariasis. Southeast Asian J. Trop. Med. Public Health 33:504-508. [PubMed] [Google Scholar]
- 5.Cohen, J. I., G. A. Fahle, M. A. Kemp, K. Apakupakul, and T. P. Margolis. 2010. Human herpesvirus 6-A, 6-B, and 7 in vitreous fluid samples. J. Med. Virol. 82:996-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dissanayake, S., A. Rocha, J. Noroes, Z. Medeiros, G. Dreyer, and W. F. Piessens. 2000. Evaluation of PCR-based methods for the diagnosis of infection in bancroftian filariasis. Trans. R. Soc. Trop. Med. Hyg. 94:526-530. [DOI] [PubMed] [Google Scholar]
- 7.Fischer, P., T. Supali, H. Wibowo, I. Bonow, and S. A. Williams. 2000. Detection of DNA of nocturnally periodic Brugia malayi in night and day blood samples by a polymerase chain reaction-ELISA-based method using an internal control DNA. Am. J. Trop. Med. Hyg. 62:291-296. [DOI] [PubMed] [Google Scholar]
- 8.Hassan, M., M. M. Sanad, I. el-Karamany, M. Abdel-Tawab, M. Shalaby, A. el-Dairouty, K. Assal, M. K. Gamal-Edin, and M. Adel el-Kadi. 2005. Detection of DNA of W. bancrofti in blood samples by QC-PCR-ELISA-based. J. Egypt. Soc. Parasitol. 35:963-970. [PubMed] [Google Scholar]
- 9.Hua, Z., J. L. Rouse, A. E. Eckhardt, V. Srinivasan, V. K. Pamula, W. A. Schell, J. L. Benton, T. G. Mitchell, and M. G. Pollack. 2010. Multiplexed real-time polymerase chain reaction on a digital microfluidic platform. Anal. Chem. 82:2310-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klion, A. D., N. Raghavan, P. J. Brindley, and T. B. Nutman. 1991. Cloning and characterization of a species-specific repetitive DNA sequence from Loa loa. Mol. Biochem. Parasitol. 45:297-305. [DOI] [PubMed] [Google Scholar]
- 11.Kluber, S., T. Supali, S. A. Williams, E. Liebau, and P. Fischer. 2001. Rapid PCR-based detection of Brugia malayi DNA from blood spots by DNA detection test strips. Trans. R. Soc. Trop. Med. Hyg. 95:169-170. [DOI] [PubMed] [Google Scholar]
- 12.Lal, R. B., and E. A. Ottesen. 1988. Enhanced diagnostic specificity in human filariasis by IgG4 antibody assessment. J. Infect. Dis. 158:1034-1037. [DOI] [PubMed] [Google Scholar]
- 13.Lizotte, M. R., T. Supali, F. Partono, and S. A. Williams. 1994. A polymerase chain reaction assay for the detection of Brugia malayi in blood. Am. J. Trop. Med. Hyg. 51:314-321. [DOI] [PubMed] [Google Scholar]
- 14.McCarthy, J. S., M. Zhong, R. Gopinath, E. A. Ottesen, S. A. Williams, and T. B. Nutman. 1996. Evaluation of a polymerase chain reaction-based assay for diagnosis of Wuchereria bancrofti infection. J. Infect. Dis. 173:1510-1514. [DOI] [PubMed] [Google Scholar]
- 15.McPherson, T., and T. B. Nutman. 2007. Filarial nematodes. In P. R. Murray, et al. (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 16.Nutman, T. B., P. A. Zimmerman, J. Kubofcik, and D. D. Kostyu. 1994. A universally applicable diagnostic approach to filarial and other infections. Parasitol. Today 10:239-243. [DOI] [PubMed] [Google Scholar]
- 17.Pipper, J., M. Inoue, L. F. Ng, P. Neuzil, Y. Zhang, and L. Novak. 2007. Catching bird flu in a droplet. Nat. Med. 13:1259-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahmah, N., A. N. Ashikin, A. K. Anuar, R. H. Ariff, B. Abdullah, G. T. Chan, and S. A. Williams. 1998. PCR-ELISA for the detection of Brugia malayi infection using finger-prick blood. Trans. R. Soc. Trop. Med. Hyg. 92:404-406. [DOI] [PubMed] [Google Scholar]
- 19.Ramzy, R. M. 2002. Field application of PCR-based assays for monitoring Wuchereria bancrofti infection in Africa. Ann. Trop. Med. Parasitol. 96(Suppl. 2):S55-S59. [DOI] [PubMed] [Google Scholar]
