Abstract
Initial in vitro studies of bevirimat resistance failed to observe mutations in the clinically significant QVT motif in SP1 of HIV-1 gag. This study presents a novel screening method involving mixed, clinically derived gag-protease recombinant HIV-1 samples to more accurately mimic the selection of resistance seen in vivo. Bevirimat resistance was investigated via population-based sequencing performed with a large, initially antiretroviral-naïve cohort before (n = 805) and after (n = 355) standard HIV therapy (without bevirimat). The prevalence of any polymorphism in the motif comprising Q, V, and T was ∼6%, 29%, and 12%, respectively, and did not change appreciably over the course of therapy. From these samples, three groups of 10 samples whose bulk sequences were wild type at the QVT motif were used to generate gag-protease recombinant viruses that captured the existing diversity. Groups were mixed and passaged with various bevirimat concentrations for 9 weeks. gag variations were assessed by amplicon-based “deep” sequencing using a GS FLX sequencer (Roche). Unscreened mutations were present in all groups, and a V370A minority not originally detected by bulk sequencing was present in one group. V370A, occurring together with another preexisting, unscreened resistance mutation, was selected in all groups in the presence of a bevirimat concentration above 0.1 μM. For the two groups with V370A levels below consistent detectability by deep sequencing, the initial selection of V370A required 3 to 4 weeks of exposure to a narrow range of bevirimat concentrations, whereas for the group with the V370A minority, selection occurred immediately. This approach provides quasispecies diversity that facilitates the selection of mutations observed in clinical trials and, coupled with deep sequencing, could represent an efficient in vitro screening method for detecting resistance mutations.
Bevirimat [3-O-(3′,3′-dimethylsuccinyl)betulinic acid], previously known as PA-457, was the first of a new generation of HIV-1 drugs specifically targeting HIV-1 virion maturation (3). Bevirimat acts by inhibiting cleavage of the CA/SP1 site in the gag polyprotein (14). This prevents the processing of p25 to p24 and p2, resulting in aberrant virion morphology (14). The low 50% inhibitory concentration (IC50), comparable to the IC50 of other approved drugs, combined with the benefits of oral administration, low probability of drug interactions, and long plasma half-life made bevirimat appear to be a promising new drug candidate (14, 24).
Initial in vitro studies demonstrated the presence of a number of single nucleotide polymorphisms, including H358Y, L363F/M, A364V, and A366T/V, in the CA/SP1 cleavage site that resulted in resistance to bevirimat (1, 19). Mutations at these sites were not, however, detected in the phase I and II clinical trials for bevirimat, even in nonresponders (23). Instead, mutations in the QVT motif of the SP1 peptide (Gag positions 369 to 371) were the primary predictors of failure of response to bevirimat (17, 20, 26). The effect of these mutations has been repeatedly demonstrated (17, 20, 26). Polymorphisms in this region have further been shown to occur at a relatively high prevalence in existing populations, raising concerns about the general applicability of bevirimat in such treatments (23). In addition to those mutations, the V362I mutation has been demonstrated to confer strong resistance to bevirimat (17) whereas the S373P and I376V mutations may confer low-level resistance (17, 26). These problems eventually led to the cessation of clinical development of bevirimat. The inability of the bevirimat development plan to detect resistance mutations in the clinically important QVT motif despite the presumed requirement of only a single nucleotide change from the wild-type NL4-3 sequence underscores the need for additional methods of detecting resistance.
Since bevirimat targets the CA/SP1 cleavage site, a further complication in the use of bevirimat arises from the fact that it would likely be used in treatment of protease inhibitor (PI)-resistant patients (2). With the exception of A364V, mutations in the CA/SP1 cleavage site have been shown to result in fitness deficits when combined with PI resistance, suggesting that these mutations may be slow to develop (2). This is consistent with findings in studies investigating clinical isolates from PI-experienced patients that found limited variability in amino acids flanking the CA/SP1 cleavage site (15, 17). However, it has been shown that PI resistance can result in an increase in the prevalence of mutations within the downstream QVT motif (27).
The aims of this study were to demonstrate the feasibility of combining coculture of recombinant HIV-1 samples derived from patient isolates with “deep” sequencing an amplicon-based sequencing method which allows the parallel sequencing of thousands of clones from a single sample) as a novel and efficient screening approach for monitoring the in vitro selection of multiple clinically derived virus variants and to investigate the nature and kinetics of bevirimat resistance. This was accomplished by genotypic observation of a large, initially antiretroviral naïve cohort and by coculturing and deep sequencing of multiple initially QVT-wild-type gag-protease recombinant viruses derived from clinical isolates in the presence of various concentrations of bevirimat.
