Abstract
Mupirocin is a topical antimicrobial used to eradicate methicillin-resistant Staphylococcus aureus (MRSA) colonization, usually in the absence of susceptibility testing. We hypothesized that high-level (HL) mupirocin resistance was associated with multidrug resistance (MDR). To this end, unique patient isolates identified at our institution during 2008 were stratified into those resistant to ≥3 non-β-lactam antimicrobial classes (MDR) and non-MDR MRSA. HL mupirocin resistance was screened by mupA PCR on all MDR isolates (n = 191) and a 20% random sample (n = 130) of non-MDR isolates; E-testing confirmed HL resistance. We found that among MDR isolates, 13 (6.8%) carried mupA, whereas none of the non-MDR isolates did (P = 0.001). Thus, although the overall prevalence of HL mupirocin resistance is low among MRSA isolates at our institution, an association exists between mupA carriage and MDR. Using genotyping and antimicrobial susceptibility profiling, we identified nine HL mupirocin-resistant clones. Whereas the majority of mupA-negative MDR isolates had a health care-associated MRSA (HA-MRSA) genotype (multilocus sequence type 5 [ST5] or SCCmec type II), the majority of mupA-positive MDR isolates had a community-associated MRSA (CA-MRSA) genotype (ST8 or SCCmec type IV). However, CA- and HA-MRSA genotypes were more evenly distributed among mupA-positive isolates compared to mupA-negative MDR isolates. Thus, in Chicago, mupA is circulating among both CA- and HA-MRSA backgrounds.
Methicillin-resistant Staphylococcus aureus (MRSA) has become a significant problem in both community and health care settings (9, 15, 22). In an effort to decrease the risk of acquiring MRSA infection in hospitals, laws mandating screening for MRSA colonization among high-risk patients (such as those admitted to intensive care units [ICUs]) have been passed in several states in the United States, including Illinois (1). Patients who test positive for MRSA colonization are usually placed in isolation, and contact precautions are instituted. Some institutions or practitioners choose to use antimicrobial decolonization regimens for identified carriers.
Mupirocin (pseudomonic acid A) has in vitro activity against most Gram-positive organisms and a few Gram-negative organisms (12). It is approved for decolonization of S. aureus nasal carriage in patients >12 years of age (16, 29). The prevalence of mupirocin resistance in S. aureus varies greatly, with most studies of clinical isolates reporting a rate of <10% (3-5, 10, 20, 21, 24, 27, 28, 30-33).
Mupirocin acts by competitively binding isoleucyl tRNA synthetase (ileS), thereby disrupting protein synthesis (14). Low-level (LL) resistance (MIC = 8 to 256 μg/ml) usually results from a mutation in the chromosomal ileS gene (35). High-level (HL) resistance (MIC ≥ 512 μg/ml) occurs with acquisition of a novel ileS gene (mupA or ileS2) on a transferable plasmid (29).
Since the early reports documenting community-associated MRSA (CA-MRSA) infection among healthy children lacking risk factors for health care-associated MRSA (HA-MRSA) infection (18), several criteria (temporal, health care exposure risk factors, antimicrobial susceptibility, and molecular signatures) have been applied to distinguish CA- and HA-MRSA (6, 9). Because one criterion for defining a CA-MRSA isolate has been non-multidrug resistance, it was concerning when multidrug-resistant (MDR) clones of CA-MRSA were documented in Taipei, Taiwan (strain USA1000, sequence type 59 [ST59]) (7), and in San Francisco and Boston (strain USA300, ST8) (11, 17). The USA300 MDR isolates were all mupirocin resistant. The MDR strain of USA300 was especially concerning because USA300 isolates are virulent and readily transmissible. Therefore, they are likely to spread to other locales. Also, data from an MRSA surveillance network in the United States suggested that resistance to mupirocin and other non-β-lactams is emerging among USA300 isolates (26).
To define the epidemiology of mupirocin resistance among MRSA isolates in the era of epidemic CA-MRSA, we screened clinical MRSA isolates from the University of Chicago Medical Center (UCMC) prospectively collected in 2008; 3 (1.8%) of the 168 isolates carried mupA (8). Because all three were resistant to ≥3 classes of non-β-lactam antimicrobials, defined in the present study as MDR, we sought to more rigorously test the hypothesis that mupA was associated with MDR. Therefore, we screened for the prevalence of mupA carriage among a stratified sample of non-MDR and MDR MRSA isolates at our institution during 2008. To assess whether the mupA-positive, MDR USA300 strain reported in San Francisco and Boston was also prevalent at our institution in Chicago, we characterized the mupA-positive isolates by SCCmec typing, multilocus sequence type (MLST) and antibiotic resistance profiling.
