Abstract
Sepsis is among the top 10 causes of mortality in the United States. Rapid administration of antibiotics is one of the most important contributors to patient survival, yet only a limited number of methods exist for rapid identification of microbes cultivated from bloodstream infections, which can lead to sepsis. While traditional single-target molecular methods have been shown to greatly improve survival for septic patients by enabling rapid deescalation of broad-spectrum antibiotics, multiplex methods offer even greater possibilities. A novel multiplex method, PCR coupled to electrospray ionization mass spectrometry (PCR/ESI-MS), was used to identify the genus and species of microorganisms found to cause human bloodstream infections. DNA was directly extracted from 234 BacT-Alert blood culture bottles, and results were compared to those obtained by clinical reference standard methods. The study results demonstrated 98.7% and 96.6% concordance at the genus and species levels, respectively. Mixtures of microbes were identified in 29 blood culture bottles, including mixed species of the same genus, as well as mixtures containing Gram-positive and Gram-negative organisms, exemplifying the PCR/ESI-MS capability to identify multiple organisms simultaneously without the need for cultivation. This study demonstrates high analytical accuracy in comparison to routine subculture of blood culture bottles and phenotypic identification of microbes. Without foreknowledge of the microorganisms potentially present, the PCR/ESI-MS methods can deliver accurate results in as little as 5 to 6 h after a positive alarm from the automated blood culture system; however, current batch mode testing limits the method's clinical utility at this time.
The ability to rapidly identify the causative agent of bloodstream infections (BSIs) is of paramount importance to clinical microbiology laboratories. Rapid identification significantly reduces the rates of patient mortality, reduces the use of unnecessary antibiotics, and lowers costs to the hospital (6, 7, 15). If untreated, BSIs can lead to sepsis or progress to severe sepsis, with one or more organ dysfunctions, which can ultimately result in septic shock and/or death.
Death due to septic shock is currently the 10th leading cause of mortality in the United States (28). More notably, the rate of sepsis-related death is on the rise. In 2000, sepsis was ranked as the 13th leading cause of death (5), symbolizing a 139% increase in incidence over a 10-year period (8). It is estimated that approximately 215,000 deaths per year are sepsis related, and the costs associated with sepsis are exceedingly high, approximately $17 billion each year and rising, due to the length of hospital stay required (1).
It is estimated that the survival increases by 7 to 10% for every hour of earlier administration of targeted antibiotics (34). Ideally, bacteria in the bloodstream should be identified as quickly as possible in order to administer the most appropriate antibiotics, as opposed to use of broad-spectrum antibiotics. This approach requires direct evaluation of bacteria from positive blood culture bottles (BCBs) or whole blood. Unfortunately, current routine clinical testing involves time-consuming cultivation of microorganisms prior to identification via colony and cell morphology and biochemical profiling. This routine identification may take several days and can result in false-negative results, particularly when antibiotic treatment has already begun (24, 36).
Studies using phenotypic testing systems directly on broth from positive blood culture bottles report a lack of accuracy for certain species and a high rate of nonidentification of species (9, 14, 35). Subsequently, several molecular testing methods are currently used in clinical laboratories for identification of microbes directly from blood culture bottles.
FDA-approved peptide nucleic acid fluorescent in situ hybridization (PNA-FISH) methods are available from bioMérieux (Durham, NC) and AdvanDx (Woburn, MA) and demonstrate successful identification of certain bacteria and yeasts directly from blood culture bottles. Evidence based on intervention studies emphasizes the potential advantages of such a rapid diagnostic method (21-23, 37), but PNA-FISH is limited because it can confirm identity of only a few organisms, and kit selection relies solely on Gram stain, which could bias results toward the identification of the predominant microbial species.
Additionally, the Xpert MRSA/SA blood culture assay (Cepheid, Sunnyvale, CA) is a real-time PCR and is FDA approved for identification of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) from blood culture bottles. GeneXpert and other PCR systems can greatly diminish the delay from blood draw to actionable results (48), but, like PNA-FISH, the method is limited to identification of only a few genetic targets.