MATERIALS AND METHODS
Resistance mutation prevalence and in vivo variability.
Plasma HIV RNA gag sequences were obtained from the British Columbia Highly Active Antiretroviral Therapy (HAART) Observational Medical Evaluation and Research (HOMER) cohort, consisting of individuals initiating standard HAART between 1996 and 1999 from whom baseline (median, 36 days prior to HAART initiation; interquartile range [IQR], 24 to 57 days; n = 805) and/or posttherapy (median, ∼4 years after HAART initiation; IQR = 2.16 to 7.15 years; n = 355) gag sequences were available (4). Each sample was sequenced in the 5′ and 3′ directions using an ABI 3700 or 3730xl DNA analyzer (Applied Biosystems, Foster, CA). Sequences were analyzed using the custom Recall program (10), with nucleotide mixtures being called when the minority peak area exceeded 25% of the majority peak area.
Bevirimat-related mutations were defined according to data from previous studies. The following mutations were considered: G357S, H358Y, V362L/I, L363M, A364V, A366V, Q369H, V370A/M/deletion (del), T371A/del, S373P, I376V, and R380K (1, 17, 19, 20, 26). The prevalence of each of the defined mutant amino acids was calculated for each location at baseline and following therapy, with mixtures counted as belonging to both groups. Prevalence values for all other mutations were calculated together, and those results were classified as “other.” Patient results were also scored for any changes that occurred in the QVT motif over the course of therapy.
Generation and coculture of recombinant viruses.
Thirty individuals from the HOMER cohort whom baseline population-based plasma HIV gag sequences exhibited the presence of wild-type amino acids in the QVT motif were identified. Samples from these individuals were randomly assigned to form three groups of 10 samples each. Ten samples were assigned per group in order to provide sufficient diversity for the selection of resistant virus while keeping the groups small enough to permit the easy quantification of the fate of each individual population. Three independent groups were used to give an indication of assay reproducibility. The original gag-protease RT-PCR amplicons used to generate the bulk sequence were used as a template to perform a second round of PCR and to generate recombinant virus as previously described (4). The resulting recombinant viruses generally reflected the quasispecies diversity present in the original patient with respect to the gag-protease region. Titers of the recombinant viruses from each group were determined as described previously (21, 22), and three mixed viral inocula were created by combining the 10 recombinant virus stocks from each group in equal concentrations. Small aliquots of the initial inocula were preserved for deep sequencing, and the remaining aliquots were used to infect parallel cultures of MAGI-CCR5 cells, which also express CXCR4 (7) (AIDS Research and Reference Reagent Program of the National Institutes of Health). During infection, MAGI-CCR5 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin and were placed in a plate with six wells containing 0 μM, 0.01 μM, 0.1 μM, 1 μM, and 10 μM bevirimat, respectively. The remaining well was not sequenced. Cultures were passaged weekly for 9 weeks under conditions in which 1 ml of supernatant from the previous culture was used to infect a new culture that had been seeded the previous day with 3 × 105 MAGI-CCR5 cells. Culture supernatants were sampled at each passage and nucleic acids extracted manually using a guanidinium-based buffer followed by isopropanol and ethanol washes as described in reference 11.
Replication capacity assay.
Replication capacity assays of individual recombinant virus stocks were performed as previously described (4). Assays were initiated at a multiplicity of infection of 0.003 and included six negative and NL4-3-infected controls. The natural log slope of the percentage of green fluorescent protein (GFP)-positive cells was calculated during the exponential phase of virus replication (days 3 to 6) and the result divided by the mean rate of spread of wild-type NL4-3 to give a normalized measure of replication capacity. This experiment was independently repeated two or three times, and the average of the results was used for subsequent determinations.
Amplification and deep sequencing.