(This research was presented in part at the 49th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, on 12 to 15 September 2009, Abstr. C2-140.)
MATERIALS AND METHODS
Setting.
The UCMC is a tertiary care facility located in Chicago, IL, that provides inpatient and outpatient services for patients from Chicago and surrounding regions. UCMC had an average of 26,000 admissions, 380,000 outpatient visits, and more than 80,000 emergency room visits in 2007 and 2008. The UCMC Clinical Microbiology Laboratory saves only bacterial isolates from blood. To expand this collection to include all types of infection isolates, starting in November 2003, we have prospectively collected consecutive MRSA clinical isolates from the UCMC Clinical Microbiology Laboratory as described previously (9). The Institutional Review Board of the Biological Sciences Division at the University of Chicago has approved the present study.
Antibiotic susceptibility testing.
All clinical isolates obtained during 2008 were identified by the Clinical Microbiology Laboratory at UCMC according to Clinical and Laboratory Standards Institute (CLSI) guidelines (37). Antibiotic susceptibility testing (AST) was performed by using the Vitek 2 instrument (bioMérieux, Raleigh, NC) with the AST-GP66 card. Intermediate susceptibility was categorized as resistant. Isolates resistant to erythromycin (ERY) but susceptible to clindamycin (CLI) were tested for inducible CLI resistance by using the D-zone test according to CLSI protocols; isolates with inducible resistance were categorized as CLI resistant. An Etest (AB Biodisk, Sweden) was used to determine the MIC of mupirocin, according to the manufacturer's instructions. Briefly, Etest strips were applied onto Bacto tryptic soy agar (Difco, Sparks, MD) that was inoculated with a suspension of cells adjusted to the optical density of a 0.5 McFarland standard (Remel, Lenexa, KS).
Bacterial isolates.
We collected 1,189 clinical MRSA isolates from the UCMC Clinical Microbiology Laboratory between 1 January and 31 December 2008. Of these, only the first isolate from each patient (n = 837) was included in the present study (unique patient isolates). Isolates were confirmed as S. aureus using the Staphaurex rapid latex test (Remel).
Molecular characterization of isolates.
Genomic DNA was extracted from each isolate by using a Qiagen DNeasy blood and tissue kit according to the manufacturer's instructions, modified by incubation with lysostaphin in the resuspension buffer (at 37°C for 30 min) to facilitate S. aureus lysis. Confirmation of S. aureus speciation was accomplished by a PCR assay specific for spa (encoding protein A) (23).
MRSA isolates were characterized by MLST to determine the genetic background and, by typing of the SCCmec element, a mobile genetic element that carries mecA into the chromosome of S. aureus (13, 19). SCCmec typing was performed by PCR (7) with type assignments using published guidelines (19). Detection of genes encoding the Panton-Valentine leukocidin (PVL) was performed as described previously (25).
For detection of mupA, we designed primers (5′-ATTGATAGACTCCCTATCAGAGTATGATAAAAAAAG-3′ and 5′-CTATAACATTTAAGTGTATATTTTTAATCAGCAAA-3′) corresponding to a region of high variability between chromosomal ileS and mupA. For the positive control, we used genomic DNA from strain NRS107 (obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus [NARSA]).
Definitions of CA- and HA-MRSA.
Four major criteria have been variably used for the classification of CA- and HA-MRSA: the interval between healthcare exposure and disease onset (temporal), risk factors, genotype, and antimicrobial susceptibility (9). We defined the CA-MRSA genotype as an isolate that carried the type IV SCCmec element or belonged to multilocus sequence type 8 (ST8). Isolates harboring type II SCCmec or belonging to ST5 were considered to have an HA-MRSA genotype. Those with other SCCmec element types and other STs were not classified. By the temporal definition, community onset is defined as an infection with cultures obtained from outpatients or from inpatients within 3 days of hospital admission. Health care onset is assigned to infections for which cultures were obtained from inpatients thereafter.
Statistical analysis.