- 20.Rao, K. V., M. Eswaran, V. Ravi, B. Gnanasekhar, R. B. Narayanan, P. Kaliraj, K. Jayaraman, A. Marson, N. Raghavan, and A. L. Scott. 2000. The Wuchereria bancrofti orthologue of Brugia malayi SXP1 and the diagnosis of bancroftian filariasis. Mol. Biochem. Parasitol. 107:71-80. [DOI] [PubMed] [Google Scholar]
- 21.Rao, R. U., L. J. Atkinson, R. M. Ramzy, H. Helmy, H. A. Farid, M. J. Bockarie, M. Susapu, S. J. Laney, S. A. Williams, and G. J. Weil. 2006. A real-time PCR-based assay for detection of Wuchereria bancrofti DNA in blood and mosquitoes. Am. J. Trop. Med. Hyg. 74:826-832. [PMC free article] [PubMed] [Google Scholar]
- 22.Rao, R. U., G. J. Weil, K. Fischer, T. Supali, and P. Fischer. 2006. Detection of Brugia parasite DNA in human blood by real-time PCR. J. Clin. Microbiol. 44:3887-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor, H. R., B. Munoz, E. Keyvan-Larijani, and B. M. Greene. 1989. Reliability of detection of microfilariae in skin snips in the diagnosis of onchocerciasis. Am. J. Trop. Med. Hyg. 41:467-471. [DOI] [PubMed] [Google Scholar]
- 24.Toe, L., B. A. Boatin, A. Adjami, C. Back, A. Merriweather, and T. R. Unnasch. 1998. Detection of Onchocerca volvulus infection by O-150 polymerase chain reaction analysis of skin scratches. J. Infect. Dis. 178:282-285. [DOI] [PubMed] [Google Scholar]
- 25.Toure, F. S., O. Bain, E. Nerrienet, P. Millet, G. Wahl, Y. Toure, O. Doumbo, L. Nicolas, A. J. Georges, L. A. McReynolds, and T. G. Egwang. 1997. Detection of Loa loa-specific DNA in blood from occult-infected individuals. Exp. Parasitol. 86:163-170. [DOI] [PubMed] [Google Scholar]
- 26.Toure, F. S., L. Kassambara, T. Williams, P. Millet, O. Bain, A. J. Georges, and T. G. Egwang. 1998. Human occult loiasis: improvement in diagnostic sensitivity by the use of a nested polymerase chain reaction. Am. J. Trop. Med. Hyg. 59:144-149. [DOI] [PubMed] [Google Scholar]
- 27.Vincent, J. A., S. Lustigman, S. Zhang, and G. J. Weil. 2000. A comparison of newer tests for the diagnosis of onchocerciasis. Ann. Trop. Med. Parasitol. 94:253-258. [DOI] [PubMed] [Google Scholar]
- 28.Weil, G. J., C. Steel, F. Liftis, B. W. Li, G. Mearns, E. Lobos, and T. B. Nutman. 2000. A rapid-format antibody card test for diagnosis of onchocerciasis. J. Infect. Dis. 182:1796-1799. [DOI] [PubMed] [Google Scholar]
- 29.Wulff-Burchfield, E., W. A. Schell, A. E. Eckhardt, M. G. Pollack, Z. Hua, J. L. Rouse, V. K. Pamula, V. Srinivasan, J. L. Benton, B. D. Alexander, D. A. Wilfret, M. Kraft, C. B. Cairns, J. R. Perfect, and T. G. Mitchell. 2010. Microfluidic platform versus conventional real-time polymerase chain reaction for the detection of Mycoplasma pneumoniae in respiratory specimens. Diagn. Microbiol. Infect. Dis. 67:22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeung, S. S., T. M. Lee, and I. M. Hsing. 2008. Electrochemistry-based real-time PCR on a microchip. Anal. Chem. 80:363-368. [DOI] [PubMed] [Google Scholar]
- 31.Zhong, M., J. McCarthy, L. Bierwert, M. Lizotte-Waniewski, S. Chanteau, T. B. Nutman, E. A. Ottesen, and S. A. Williams. 1996. A polymerase chain reaction assay for detection of the parasite Wuchereria bancrofti in human blood samples. Am. J. Trop. Med. Hyg. 54:357-363. [DOI] [PubMed] [Google Scholar]
- 32.Zimmerman, P. A., R. H. Guderian, E. Aruajo, L. Elson, P. Phadke, J. Kubofcik, and T. B. Nutman. 1994. Polymerase chain reaction-based diagnosis of Onchocerca volvulus infection: improved detection of patients with onchocerciasis. J. Infect. Dis. 169:686-689. [DOI] [PubMed] [Google Scholar]