Nucleic acids extracted from initial inocula (i.e., the input mixtures of recombinant viruses for each group) and from passaged samples from all time points were amplified in triplicate to minimize error introduced by amplification. Multiplex identifier (MID)-tagged primers (as shown in Table S1 in the supplemental material) that cover HxB2 nucleotides 1759 to 1982 were used, allowing multiple samples to be processed in the same region (an enclosed area on the sequencing plate). All triplicate amplifications of the same sample were performed using the same MID. Triplicate amplifications were quantified using an Invitrogen Quant-iT PicoGreen double-stranded DNA (dsDNA) reagent assay and a DTX 880 multimode detector (Beckman Coulter, Fullerton, CA), and the quantified volumes were combined in equal concentrations. Combined triplicate samples were then pooled at equal concentrations with other samples which had been amplified using different MIDs such that each MID was represented only once in each pool (12 samples per pool). The amplified product from each pool was then added to DNA capture beads in a ratio of between 0.6 and 0.7 molecules of amplified product per capture bead. These combinations were then split into 3 emulsion PCR reaction mixtures to ensure sufficient DNA recovery. Emulsion PCR and DNA bead enrichment were then carried according to the genome sequencer (GS) FLX amplicon manual (Roche, Basel, Switzerland). Beads were then processed with a 454 (GS FLX) sequencer (Roche, Basel, Switzerland) according to manufacturer instructions.
Data analysis.
Unique sequences were aligned within the sequencing run by the use of a modified Smith-Waterman algorithm (25). Sequences from the initial mixtures and from each concentration that was amplifiable at the final time point were checked for recombination by the recombination detection program (RDP) method (18). The prevalence of each sequence was calculated as the number of reads for that particular sequence divided by the total number of reads passing the quality controls for that sample. Each sequence was then scored for the presence or absence of the defined bevirimat-relevant mutations. Unique combinations of resistance mutations with 2% prevalence or greater for at least one time point were plotted over time. To these data, we fit a deterministic two-locus two-allele model with 3 selection parameters (allowing for epistasis, i.e., a nonadditive fitness advantage in the double mutant) and a recombination rate parameter with nonoverlapping discrete generations (12). This genetic population model determined fitness by ordinary least-squares estimation using the Davidson-Fletcher-Powell optimization function in the Bhat R package (9). Wild-type fitness was fixed at a value of 1. This model assumes that the population size is effectively infinitely large and that the rate of mutation is effectively zero, i.e., that the influx of new mutations is negligible relative to the substantial frequencies of mutant alleles already present in the population at time 0. Although, in the case of HIV-1, generations undoubtedly overlap in continuous time, sampling of allele frequencies at regular intervals makes this model a convenient approximation. Fitness of a given allelic combination relative to the wild type was estimated as 1 plus the selection coefficient.
In order to determine the patient of origin for each virus, mixtures in the population sequences from each patient were resolved into all possible permutations. A Basic Local Alignment Search Tool (BLAST) database (6) was made for each group, containing all possible input sequences for that group. A BLAST search (6) for sequences from each group was then performed using the corresponding database, and the top hit was designated as representing the patient of origin for that particular sequence. The prevalences for each sequence mapping to a particular patient of origin within each given condition were summed and the results plotted. Finally, mean nucleotide entropy was calculated for each set of conditions as a measure of viral diversity by the method of Korber et al. (13).
RESULTS
Prevalence and variability of bevirimat resistance mutations in vivo.
For the HOMER cohort at baseline (n = 805), the approximate prevalences of any polymorphism in the QVT motif were 6%, 29%, or 12% for codons 369, 370, and 371, respectively. This remained essentially unchanged over the course of therapy (Table 1). The most commonly observed QVT polymorphism was V370A, which occurred in 19.9% of the population, followed by T371del, which occurred at 5.3% of the population at baseline (Table 1). Remarkably little variation was observed in amino acids at the CA/SP1 cleavage site (amino acids 363, 364, and 366) (Table 1). Other sites analyzed showed a moderate to high prevalence of polymorphisms (Table 1). Of the total of 182 variants with G357S, 106 also had a mutation in the QVT motif, a significantly higher proportion than would be expected from the independent prevalences of the two mutations (two-sided binomial test; P = 0.002).
TABLE 1.