Pyrosequencing is another genetic method that shows promise for detecting bacteria directly from blood culture bottles. Accurate species identification is documented; however, the method is not yet FDA approved (31, 32). Moreover, sequence-based approaches require a relatively complex skill set, compounded by labor intensity, for assay performance and interpretation of sequences. In addition, sequencing methods have difficulty with microbial mixtures, which can occur in blood culture broth.
In addition to molecular-based methods, protein-based identification methods that rely on matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) are being used for identification of bacterial species. Methods that use Bruker and Shimadzu mass spectrometers report high concordance and utility in clinical laboratories (10). While these methods are developing for direct testing from blood culture bottles (42), they are not yet FDA approved.
As an alternative to other molecular methods, we describe the use of the PCR coupled to electrospray ionization mass spectrometry (PCR/ESI-MS) for rapid and high-throughput genotypic characterization of bacteria directly from positive blood culture bottles (17, 18, 29, 47). PCR/ESI-MS uses broad-range primers that are specific for groups of microbes rather than for any one particular species. Primers amplify across a region that is variable between species and strains. These broad-range primers are used in conjunction with species- or strain-specific primers. Additional primer pairs target identifiable genes for antibiotic resistance or particular pathogenic features. General methods for genotypic identification of bacteria and fungi by PCR/ESI-MS have been previously described (4, 16, 17, 26, 47).
The mass/charge (m/z) ratio of the PCR amplicon is measured via mass spectrometry, resulting in a calculated molecular mass for the forward and reverse strand of each amplicon, which is then correlated to the amplicon base composition, as illustrated in Fig. 1. Measurement is possible with high mass accuracy, and resolution is attained by internal calibration combined with the added specificity achieved due to the reverse complementarities of the genetic material. For each amplicon, data can be used in tandem with those from products amplified by several other primers to identify the bacterial species and strains present. Finally, the resulting genetic fingerprints are compared to those in a database of known DNA base compositions.
FIG. 1.
Determining the base composition from mass spectral data: example using BCB115. (A) The spectra are collected and displayed as a 96-well plate format. Mass spectra can be viewed independently for each of the 96 wells. (B) Electrospray-generated mass spectra. This sample contains the PCR amplicons from two different organisms: Bacteroides thetaiotaomicron (top) and Staphylococcus aureus (bottom). (C) Deconvoluted mass spectra showing molecular masses of the forward and reverse strands of each PCR amplicon. An algorithm is used to calculate the base composition from the molecular mass and was calculated to be 30A-27G-23C-19T (top) and 27A-30G-21C-21T (bottom).
A major advantage to this method is the ability to characterize an organism without prior knowledge. The PCR/ESI-MS technique has the potential to detect virtually all bloodstream infections in a single, rapid assay. This study was designed to demonstrate the ability of PCR/ESI-MS to detect bacterial and fungal pathogens directly from positive blood culture bottles, showing the potential for faster microbial identification that could hasten administration of targeted antimicrobial therapy aimed to decrease the mortality, morbidity, and length of patient hospitalizations.
(Results of the study were reported in part as Proceedings of the Association of Molecular Pathology, 15th Annual Meeting, Kissimmee, FL, 20 November 2009.)
MATERIALS AND METHODS
Patients and specimens.
The study design was a preclinical comparison of the Abbott Sterile Fluid Bacteria and Candida assay (now marketed as the BAC detection [BAC] assay; Abbott Molecular, Des Plains, IL). The BAC assay was compared to phenotypic reference “gold standard” methods. Subjects included individuals at the University Medical Center (UMC), in Tucson, AZ, whose routine care included a blood culture. One hundred eighty-nine positive blood culture broth samples and 45 broth samples negative after 5 days of incubation were tested.
Sample selection.
Procedures were performed in accordance with ethical standards, as reviewed by the University of Arizona Human Subjects Protection Program. Remnant specimens were obtained from the University of Arizona Infectious Disease Research Core's microbial biorepository. Samples were selected from among those collected between 29 June 2008 and 23 October 2008.
The positive and negative cases, as determined by the reference standard culture methods, were selectively chosen by the investigator prior to PCR/ESI-MS testing to create a high-prevalence sample set that contained a broad diversity of microbial species and to avoid redundancy of the sampling for particular high-prevalence microorganisms. Samples were blinded prior to evaluation with PCR/ESI-MS. Mixed specimens were purposefully selected as challenge samples to test the accuracy of the PCR/ESI-MS assay on patient specimens containing multiple microbes.