Statistical analyses were performed using STATA/SE 10 (College Station, TX). Two-tailed Fisher exact tests and logistic regression analyses were used when appropriate. P values of <0.05 were deemed statistically significant. Our strategy for selecting a random sample of isolates consisted in selecting all clinical isolates from patients whose UCMC medical record number ended with 0, followed by those ending with 1, then with 2, then with 3, etc., until the desired number of isolates was reached.
RESULTS
Screening for mupA.
The 837 unique patient MRSA isolates from 2008 were stratified into MDR and non-MDR. The majority of the isolates (77% [646/837]) were non-MDR, with most (52% [436/837]) exhibiting resistance to only one non-β-lactam antimicrobial class, suggesting a large proportion of CA-MRSA was in the sample. Thus, 23% (191/837) of the isolates were MDR.
To compare the prevalence of mupA carriage in MDR and non-MDR MRSA isolates, the mupA PCR assay was performed on all 191 MDR isolates and a 20% randomly selected sample (130/646) of the non-MDR MRSA isolates (hereafter referred to as the random sample). We found that 6.8% (13/191) of the MDR MRSA isolates carried mupA, whereas none of the 130 non-MDR isolates did (P = 0.001).
All 13 mupA-positive isolates demonstrated HL mupirocin resistance, with an MIC of >1,024 μg/ml, the same as the positive control strain, NRS107. A mupA-negative control isolate had an MIC of 0.38 μg/ml.
Sites of isolation.
The distribution of the sites of isolation from which the MRSA isolates were obtained differed significantly when comparing the random non-MDR sample to the MDR MRSA isolates (P < 0.0001) (Table 1). This result was not due to a selection bias since the random sample of non-MDR MRSA isolates was from sites of isolation similar to those of the total non-MDR sample (P = 0.09). No difference in sites of isolation was found between the mupA-positive and mupA-negative MDR isolates (P = 0.2).
TABLE 1.
Patient age, sex, and site of isolation for unique patient MRSA isolates from UCMCa
| Parameter | No. of isolates (%) |
|||
|---|---|---|---|---|
| Non-MDRc (n = 130) | MDR |
|||
| Total (n = 191) | mupA positive (n = 13) | mupA negative (n = 178) | ||
| Patient demographics | ||||
| Overall age range | 5 mo to 81 yrs | 3 wks to 89 yrs | 9 to 72 yrs | 3 wks to 89 yrs |
| No. ≤18 yrs old | 63 (48.5) | 18 (9.4) | 1 (7.7) | 17 (9.6) |
| No. male | 63 (48.5) | 91 (47.6) | 7 (53.8) | 84 (47.2) |
| Sites of isolationb | ||||
| Skin or soft tissue | 112 (86.2) | 88 (46.1) | 9 (69.2) | 79 (44.4) |
| Blood | 4 (3.1) | 15 (7.9) | 0 (0) | 15 (8.4) |
| Bone | 0 (0) | 2 (1) | 1 (7.7) | 1 (0.6) |
| Ear or eye | 0 (0) | 2 (1) | 0 (0) | 2 (1.1) |
| Urine | 1 (0.8) | 18 (9.4) | 0 (0) | 18 (10.1) |
| Respiratory tract | 10 (7.7) | 10 (7.7) | 2 (15.4) | 49 (27.5) |
| Other | 3 (2.3) | 3 (2.3) | 1 (7.7) | 14 (7.9) |
Isolates were obtained from 1 January 2008 to 31 December 2008. Values are expressed as the number of isolates (%) except as noted for the demographic data.
The site of isolation is based on the microbiology order request.
This value represents a 20% random sample of the non-MDR isolates.
Genotypic characteristics of MDR MRSA isolates.
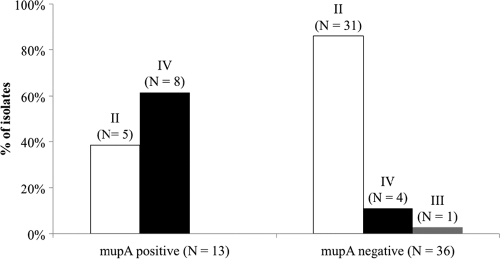
The genotypes of a sample of the MDR MRSA isolates that lacked mupA (n = 36) and the MDR MRSA isolates that carried mupA (n = 13) were determined. Among the mupA-positive MRSA isolates, 9 patterns (A to I) were distinguished by combining the genotypic and antimicrobial susceptibility profiles (Table 2). As shown in Fig. 1, the distribution of SCCmec types II and IV differed significantly between mupA-positive and mupA-negative MDR isolates (P = 0.001). Although a minority of mupA-negative MDR isolates carried SCCmec type IV (11%), it was carried by a majority (62%) of mupA-positive MDR isolates. Also, SCCmec types II and IV were more evenly distributed among the mupA-positive MDR isolates (38 and 62%, respectively) compared to the mupA-negative MDR isolates (86 and 11%, respectively).