Prevalence of bevirimat-associated resistance mutations in HIV-1 gag in the HOMER cohort
| Codon | Amino acid | Prevalencea (%) |
|
|---|---|---|---|
| Baseline (n = 805) | Posttherapy (n = 355) | ||
| 357 | S | 22.6 | 22.0 |
| Other | 1.0 | 1.7 | |
| 358 | Y | 0.0 | 0.0 |
| Other | 0.0 | 0.0 | |
| 362 | L | 0.0 | 0.0 |
| I | 15.9 | 13.8 | |
| Other | 0.0 | 0.0 | |
| 363 | M | 0.4 | 0.3 |
| Other | 0.0 | 0.0 | |
| 364 | V | 0.0 | 0.0 |
| Other | 0.0 | 0.0 | |
| 366 | V | 0.0 | 0.0 |
| Other | 0.0 | 0.0 | |
| 369 | H | 5.2 | 2.8 |
| Other | 0.7 | 1.1 | |
| 370 | A | 19.9 | 22.5 |
| M | 3.0 | 3.7 | |
| del | 2.0 | 1.4 | |
| Other | 3.9 | 6.5 | |
| 371 | A | 2.0 | 1.7 |
| del | 5.3 | 3.9 | |
| Other | 4.5 | 5.4 | |
| 373 | P | 26.3 | 28.2 |
| Other | 24.0 | 22.0 | |
| 376 | V | 16.0 | 16.3 |
| Other | 5.6 | 4.5 | |
| 380 | K | 44.3 | 42.3 |
| Other | 3.2 | 2.3 | |
The prevalence of analyzed mutations at each specified location is shown at baseline (median value at 36 days prior to HAART; n = 805) and posttherapy (median value at ∼4 years after HAART initiation; n = 355).
Of the individuals whose gag sequences were available both at baseline and posttherapy (n = 268), the sequences of a total of 13.1% exhibited at least one change in the QVT motif, with 8.6% developing new polymorphisms and 4.5% reverting to the wild type over a median period of approximately 4 years (IQR = 2.16 to 7.15). Of the total of 355 patients whose posttherapy sequence data were available, 305 had experienced some level of exposure to protease inhibitors. Of these, only 16 had protease inhibitor resistance mutations. Associations between protease inhibitor resistance and bevirimat-associated resistance were not tested due to the low prevalence of protease inhibitor resistance.
In vitro selection of bevirimat resistance and deep sequencing.
A median of >2,700 HIV gag sequences spanning HxB2 nucleotides 1759 to 1982 was acquired at each stage of viral passage in vitro. The RDP algorithm revealed no evidence of recombination (data not shown).
Mutations present in the initial inocula.
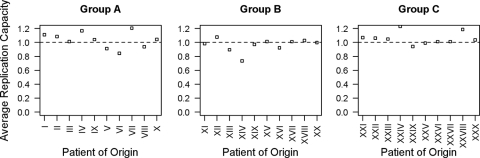
Deep sequencing revealed low levels of mutations outside the QVT motif in all groups for the initial inocula. In group A, this included an ∼18.1% prevalence of I376V. Group B had a ∼3.6% minority of I376V and a ∼2.1% minority of S373P/R380K. Group C had a number of mutations present in the initial inoculum, including ∼11.4% V362I, ∼10.5% S373P/I376V, and ∼2.1% I376V. Despite bulk-sequence-based screening, group C also had a ∼4.8% prevalence of V362I/V370A. Nearly all other analyzed mutations were detected in each group; however, these occurred below the threshold of consistent detection of ∼1% total prevalence. G357S was not present above the 1% threshold despite being at 22.6% prevalence in the general population, since it tended to cooccur with mutations in the QVT motif and thus was screened out of the input population. Some variations in the replication of the input viruses compared to that of NL4-3 did exist in monocultures, with group A having a median replication capacity of 1.04 (IQR = 0.96 to 1.10), group B having a median replication capacity of 0.99 (IQR = 0.94 to 1.01), and group C having a median replication capacity of 1.04 (IQR = 1.01 to 1.07) (Fig. 1). Variations in the proportions of virus derived from each patient also existed in the initial inoculum, including one sample from group B in which virus could not be detected.
FIG. 1.
Average replication capacities in monocultures in the absence of drug. Average replication capacities normalized to NL4-3 are shown for recombinant viruses from each individual patient. Replication capacities were derived as described in reference 4. The dashed horizontal line represents the reference replication capacity (wild-type NL4-3). Patients of origin are separated by group.
Mutation profiles and patients of origin.
