Acanthamoeba keratitis (AK) is a rare corneal infection caused by the free-living amoeba Acanthamoeba (1). Diagnosis of AK is based on the culture of corneal scrapings or detection of the organism in pathology specimens (1). Culture positivity and histology may require a long time (3 to 7 days on average) and are highly dependent on the number of Acanthamoeba cysts and trophozoites recovered from the cornea.
Early diagnosis of AK is one of the most important factors determining the final outcome (1). Molecular detection of Acanthamoeba by PCR provides a rapid and sensitive method for diagnosis (1). However, DNA extraction from Acanthamoeba often requires time-consuming and expensive commercial kits, as demonstrated in previous studies (2). Acanthamoeba cysts are resistant to physical, mechanical, or enzymatic lysis, and the presence of inhibitors to Taq polymerase can compromise the efficiency of DNA amplification (2).
We report a simple, fast, and inexpensive technique for Acanthamoeba DNA extraction with Chelex resin (MB Chelex-100 resin; Bio-Rad Laboratories, Hercules, CA), a commercially available polystyrene-divinylbenzene iminodiacetate material that has been used extensively in DNA extraction for forensic and microbiological purposes (3, 5, 6).
Our protocol consisted of addition of 200 μl of Chelex solution (10% [wt/vol]) in 0.1% Triton X-100 and 10 mM Tris buffer (pH 8.0) to cyst suspensions (approximately 104 cysts/ml in balanced salt solution). The material was vortexed for 10 s, centrifuged at 10,000 × g for 10 s, and then heated at 95°C for 20 min and finally centrifuged at 10,000 × g for 20 s. The supernatant was used as the substrate for a PCR (4 μl). The following Acanthamoeba genus-specific primers were used: forward primer 5′-TCTCACAAGCTGCTAGGGCGTCA-3′ and reverse primer 5′-GTCAGAGGTGAAATTCTTGG-3′, which gave a 250-bp product (4).
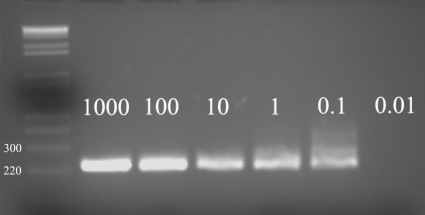
To test the detection limits of our assay, Chelex extraction was performed on serial dilutions of Acanthamoeba castellanii (T4 genotype; ATCC) cysts in saline solution. As shown in Fig. 1, PCR run after Chelex extraction was able to amplify DNA equivalent to a minimal concentration of 0.1 cyst per PCR.
FIG. 1.
One percent agarose gel showing detection limits of Chelex DNA extraction. The numbers to the left of the lanes represent molecular masses in kilodaltons.
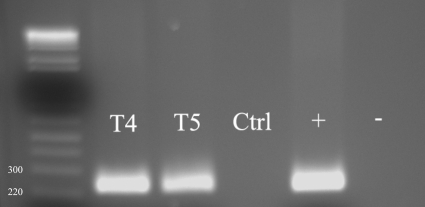
To assess the potential use of Chelex extraction in the diagnosis of AK, cysts were also injected into the corneas of eye bank eyes. The corneas of two whole eyes obtained from the local eye bank were injected intrastromally with 0.2 ml of a suspension of 104/ml A. castellanii and Acanthamoeba lenticulata (T5 genotype; ATCC) cysts, respectively, in saline solution; a third eye was injected with saline and used as a negative control. The eyes were stored in Optisol at 35°C for 24 to 48 h before the corneas were scraped with a crescent blade. Chelex solution was added to the corneal scraping, and DNA extraction and PCR were performed as described. As shown in Fig. 2, the presence of A. castellanii (T4) and A. lenticulata (T5) was detectable in the corneal scrapings obtained from both eyes.
FIG. 2.
One percent agarose gel showing Acanthamoeba DNA recovery from in vitro-infected eyes by use of Chelex resin. The numbers to the left of the lanes represent molecular masses in kilodaltons. T4, Acanthamoeba castellanii; T5, Acanthamoeba lenticulata; Ctrl, eye injected with saline; +, positive PCR control; −, negative PCR control.
Chelex resin DNA extraction appears to be a sensitive and reliable method for Acanthamoeba DNA extraction from culture or corneal scrapings and may represent a fast and inexpensive alternative to commercial kits for rapid molecular diagnosis of AK or genotyping of Acanthamoeba strains. Further evaluations of Chelex DNA extraction in a clinical setting are needed to confirm our findings.
Acknowledgments
The authors report they have no conflicts of interest.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Dart, J. K., V. P. Saw, and S. Kilvington. 2009. Acanthamoeba keratitis: diagnosis and treatment update. Am. J. Ophthalmol. 148:487-499. [DOI] [PubMed] [Google Scholar]
- 2.Goldschmidt, P., et al. 2008. Resistance of Acanthamoeba to classic DNA extraction methods used for the diagnosis of corneal infections. Br. J. Ophthalmol. 92:112-115. [DOI] [PubMed] [Google Scholar]
- 3.Khan, N. A., and T. A. Paget. 2002. Molecular tools for speciation and epidemiological studies of Acanthamoeba. Curr. Microbiol. 44:444-449. [DOI] [PubMed] [Google Scholar]
- 4.Ledee, D. R., et al. 2009. Molecular identification of T4 and T5 genotypes in isolates from Acanthamoeba keratitis patients. J. Clin. Microbiol. 47:1458-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuchimochi, T., et al. 2002. Chelating resin-based extraction of DNA from dental pulp and sex determination from incinerated teeth with Y-chromosomal alphoid repeat and short tandem repeats. Am. J. Forensic Med. Pathol. 23:268-271. [DOI] [PubMed] [Google Scholar]
- 6.Yang, J. L., et al. 2008. A simple and rapid method for extracting bacterial DNA from intestinal microflora for ERIC-PCR detection. World J. Gastroenterol. 14:2872-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]