TABLE 2.
Susceptibility and genotypic profiles for mupA-positive MDR MRSA isolates from UCMC from 1 January 2008 to 31 December 2008a
| Clonal cluster | Pattern | n | Susceptibility profile |
No. of agents | Genotypic profile |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | CLI | CIP | GEN | RIF | TMP-SXT | VAN | SCCmec type | ST | PVL | ||||
| CC5 (n = 6) | A | 1 | R | R | R | S | S | S | S | 3 | II | 105 | NEG |
| B | 1 | R | R | R | S | S | S | S | 3 | IV | 5 (slv) | NEG | |
| C | 2 | R | R | R | R | S | S | S | 4 | II | 5 | NEG | |
| D | 2 | R | R | R | R | R | R | S | 6 | II | 5 | NEG | |
| CC8 (n = 7) | E | 1 | R | R | R | S | S | S | S | 3 | IV | 8 | POS |
| F | 2 | R | S | R | I | S | R | S | 4 | IV | 8 | NEG | |
| G | 2 | R | R | R | I | S | R | S | 5 | IV | 8 | NEG | |
| H | 1 | R | R | R | I | R | R | S | 6 | IV | 8 | NEG | |
| I | 1 | R | S | S | R | R | R | S | 4 | IV | 449 | NEG | |
The number of agents refers to the number of antimicrobial agents against which each isolate exhibited resistance. “n” indicates the number of isolates within each clone. Antibiotics: ERY, erythromycin; CLI, clindamycin; CIP, ciprofloxacin; GEN, gentamicin; RIF, rifampin; TMP-SXT, trimethoprim-sulfamethoxazole; VAN, vancomycin. Other abbreviations: R, resistant; S, susceptible; I, intermediate; ST, multilocus sequence type; CC, clonal cluster; PVL, Panton-Valentine leukocidin; NEG, negative; POS, positive; slv, single-locus variant. The patterns are based on the distinct SCCmec type and antimicrobial resistance profile within each ST.
FIG. 1.
Distribution of SCCmec types (Roman numerals above each bar) among mupA-positive and mupA-negative MDR MRSA isolates. The sample includes all mupA-positive MDR (n = 13) isolates and a 20% random sample (n = 36) of mupA-negative MDR isolates.
The distribution of sequence types was significantly different comparing mupA-positive with mupA-negative MDR isolates (P = 0.004) (Table 3). Whereas the large majority of mupA-negative MDR isolates belonged to ST5 (72%), the majority of mupA-positive MDR isolates belonged to ST8 (46%). However, the mupA-positive MDR isolates were more evenly distributed between ST5 and ST8 genetic backgrounds (31 and 46%, respectively) compared to the mupA-negative MDR isolates (72 and 8%, respectively).
TABLE 3.
Distribution of mupA-positive and mupA-negative MDR MRSA isolates according to clonal cluster and sequence typea
| Clonal cluster | ST | No. (%) |
|
|---|---|---|---|
| mupA negative | mupA positive | ||
| CC5 | 5 | 26 (72) | 4 (30.8) |
| 5slv | 0 (0) | 1 (7.7) | |
| 5dlv | 1 (2.8) | 0 (0) | |
| 231 | 2 (5.6) | 0 (0) | |
| 105 | 2 (5.6) | 1 (7.7) | |
| CC8 | 8 | 3 (8.3) | 6 (46.2) |
| 239 | 1 (2.8) | 0 (0) | |
| 449 | 0 (0) | 1 (7.7) | |
| CC22 | 22slv | 1 (2.8) | 0 (0) |
| Total | 36 (100) | 13 (100) | |
CC, clonal cluster; ST, sequence type; slv, single-locus variant; dlv, double-locus variant; MDR, multidrug resistant; MRSA, methicillin-resistant S. aureus.