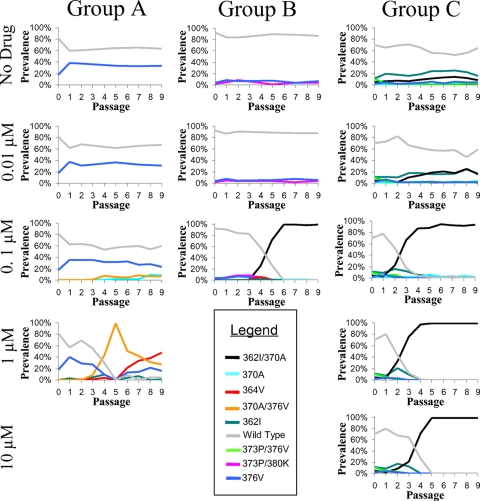
In one of the three mixed cultures, a V370A/I376V mutant emerged and took over the population by passage 5 in the presence of 1 μM bevirimat (Fig. 2, group A). This variant had an estimated relative fitness of ∼1.70 (95% confidence interval [CI], 1.03 to 2.74) compared to a fitness value of 1 for the wild type in the presence of 1 μM bevirimat according to the population genetics model and emerged from a single patient of origin (Fig. 3, group A). An A364V variant later emerged and was traced to a second independent patient of origin (Fig. 3, group A). The A364V variant grew to dominate the culture, though it did not eliminate the V370A/I376V variant (Fig. 2, group A). This variant had an estimated relative fitness of ∼3.52 (95% CI, 1.32 to 4.77) compared to the wild type in the presence of 1 μM bevirimat according to the population genetics model.
FIG. 2.
Selection of bevirimat resistance over time. Three independent mixed cultures (A, B, and C) of recombinant patient-derived viruses were passaged at the indicated concentrations of bevirimat, and the prevalences of unique combinations of bevirimat-associated resistance mutations were determined. Only drug concentrations that were amplifiable at all tested time points and only mutation profiles with at least 2% prevalence at any time point are shown.
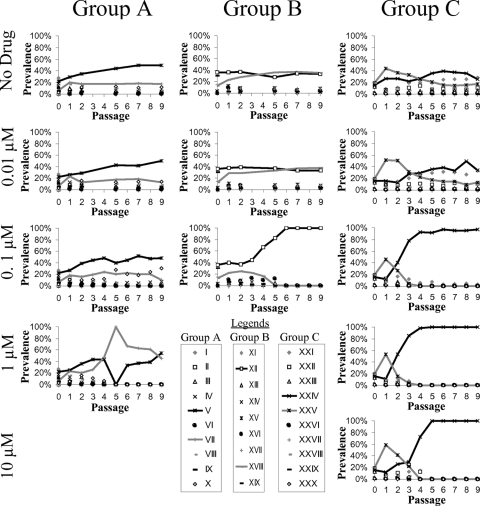
FIG. 3.
Contribution of virus from each patient of origin over time. The patients of origin of the viruses from the mixed cultures presented in Fig. 2, taken as the top hit in a BLAST search of a database containing all input sequences for each specific group, are shown for each passage. The results for the patients of origin for the two most common viruses are shown with black and gray lines. Results from each of the three groups of gag-protease recombinant HIV strains grown in competition are shown, with the concentration of bevirimat increasing from the top to bottom panels. Only the results for drug concentrations that were amplifiable at all tested time points are shown.
In a second mixed culture, in the presence of 0.1 μM bevirimat, a V362I/V370A variant emerged that was also from a single patient of origin (Fig. 3, group B) and rose to nearly 100% prevalence by passage 5 (Fig. 2, group B). Relative fitness could not be reliably estimated for this culture by the population genetics model due to the low prevalence and the variation of mutation minorities, aside from the final V362I/V370A variant. In the final mixed culture, the V362I/V370A variant was rapidly selected, reaching ∼100% prevalence at all concentrations of bevirimat ≥ 0.1 μM by passage 4 (Fig. 2, group C), with estimated relative fitness values according to the population genetics model of between 2.50 and 4.01 compared to the wild type value of 1 (Fig. 4). In all cases, the V362I/V370A mutant originated from the same patient of origin (Fig. 3, group C). At bevirimat concentrations below 0.1 μM, the V362I/V370A variant remained at a low level of prevalence (Fig. 2, group C), as would be expected from the population model fitness estimates, which were near 1 (Fig. 4). Fitness estimates for group C appear to fit the data well, with predicted proportions closely matching observed proportions over time (Fig. 4).
FIG. 4.
Population genetics model of resistance selection in mixed culture. Deterministic two-locus two-allele models fit by ordinary least-squares estimation calculated using the Davidson-Fletcher-Powell optimization function in the R package Bhat are shown for group C. The allele frequencies predicted by the model are shown by lines, while the actual measured frequencies are shown as points. The wild type is shown in black, mutation 370A is shown in red, mutation 362I is shown in blue, and the 362I/370A double mutant is shown in green. Fitness estimates with 95% confidence intervals are shown in the table in the lower right panel. In some instances, the predicted trajectory of allele frequencies overshot the boundary (at 0 and 1) due to the discrete-time approximation used in the population genetic model.