The sample set had the following characteristics: 33.8% were from inpatient wards, 22.9% were from the Emergency Department, 13.9% were from the adult intensive care unit (ICU), 11.9% were from bone marrow transplant wards, 7.0% were from pediatric inpatients, 4.5% were from the pediatric ICU, 4.5% were from other adult outpatient units, and 1.5% were from autopsy. The patients' ages ranged from 14 days to 93 years, with a median age of 50 years and a mean age of 45.3 years. Gender mix was 39.3% female and 60.7% males.
Blood culture broths.
Two milliliters of blood from FA and FN blood culture bottles (bioMérieux, Durham, NC) determined to be positive by the BacT-Alert 3D instrument (bioMérieux) was removed and placed into sterile tubes for cryostorage (Starstedt, Newton, NC). These specimens were stored at −80°C until they were utilized in this series of experiments. Negative broths were incubated for 5 days and similarly processed and stored.
Reference standard methods for identification.
Aliquots from positive blood culture bottles were subjected to Gram stain and plating on Trypticase soy agar supplemented with 5% sheep blood (TSA; Remel, Lenexa, KS). Chocolate agar (CHOC; Remel) and MacConkey agar (MAC; Remel) plates were inoculated, and Sabouraud agar (SAB; Remel) plates were also inoculated, if the result of the Gram stain indicated yeast. Postsubculture, colony morphology was used as the basis for additional phenotypic testing, based on determinative protocols described in the Manual of Clinical Microbiology (2) and in accordance with guidelines issued by the Clinical and Laboratory Standards Institute (CLSI) (11-13).
Blood cultures were examined to determine their representative Gram reaction and bacterial morphologies. Microorganisms were identified according to standard clinical laboratory methods (2), including Vitek 2 Gram-Positive identification (GP ID), Vitek 2 Gram-Negative identification (GN ID), and Vitek 2 Yeast identification (YST ID) (bioMérieux) cards.
DNA isolation from positive blood cultures.
DNA was extracted from 200 μl of remnant blood culture broth using a magnetic bead-based extraction method. The DNA Bacteria protocol was used to perform all extractions, along with either the associated DNA blood or DNA tissue kit (Qiagen, Valencia, CA) designed specifically for the BioRobot EZ1 instrument. The switch to the DNA tissue kit was undertaken after a decrease in signal intensity was noted for fungal species and some Gram-positive species in the first 98 samples. The rigid peptidoglycan encountered in Gram-positive bacteria and chitin encountered in fungi required a harsher extraction methodology in order to efficiently extract DNA from these organisms (33). The EZ1 DNA tissue kit contains proteinase K within its buffers, providing a mechanism to more efficiently degrade these rigid cell walls, resulting in enhanced DNA extraction, and was used for the remainder of the samples. Those samples known to contain Gram-positive organisms or fungi were reextracted with the tissue kit and reanalyzed. If these organisms were known not to be present, the sample was not reextracted in the interest of resources, since a reanalysis would not provide new results. A centrifugation step was not performed prior to extraction for the removal of charcoal from the blood culture bottles. Samples were processed and eluted in 60 μl elution buffer and stored until PCR/ESI-MS was performed.
PCR/ESI-MS.
Protocols provided with the Sterile Fluid Bacteria and Candida assay, now called the BAC detection assay (Abbott Molecular), were followed using six samples per plate. Briefly, DNA extracted from the blood culture bottles was diluted to 160 μl using the supplied DNA dilution buffer and distributed into the 96-well PCR plate across 16 wells per sample, such that each extract experienced 16 primer pairs targeted toward bloodstream pathogens. Of these primer pairs, ribosomal regions, antibiotic resistance genes, and bacterial group-specific primers were used. The primers and their functions are summarized in Table 1.
TABLE 1.