The carriage of pvl genes among MDR MRSA isolates was infrequent with a similar prevalence observed among mupA-positive (1/13, 7.7%) and mupA-negative (2/36, 5.6%) MDR MRSA isolates (P = 1).
Association of mupA with resistance to non-β-lactam antibiotics.
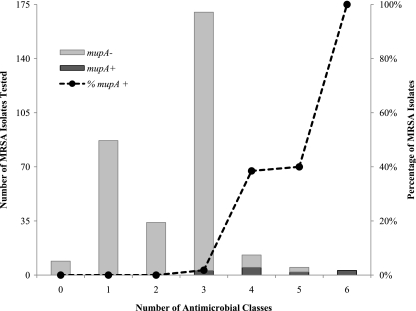
As determined by logistic regression, there was a statistically significant association between mupA carriage and a greater number of non-β-lactam antimicrobial classes to which an isolate was resistant (odds ratio = 9.83, 95% confidence interval = 4.04 to 23.9; P < 0.001). The frequency of MRSA isolates carrying mupA was highest for isolates resistant to ≥4 non-β-lactam agents, although there were too few of these isolates to detect a statistical significance (Fig. 2). The same association was detected in a subanalysis that included only antibiotics with resistance determinants that are often carried on transferable plasmids (ERY, CLI, gentamicin [GEN], and trimethoprim-sulfamethoxazole [TMP-SXT]).
FIG. 2.
Total number of MRSA isolates tested for the presence of mupA stratified by the number of non-β-lactam antimicrobial classes to which isolates were resistant (n = 321). The percentage of isolates in each group carrying mupA is shown as a dashed line, illustrating an increasing tendency of mupA carriage with an increasing number of agents to which isolates are resistant.
When considering the relationship between MRSA carriage of mupA and resistance to any single antimicrobial agent, mupA was significantly more likely to be carried by isolates resistant to GEN, rifampin (RIF), or TMP-SXT (P < 0.0001), in comparison to ERY-, CLI-, or ciprofloxacin (CIP)-resistant isolates (P = 1, P = 0.3, and P = 0.07, respectively). The association between mupA carriage and GEN or TMP-SXT might be due to the fact that these resistance determinants were carried together on the same plasmid.
However, the association between RIF resistance and mupA carriage was unexpected since RIF resistance is chromosomally mediated (2). To disprove the possibility that the RIF-resistant, mupA-positive isolates might represent a single MRSA clone, we studied the genotypic and susceptibility profiles of these isolates (Table 2). Among the four mupA-positive, RIF-resistant MRSA isolates, two belonged to ST5 and had an identical susceptibility profile (pattern D), one belonged to ST8 (pattern H), and one belonged to ST449 (pattern I). These data suggest that there were at least three mupA-positive clones among the four RIF-resistant isolates. Therefore, the association between mupA and RIF resistance was not due to the circulation of a single clone.
Relationship between the temporal and genotypic criteria of CA- and HA-MRSA among MDR MRSA isolates.
All nine MDR isolates belonging to ST8 (CA genotype) were from infections with community onset. Of the 30 isolates belonging to ST5 (HA genotype), only 3 (10%) were isolated from patients with health care-onset infections.
All 12 MDR isolates carrying SCCmec type IV (CA genotype) were from patients with community-onset infections. Of the 36 isolates carrying SCCmec type II (HA genotype), only 4 (11%) were isolated from patients with health care onset infections.
DISCUSSION
With the increasing prevalence of virulent MRSA strains in the United States, many states have mandated testing for MRSA colonization on admission to ICUs. In instances in which decolonization is to be attempted, it is important to maintain awareness of the susceptibilities of MRSA to antimicrobial agents that could be used for decolonizing. Mupirocin is often used for this purpose, although the mupirocin susceptibility of the MRSA isolate is often unknown. Although the overall rate of mupirocin resistance was low (1.8%), a 6.8% incidence of HL mupirocin resistance was found among MDR isolates when we stratified our sample. Moreover, we found a strong association of HL mupirocin resistance in isolates with resistance to ≥4 non-β-lactam antimicrobial classes. The low overall prevalence of HL mupirocin resistance might reflect the lack of a policy at our institution to use mupirocin to decolonize patients who screen positive for MRSA at admission.