In general, each unique mutation profile tended to derive from a single patient of origin (Fig. 2 and 3). Interestingly, in group A, at 1 μM bevirimat, the viruses of only two patients were represented following passage 5 (Fig. 3) despite a larger number of distinct resistance profiles (Fig. 2). At low concentrations (0.01 μM) or in the absence of bevirimat, some fitness differences were observed in the change in overall prevalences of viruses from each patient; however, no virus was greatly superior to the others in this respect (Fig. 3). This is as would be expected from the similar average monoculture replication capacity values of each of the input viruses (Fig. 1).
Viral diversity.
Viral diversity (as measured by mean nucleotide entropy over the sequenced region) remained relatively constant at bevirimat concentrations of 0.01 μM or 0 μM (data not shown). At higher concentrations (≥0.1 μM) of bevirimat in which a single resistance profile took over the population, mean nucleotide entropy decreased sharply (i.e., proportions of each nucleotide at each position shifted away from equality on average) with the rise of the resistance mutation, suggesting a corresponding drop in the overall diversity (data not shown). Following the diversity drop, a slight recovery of diversity was observed under some sets of conditions, suggesting potential diversification of the now-dominant virus (data not shown).
DISCUSSION
This study established the prevalence of preexisting bevirimat resistance in the QVT motif and its biological variability over the course of standard HAART regimens which do not include bevirimat. It also demonstrated the feasibility of using mixed populations of recombinant virus coupled with deep sequencing in order to screen for clinically relevant drug resistance mutations. Finally, this study showed the effect of bevirimat selection on viral diversity in vitro.
The data from our large treatment-naïve cohort showed a nearly 50% prevalence of preexisting bevirimat resistance in the form of QVT motif polymorphisms, as reported for other studies (20, 23, 26, 27). The prevalence of naturally occurring polymorphisms in the QVT motif was not substantially affected by exposure to long-term PI- and/or nonnucleoside reverse transcriptase inhibitor (NNRTI)-based HAART (Table 1). These results appear to agree with the relatively inconsequential differences in bevirimat resistance observed by both Margot et al. and Seclén et al. (17, 23) between treatment-experienced and treatment-naïve patients (17) and with the high degree of sequence conservation in the CA-SP1 cleavage site found by Malet et al. in PI-experienced patients (16) but appear to contradict the increased prevalence of bevirimat resistance in patients with PI resistance found by Verheyen et al. (27). While the vast majority of patients within the group observed in this study were exposed to PIs at some point during treatment, PI resistance was rare. As such, the findings of this study do not necessarily contradict those of Verheyen et al. but instead represent a more general look at the relationship between HAART exposure and naturally occurring genetic variations in gag associated with decreased susceptibility to bevirimat rather than the relationship specifically seen in the PI-resistant population.
The use of mixed recombinant virus derived from patient samples allowed a range of drug concentrations to be queried against a panel of clinically derived isolates from the target population to screen for clinically relevant resistance. It also allowed the testing of 30 different clinically derived recombinant viruses in six different drug concentrations, while requiring maintenance of only three 6-well culture plates, and was capable of yielding results in less than 3 months. The ability to rapidly detect preexisting drug resistance present in the population could allow modifications to be made to drug development procedures and thus reduce or avoid the problems that have haunted the clinical development of bevirimat. To address this issue, we tested a mixed culture system coupled with deep sequencing. As it is likely that not all drugs would have levels of preexisting mutations as high as were encountered with bevirimat, bulk sequencing was used to screen out obvious cases of preexisting resistance in this “proof of principle” study. Despite this screening, the approach was still capable of detecting commonly occurring preexisting resistance in all test groups. Had this method been used for bevirimat, it would have been obvious to the developers that, if implementation of the drug in therapy were to proceed, patients infected with virus that had the key mutations would have to be screened out. With this insight, the efforts of the developers, who spent tens of millions of dollars on the clinical development of bevirimat, would have yielded very different results.
Where V370A took over the population, it was consistently observed to arise alongside other bevirimat-associated mutations. This could occur when these other mutations are not sufficient on their own to completely overcome the effects of the drug and take over the population but allow enough replication to facilitate the development of V370A. It thus may be possible that two bevirimat resistance mutations are required to provide sufficient selective advantage for a single virus to effectively take over the population.