Primers used in the BAC detection assay and their gene targets
| Primer identifier | Gene | Target |
|---|---|---|
| 346 | 16S rRNA | All bacteria |
| 348 | 16S rRNA | All bacteria |
| 361 | 16S rRNA | All bacteria |
| 349 | 23S rRNA | All bacteria |
| 2249 | tufB | Elongation factor EF-Tu; staphylococci |
| 3350 | rplB | 50S ribosomal subunit protein L2; Firmicutes |
| 358 | valS | Valyl-tRNA synthetase; proteobacteria and gamma enterobacteria |
| 3346 | rpoB | RNA polymerase; beta- and gammaproteobacteria |
| 3921 | rpoB | RNA polymerase; beta- and gammaproteobacteria |
| mecA | mecA | Methicillin resistance |
| vanA/vanB | vanA/vanB | Vancomycin resistance |
| KPC | blaKPC | Carbapenem resistance |
| 3030 | 25S rDNA | All fungi |
| 3031 | 25S rDNA | All fungi |
| 3766 | 25S rDNA | All fungi |
| 3865 | mDNA | Mitochondrial DNA; all fungi |
In addition to target amplification components, each well contains calibrant DNA that is simultaneously amplified, allowing a positive amplification control and semiquantitative analysis. The relative intensity of the target DNA can be compared to that of the calibrant, which is present at a known concentration and provides a relative concentration of target DNA initially present.
The extracted DNA was amplified using the following PCR program on an MJ Research PTC 240 Tetrad 2 thermocycler: 95°C for 10 min; 8 cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s; 37 cycles of 95°C for 15 s, 48°C for 20 s, and 72°C for 20 s; 72°C for 2 min; and hold at 4°C. The T5000 instrument was used to desalt the amplicon with anionic exchange and to detect the mass-to-charge ratio (m/z) for each amplicon via electrospray ionization into the time-of-flight mass spectrometer.
The T5000 dedicated software package was used to unambiguously obtain the base count of each amplicon, which was compared to the count in a reference database of the amplicon expected from each primer pair. A combination of results from each primer pair was used to identify the bacterial and fungal organisms present in the blood culture bottles. The confidence of identification was determined by the software package, utilizing information such as the number of copies of DNA per well, the number of primers making the identification, and the uniqueness of that base composition. For these experiments, all data meeting a threshold of 87.5% confidence for microbial identification were reported; data with <87.5% confidence were recorded as uninterpretable.
Comparative analysis.
For this study, the results obtained by reference standard methods were defined as identification results generated using traditional phenotypic and biochemical analyses with the Vitek 2 system and additional reference standard phenotypic methods, as discussed above. Reference standard results were compared with the microorganism identifications made using the BAC assay. Results that were in agreement between the two methods were considered to be concordant, and those with documented discrepancies were further analyzed.
Retrospective culture-based analysis.
To verify the presence of additional organisms determined to be present by the PCR/ESI-MS assay, 50 μl of remnant blood culture specimens was enriched in 1 ml of either TSB with 0.5% sodium chloride Lim broth or chopped meat broth (CMB; Becton Dickinson), depending on the organism's metabolic requirements. The organisms were incubated in either ambient air, 5% CO2, or an anaerobic environment using a GasPakEZ anaerobe container system with indicator (Becton Dickinson). These enriched cultures were then subcultured on TSA, CHOC, MAC, SAB, or Columbia colistin naladixic acid (CNA) agar (Becton Dickinson) and incubated under the aforementioned conditions to isolate and identify any previously unidentified organisms. Selected organisms identified to have a discrepant genus or species by PCR/ESI-MS were isolated and cultured on TSA slants or CMB, incubated at 37°C for 24 h, and submitted for MALDI-TOF analysis using the BioTyper (version 2.0) program (Bruker Daltonics, Bremen, Germany), as previously described (10).
RESULTS
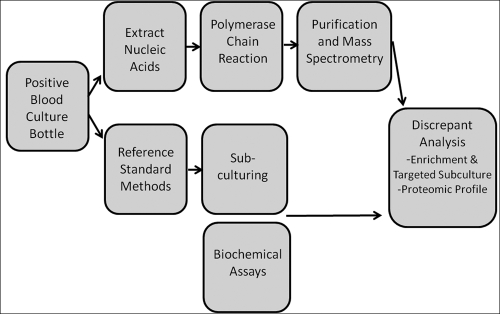
For this study, microbial identifications, made using traditional reference standard methods with biochemical analysis and phenotypic methods, were compared directly to results obtained with PCR/ESI-MS. The standardized work flow for these experiments can be found in Fig. 2. The distribution of the organisms selected from positive blood culture bottles at UMC during the evaluation period can be found in Table 2. Table 2 illustrates the number of times that each of these organisms was identified in the context of a mixture. In several instances (15/234), the PCR/ESI-MS technique identified organisms that were not isolated in the original blood culture but that were later confirmed from targeted subculturing procedures using selective and differential media to allow outgrowth and identification of slower-growing microbes or organisms present at a low density. The resulting data are discussed below with increasing levels of complexity, first examining the identification concordance as a molecular Gram stain, then genus concordance, and finally, species concordance. Figure 3 illustrates the base composition results from each primer in the PCR/ESI-MS analysis for one representative sample.