The epidemiology of our HL mupirocin-resistant isolates contrasts with other geographic regions. A surgical ICU in St. Louis (2002 to 2004) (20) and a study from Canada (1995 to 2004) (34) reported that HL mupirocin resistance was found mostly in MRSA isolates with an HA-MRSA genotype. At the other end of the spectrum, a high prevalence of a single HL mupirocin-resistant MDR clone of CA-MRSA strain, USA300/ST8, was circulating among men who have sex with men (MSM) in San Francisco and Boston during 2004 to 2006 (11, 17). In contrast to San Francisco and Boston, we found at least nine HL mupirocin-resistant MDR clones, distinguished by an analysis of the SCCmec type, ST, and antibiotic susceptibility profiles. Moreover, four clones were found among the mupirocin-resistant MDR isolates that had a CA-MRSA genotype, despite their small number. Another notable difference is that our HL mupirocin-resistant MDR CA-MRSA isolates were not primarily circulating among MSM, since half of our mupA-positive ST8 isolates were obtained from females. Although we found CA-MRSA MDR isolates, they were not disseminated widely in our region, and more than one clone exists.
A unique characteristic of our study was that the distribution of CA- and HA-MRSA genotypes was significantly different when mupA-negative and mupA-positive MDR MRSA isolates were compared. Whereas a great majority of the mupA-negative MDR isolates had a HA-MRSA genotype (as would be expected for HA-MRSA), the majority of mupA-positive MDR MRSA had a CA-MRSA genotype. However, the mupA-positive isolates were more evenly distributed between both CA- and HA-MRSA genotypes compared to the mupA-negative MDR isolates. This suggests that MDR plasmids harboring mupA are being transferred readily into CA- and HA-MRSA genotypes in Chicago. These data support the idea that CA-MRSA isolates have been increasing in resistance to non-β-lactam agents since the emergence of CA-MRSA strains (9, 18). In support of this, McDougal et al. recently reported the emergence of non-β-lactam antimicrobial resistance among clinical USA300 CA-MRSA isolates in several U.S. locations (26). However, in that study, prevalence of isolates with resistance to ≥3 non-β-lactam agents was not reported; the study only focused on invasive isolates and was not centered in the Midwest as it was here.
Our study provides an estimate of the prevalence of HL mupirocin resistance among MRSA isolates in a large metropolitan medical center in the midwestern United States where data are lacking. Previous determinations of the prevalence of HL mupirocin resistance on unselected isolates are few, and study designs and approaches to characterizing the isolates have varied widely (3, 20, 27, 36). The current study included an unbiased sample of unique patient consecutive MRSA isolates obtained from any cultured body site over a 1-year period, thereby allowing for the inclusion of both CA- and HA-MRSA genotypes. Moreover, the methods we used to genotype the isolates can be reproducibly performed by other investigators, which allows our results to be readily compared to other studies. Furthermore, by stratifying the sample into MDR and non-MDR, we were able to identify an association between mupA carriage and MDR among both CA- and HA-MRSA genotypes. By genotyping the isolates, we also showed that the CA-MRSA genotypes correlated closely with community-onset infection, whereas strains with the HA genotype had poor concordance with health care onset.
A limitation of the present study is that we did not assess risk factors for previous health care exposure. We also examined isolates from only one medical center. Nevertheless, we serve an inner city population in a major metropolitan area that is highly affected by MRSA. The information we report here may also apply to other large medical centers in our city as well as in other cities of the United States where data are lacking.
In conclusion, the association between mupirocin resistance and MDR has important clinical implications. First, mupirocin susceptibility testing should be considered when decolonization with mupirocin is planned for patients colonized with MDR MRSA isolates. Second, given that mupA is usually physically linked to multiple determinants of antimicrobial resistance on a conjugal plasmid, widespread use of mupirocin has the potential to increase the prevalence of MDR among CA- and HA-MRSA isolates alike by selecting for MDR isolates. Third, although the MDR USA300 strains have not yet become widespread in our region, the presence of such strains in our collection underscores the need for diligence in continued surveillance.
Acknowledgments
This research was supported by the Children's Research Foundation Fund (A.C.). Pfizer (R.S.D.) provided support for the prospective surveillance of MRSA. Isolate NRS107 was obtained from NARSA under NIAID/NIH contract HHSN272200700055C.
None of the authors have any conflict of interest relating to this project.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Association for Professionals in Infection Control and Epidemiology. 2010. MRSA laws and pending legislation. APIC, Washington, DC. http://www.apic.org/downloads/legislation/MRSA_map.gif.