The emergence of a single dominant resistance profile was associated with decreased viral diversity, which is consistent with the observation that resistance arose from a single patient strain rather than from recombination. This would be as expected from a particular virus subpopulation gaining a mutation and arising to take over the population, as was suggested by the finding of a single patient of origin of each dominant resistance profile. The lack of an apparently dominant virus under conditions in which a low concentration of or no drug was present is consistent with the comparable replication capacities of the input viruses and suggests that the competitive advantage displayed by patient viruses that are dominant at higher concentrations is dependent on the drug rather than an intrinsic property of the viruses themselves.
Mixtures of gag-protease recombinant viruses derived from clinical samples gave a closer approximation of the development of resistance seen in vivo than have previous in vitro resistance assays that failed to detect mutations in the clinically important QVT motif (1, 14, 28). The use of deep sequencing allowed these mutations to be traced back to the individual mutational background and to the specific patient of origin, allowing a better view of the interactions between different mutations as well as identifying specific patients likely to develop resistance when receiving bevirimat therapy. The benefits of this methodology are likely due to a combination of mixtures of virus genotypes directly providing resistance and/or a background of greater diversity in which resistance can emerge. The inability of the RDP algorithm to detect recombination could have been due to a lack of power resulting from the relatively small region of gag that was analyzed rather than to a true absence of recombination, particularly because some recombination was predicted by the population genetics model. If recombination were occurring at a high frequency, however, it would likely have been detected.
The use of several different constant drug concentrations rather than a single concentration as used by Adamson et al. (1) or gradually increasing concentrations as used by Li et al. and Zhou et al. (14, 28) may have also contributed the ability of the present method to select V370A despite the failure of previous methods to do so (1, 14, 28). In the study by Adamson et al., selection was performed using 0.0766 μM bevirimat (1), a concentration below the NL4-3-specific IC50 (0.204 μM) of bevirimat (8); thus, that experiment may not have provided sufficient drug pressure to select for resistant variants in the QVT motif. In studies by Li et al. and Zhou et al., initial selection for resistance was performed at a set concentration that was maintained until evidence of resistance was observed, at which time the drug concentrations were gradually increased (14, 28). As was seen in this study, however, V370A on its own may not provide a sufficient replication advantage to emerge at the concentrations of 0.12 μM (28) and 2.45 μM (14) that were used for initial selection in the studies by Li et al. and Zhou et al. or at the concentration of 0.0766 μM that was used by Adamson et al. (1). The genetic barrier to generation of V370A mutations in a background without other resistance mutations may thus be higher than the genetic barrier to generation of mutations in the CA/SP1 site, as it would require the mutation of two nucleotides (one at 370 and one at another resistance site) rather than one, thus making the generation of mutations in the CA/SP1 sites more likely to occur. This would be consistent with the resistance patterns observed in those previous studies (1, 14, 28).
While the method used in this study allowed the development of resistance similar to that seen in vivo, it had limitations in that it did not control for drug-independent differences in replication capacity among recombinant viruses combined in the initial inoculum. It should be noted that reproducibility was not perfect when groups of 10 samples were used. The number of samples per group or the total number of groups could be increased to provide a more reproducible estimate of even rare clinically relevant mutations. Also, the use of recombinant viruses and deep sequencing restricts analysis to a specific region of interest, thus causing any mutations outside the analyzed area to be missed. This could have affected the fitness of specific variants if additional compensatory mutations were present outside the analyzed area.
This study demonstrated that the prevalence of polymorphisms in the QVT motif conferring bevirimat resistance was not strongly affected by long-term PI- and/or NNRTI-based HAART and showed little variability over several years of observation, suggesting that genotypic screening is feasible, though it may require greater sensitivity than current population-based sequencing methods are capable of providing. In addition to this, a minimum concentration of 0.1 μM bevirimat was required for the selection of resistance mutations in vitro, a concentration below which no major selective forces were observed. Finally, even at relatively low levels of baseline prevalence, viruses containing polymorphisms in the QVT motif were rapidly selected. This confirms that preexisting resistance mutations at low levels in a population would be rapidly selected in mixed cultures. These results further confirm the applicability of the use of mixtures of recombinant viruses derived from clinical isolates coupled with deep sequencing-based observation as an efficient in vitro method for screening preexisting resistance mutations for bevirimat and other emerging drugs.
Supplementary Material
Acknowledgments
We thank Jennifer Sela and Pamela Rosato for assistance with recombinant virus generation.