FIG. 2.
Work flow for this study.
TABLE 2.
Breakdown of hematopathogens investigated in positive blood culture bottles in this study, as identified by culture-based methods
| Organism | No. of samplesa | No. in mixed samplesb |
|---|---|---|
| Abiotrophia sp. | 1 | 0 |
| Acinetobacter baumannii | 5 | 1 |
| Acinetobacter lwolfii | 1 | 0 |
| Actinomyces israelii | 1 | 0 |
| Anaerobic Gram-positive cocci | 3 | 1 |
| Anaerobic Gram-negative rods | 1 | 0 |
| Anaerobic diphtheroids | 1 | 0 |
| Bacillus (not B. anthracis) | 3 | 0 |
| Beta-hemolytic streptococci | 11 | 5 |
| Bacteroides thetaiotaomicron | 4 | 1 |
| Candida albicans | 5 | 0 |
| Candida spp. | 3 | 0 |
| Citrobacter freundii | 1 | 0 |
| Clostridium perfringens | 2 | 1 |
| Clostridium septicum | 1 | 0 |
| Coagulase-negative staphylococci | 23 | 10 |
| Corynebacterium spp. | 3 | 0 |
| Enterobacter aerogenes | 1 | 0 |
| Enterobacter cloacae | 9 | 0 |
| Enterococcus faecalis | 14 | 3 |
| Enterococcus faecium | 16 | 1 |
| Escherichia coli | 11 | 0 |
| Fusobacterium sp. | 1 | 0 |
| Haemophilus influenzae | 1 | 0 |
| Klebsiella oxytoca | 3 | 2 |
| Klebsiella pneumoniae | 8 | 0 |
| Lactobacillus spp. | 2 | 0 |
| Micrococcus spp. | 7 | 0 |
| Nonhemolytic streptococci | 2 | 0 |
| Pantoae sp. | 1 | 0 |
| Proteus mirabilis | 1 | 0 |
| Pseudomonas aeruginosa | 6 | 0 |
| Pseudomonas stutzeri | 1 | 0 |
| Salmonella sp. | 1 | 0 |
| Staphylococcus aureus | 41 | 7 |
| Stenotrophomonas maltophilia | 1 | 0 |
| Stomatococcus mucilaginosus | 1 | 1 |
| Streptococcus gordonii | 1 | 0 |
| Streptococcus pneumoniae | 1 | 0 |
| Viridans group streptococci | 12 | 3 |
| Total | 211 | 34 |
| Total pathogenic | 177 | |
| Total nonpathogenic | 34 |
The total number of occurrences of each organism is listed.
The frequency in which that organism occurred in a mixture with other organisms.
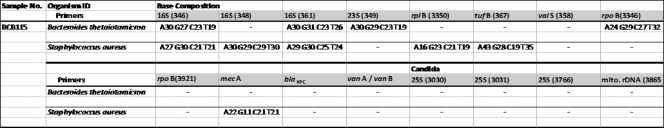
FIG. 3.
Base composition data obtained from sample BCB115, which contained a mixture of the Gram-negative anaerobe Bacteroides thetaiotaomicron and the Gram-positive methicillin-resistant Staphylococcus aureus. The composition of the PCR amplicon produced by each primer is shown.
Gram stain concordance.
The result obtained from a Gram stain is the preliminary information that is used to guide antibiotic treatment in clinical patients (25, 41). The first step in evaluating the PCR/ESI-MS capability for identifying bloodstream infections was to determine that the organism that was identified is in agreement with the Gram stain result reported by the reference standard methods. During this study, we found no errors in designation of the correct Gram stain category by the PCR/ESI-MS.