- 2.Aubry-Damon, H., C. J. Soussy, and P. Courvalin. 1998. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 42:2590-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babu, T., V. Rekasius, J. P. Parada, P. Schreckenberger, and M. Challapalli. 2009. Mupirocin resistance among methicillin-resistant Staphylococcus aureus-colonized patients at admission to a tertiary care medical center. J. Clin. Microbiol. 47:2279-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, W. E., M. L. Wallis, K. B. Tucker, B. A. Reboussin, and R. J. Sherertz. 2004. Staphylococcus aureus nasal carriage in a student community: prevalence, clonal relationships, and risk factors. Infect. Control Hosp. Epidemiol. 25:485-491. [DOI] [PubMed] [Google Scholar]
- 5.Bowler, W. A., J. Bresnahan, A. Bradfish, and C. Fernandez. 2010. An integrated approach to methicillin-resistant Staphylococcus aureus control in a rural, regional-referral healthcare setting. Infect. Control Hosp. Epidemiol. 31:269-275. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., and R. S. Daum. 2007. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine leukocidin. Lab. Invest. 87:3-9. [DOI] [PubMed] [Google Scholar]
- 7.Boyle-Vavra, S., B. Ereshefsky, C. C. Wang, and R. S. Daum. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 43:4719-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadilla, A., R. S. Daum, and S. Boyle-Vavra. 2009. Mupirocin resistance associated with multi-drug resistant MRSA. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-140.
- 9.David, M. Z., D. Glikman, S. E. Crawford, J. Peng, K. J. King, M. A. Hostetler, S. Boyle-Vavra, and R. S. Daum. 2008. What is community-associated methicillin-resistant Staphylococcus aureus? J. Infect. Dis. 197:1235-1243. [DOI] [PubMed] [Google Scholar]
- 10.Deshpande, L. M., A. M. Fix, M. A. Pfaller, and R. N. Jones. 2002. Emerging elevated mupirocin resistance rates among staphylococcal isolates in the SENTRY Antimicrobial Surveillance Program (2000): correlations of results from disk diffusion, Etest and reference dilution methods. Diagn. Microbiol. Infect. Dis. 42:283-290. [DOI] [PubMed] [Google Scholar]
- 11.Diep, B. A., H. F. Chambers, C. J. Graber, J. D. Szumowski, L. G. Miller, L. L. Han, J. H. Chen, F. Lin, J. Lin, T. H. Phan, H. A. Carleton, L. K. McDougal, F. C. Tenover, D. E. Cohen, K. H. Mayer, G. F. Sensabaugh, and F. Perdreau-Remington. 2008. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann. Intern. Med. 148:249-257. [DOI] [PubMed] [Google Scholar]
- 12.Eltringham, I. 1997. Mupirocin resistance and methicillin-resistant Staphylococcus aureus (MRSA). J. Hosp. Infect. 35:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farmer, T. H., J. Gilbart, and S. W. Elson. 1992. Biochemical basis of mupirocin resistance in strains of Staphylococcus aureus. J. Antimicrob. Chemother. 30:587-596. [DOI] [PubMed] [Google Scholar]
- 15.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 16.GlaxoSmithKline. 2009. Bactroban nasal prescribing information. GlaxoSmithKline, Brentford, United Kingdom. http://us.gsk.com/products/assets/us_bactroban_nasal.pdf.