Art F. Y. Poon is funded by a Canadian Institutes of Health Research (CIHR) Fellowships Award in HIV/AIDS Research (200802HFE). P. Richard Harrigan is a CIHR-GSK Chair. The project was also supported in part by a grant from the CIHR to Zabrina L. Brumme and Mark A. Brockman. Zabrina L. Brumme is a CIHR New Investigator Award recipient.
Footnotes
Published ahead of print on 17 November 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Adamson, C. S., et al. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (bevirimat). J. Virol. 80:10957-10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, C. S., K. Waki, S. D. Ablan, K. Salzwedel, and E. O. Freed. 2009. Impact of human immunodeficiency virus type 1 resistance to protease inhibitors on evolution of resistance to the maturation inhibitor bevirimat (PA-457). J. Virol. 83:4884-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blair, W. S., et al. 2009. New small-molecule inhibitor class targeting human immunodeficiency virus type 1 virion maturation. Antimicrob. Agents Chemother. 53:5080-5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockman, M. A., et al. 2010. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity but may be largely compensated in chronic infection. J. Virol. 84:11937-11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Camacho, C., et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. HIV-1 coreceptors participate in postentry stages of the virus replication cycle and function in SIV infection. J. Virol. 71:3932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe, S. S., et al. 2008. Measurement of maturation inhibitor susceptibility using the PhenoSense HIV assay. Abstr. XVth Conf. Retrovir. Opportunistic Infect., abstr. 880.
- 9.Fletcher, R. 1970. A new approach to variable metric algorithms. Comp. J. 13:317-322. [Google Scholar]
- 10.Harrigan, P. R., et al. 2002. Performance of RE_Call basecalling software for high-throughput HIV drug resistance basecalling using “in-house” methods. Abstr. XIV Int. AIDS Conf., abstr. TuPeB4598.
- 11.Harrigan, P. R., et al. 2005. Predictors of HIV drug-resistance mutations in a large antiretroviral-naïve cohort initiating triple antiretroviral therapy. J. Infect. Dis. 191:339-347. [DOI] [PubMed] [Google Scholar]
- 12.Kimura, M. 1956. Rules for testing stability of a selective polymorphism. Proc. Natl. Acad. Sci. U. S. A. 42:336-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korber, B. T., et al. 1994. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J. Virol. 68:7467-7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, F., et al. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. U. S. A. 100:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malet, I., et al. 2007. Association of Gag cleavage sites to protease mutations and to virological response in HIV-1 treated patients. J. Infect. 54:367-374. [DOI] [PubMed] [Google Scholar]
- 16.Malet, I., et al. 2007. Primary genotypic resistance of HIV-1 to the maturation inhibitor PA-457 in protease inhibitor-experienced patients. AIDS 21:871-873. [DOI] [PubMed] [Google Scholar]
- 17.Margot, N. A., C. S. Gibbs, and M. D. Miller. 2010. Phenotypic susceptibility to bevirimat among HIV-1-infected patient isolates without prior exposure to bevirimat. Antimicrob. Agents Chemother. 54:2345-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, D., and E. Rybicki. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562-563. [DOI] [PubMed] [Google Scholar]
- 19.Martin, D. E., K. Salzwedel, and G. P. Allaway. 2008. Bevirimat: a novel maturation inhibitor for the treatment of HIV-1 infection. Antivir. Chem. Chemother. 19:107-113. [DOI] [PubMed] [Google Scholar]
- 20.McCallister, S., et al. 2008. HIV-1 Gag polymorphisms determine treatment response to bevirimat (PA-457). Antivir. Ther. 13(Suppl. 3):A10. [Google Scholar]
- 21.Miura, T., et al. 2009. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J. Virol. 83:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura, T., et al. 2010. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J. Virol. 84:7581-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seclén, E., et al. 2010. High prevalence of natural polymorphisms in Gag (CA-SP1) associated with reduced response to bevirimat, an HIV-1 maturation inhibitor. AIDS 24:467-469. [DOI] [PubMed] [Google Scholar]
- 24.Smith, P. F., et al. 2007. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-O-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob. Agents Chemother. 51:3574-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 26.Van Baelen, K., et al. 2009. Susceptibility of human immunodeficiency virus type 1 to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag spacer peptide 1. Antimicrob. Agents Chemother. 53:2185-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verheyen, J., et al. 2010. High prevalence of bevirimat resistance mutations in protease inhibitor-resistant HIV isolates. AIDS 24:669-673. [DOI] [PubMed] [Google Scholar]
- 28.Zhou, J., et al. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J. Virol. 78:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.