- 17.Han, L. L., L. K. McDougal, R. J. Gorwitz, K. H. Mayer, J. B. Patel, J. M. Sennott, and J. L. Fontana. 2007. High frequencies of clindamycin and tetracycline resistance in methicillin-resistant Staphylococcus aureus pulsed-field type USA300 isolates collected at a Boston ambulatory health center. J. Clin. Microbiol. 45:1350-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herold, B. C., L. C. Immergluck, M. C. Maranan, D. S. Lauderdale, R. E. Gaskin, S. Boyle-Vavra, C. D. Leitch, and R. S. Daum. 1998. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA 279:593-598. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, J. C., T. J. Rogers, P. Brookmeyer, W. M. Dunne, Jr., G. A. Storch, C. M. Coopersmith, V. J. Fraser, and D. K. Warren. 2007. Mupirocin resistance in patients colonized with methicillin-resistant Staphylococcus aureus in a surgical intensive care unit. Clin. Infect. Dis. 45:541-547. [DOI] [PubMed] [Google Scholar]
- 21.Jones, P. G., T. Sura, M. Harris, and A. Strother. 2003. Mupirocin resistance in clinical isolates of Staphylococcus aureus. Infect. Control Hosp. Epidemiol. 24:300-301. [DOI] [PubMed] [Google Scholar]
- 22.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763-1771. [DOI] [PubMed] [Google Scholar]
- 23.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaPlante, K. L. 2007. In vitro activity of lysostaphin, mupirocin, and tea tree oil against clinical methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 57:413-418. [DOI] [PubMed] [Google Scholar]
- 25.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 26.McDougal, L. K., G. E. Fosheim, A. Nicholson, S. N. Bulens, B. M. Limbago, J. E. Shearer, A. O. Summers, and J. B. Patel. 2010. Emergence of resistance among USA300 methicillin-resistant Staphylococcus aureus isolates causing invasive disease in the United States. Antimicrob. Agents Chemother. 54:3804-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mongkolrattanothai, K., P. Mankin, V. Raju, and B. Gray. 2008. Surveillance for mupirocin resistance among methicillin-resistant Staphylococcus aureus clinical isolates. Infect. Control Hosp. Epidemiol. 29:993-994. [DOI] [PubMed] [Google Scholar]
- 28.O'Shea, S., L. Cotter, S. Creagh, S. Lydon, and B. Lucey. 2009. Mupirocin resistance among staphylococci: trends in the southern region of Ireland. J. Antimicrob. Chemother. 64:649-650. [DOI] [PubMed] [Google Scholar]
- 29.Patel, J. B., R. J. Gorwitz, and J. A. Jernigan. 2009. Mupirocin resistance. Clin. Infect. Dis. 49:935-941. [DOI] [PubMed] [Google Scholar]
- 30.Paterson, D. L., J. D. Rihs, C. Squier, T. Gayowski, A. Sagnimeni, and N. Singh. 2003. Lack of efficacy of mupirocin in the prevention of infections with Staphylococcus aureus in liver transplant recipients and candidates. Transplantation 75:194-198. [DOI] [PubMed] [Google Scholar]
- 31.Perl, T. M., J. J. Cullen, R. P. Wenzel, M. B. Zimmerman, M. A. Pfaller, D. Sheppard, J. Twombley, P. P. French, and L. A. Herwaldt. 2002. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N. Engl. J. Med. 346:1871-1877. [DOI] [PubMed] [Google Scholar]
- 32.Ridenour, G., R. Lampen, J. Federspiel, S. Kritchevsky, E. Wong, and M. Climo. 2007. Selective use of intranasal mupirocin and chlorhexidine bathing and the incidence of methicillin-resistant Staphylococcus aureus colonization and infection among intensive care unit patients. Infect. Control Hosp. Epidemiol. 28:1155-1161. [DOI] [PubMed] [Google Scholar]
- 33.Rotger, M., A. Trampuz, K. E. Piper, J. M. Steckelberg, and R. Patel. 2005. Phenotypic and genotypic mupirocin resistance among staphylococci causing prosthetic joint infection. J. Clin. Microbiol. 43:4266-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simor, A. E., T. L. Stuart, L. Louie, C. Watt, M. Ofner-Agostini, D. Gravel, M. Mulvey, M. Loeb, A. McGeer, E. Bryce, and A. Matlow. 2007. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus strains in Canadian hospitals. Antimicrob. Agents Chemother. 51:3880-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Udo, E. E., L. E. Jacob, and B. Mathew. 2001. Genetic analysis of methicillin-resistant Staphylococcus aureus expressing high- and low-level mupirocin resistance. J. Med. Microbiol. 50:909-915. [DOI] [PubMed] [Google Scholar]
- 36.Vasquez, J. E., E. S. Walker, B. W. Franzus, B. K. Overbay, D. R. Reagan, and F. A. Sarubbi. 2000. The epidemiology of mupirocin resistance among methicillin-resistant Staphylococcus aureus at a Veterans' Affairs hospital. Infect. Control Hosp. Epidemiol. 21:459-464. [DOI] [PubMed] [Google Scholar]
- 37.Wikler, M. A. 2008. Performance standards for antimicrobial susceptibility testing: 18th informational supplement, vol. 28. Clinical and Laboratory Standards Institute, Wayne, PA.