Genus concordance.
Concordance on the genus level is more relevant than a simple Gram stain concordance and is one of the fundamental assay parameters that could support targeted antimicrobial therapy. Of the 211 organisms contained in the 234 blood culture bottles, a 98.72% combined genus concordance was observed, as illustrated in Table 3. This high concordance value takes into account those samples in which one organism of each sample was correctly identified but a false-negative or false-positive result was also observed, in addition to the concordant result.
TABLE 3.
Concordance of PCR-ESI/MS with reference standard identification
| Parameter | Genus |
Species |
||||
|---|---|---|---|---|---|---|
| Frequency (%) | Sample proportionc | Summed concordance (%) | Frequency (%) | Sample proportionc | Summed concordance (%) | |
| Concordant | 93.59 | 219/234 | 86.75 | 203/234 | ||
| False negativea | 3.42 | 8/234 | 98.72 | 4.27 | 10/234 | 94.01 |
| False positivea | 1.71 | 4/234 | 2.99 | 7/234 | ||
| False negative | 0.43 | 1/234 | 0.43 | 1/234 | ||
| False positive | 0.43 | 1/234 | 0.43 | 1/234 | ||
| Discordant | 0.43 | 1/234 | 2.56 | 6/234 | ||
| Discordant, true positiveb | 0 | 0/234 | 2.56 | 6/234 | ||
A result was false negative or false positive when one organism of a mixture was correctly identified.
A discordant result with the reference standard identification but found to be true by discrepant analysis.
Data represent the number of samples with the indicated result divided by the total number of samples tested.
Species concordance.
An additional level of identity is achievable using the PCR/ESI-MS technique to obtain organism identification on the species level. Of the 234 blood culture bottles that were evaluated and the 211 organisms contained within those bottles, a 96.58% species combined concordance was observed, as illustrated in Table 3. This value takes into account those samples that contained additional organisms and mixed organisms where one organism of each sample was correctly identified.
Species identification is clinically relevant in a variety of situations, particularly when nonpathogenic skin flora need to be distinguished from true pathogens, such as viridans group streptococci from Streptococcus pneumoniae and coagulase-negative staphylococci (CoNS) from Staphylococcus aureus, which may appear to be nonhemolytic colonies under certain circumstances. In this data set, there were six incidences where S. pneumoniae was identified in a mixture with viridans group streptococci by PCR/ESI-MS but was characterized as only viridans group streptococci by standard hospital testing of BCBs 106, 110, 138, 176, 296, and 365. In each of these cases, additional testing proved the additional presence of S. pneumoniae, which was otherwise missed by reference methods.
Mixtures.
This study analyzed a select set of patient samples that contained mixtures of organisms to demonstrate the ability of PCR/ESI-MS to identify coinfections. A tabulated list of mixtures determined by PCR/ESI-MS can be found in Table 4. Of the 29 mixtures investigated, 19 contained mixtures of two or more Gram-positive organisms, 3 contained mixtures of two or more Gram-negative organisms, 2 contained a mixture of fungi with Gram-positive/Gram-negative bacteria, and 5 contained mixtures of a Gram-positive organism with a Gram-negative organism. Of these mixtures, 7 were found to have a false-negative result by PCR/ESI-MS, where one organism of each sample was correctly identified but the second organism was not, resulting in a 24% error rate for mixtures. A rationale for this occurrence is further addressed in the Discussion section.
TABLE 4.
Results for samples containing mixtures of organisms
| Sample no. | Organisms contained in mixture | False negativea | Gram stain result |
|---|---|---|---|
| BCB028 | Staphylococcus capitis | Positive | |
| Staphylococcus epidermidis | Positive | ||
| Staphylococcus warneri | Positive | ||
| BCB089 | Coagulase-negative staphylococcus sp. | FN | Positive |
| Beta-hemolytic streptococci | Positive | ||
| BCB095 | Staphylococcus hominis | Positive | |
| Pseudomonas stutzeri | Negative | ||
| BCB110 | Streptococcus dysgalactiae | Positive | |
| Streptococcus pneumoniae | Positive | ||
| BCB115 | Bacteroides thetaiotaomicron | Negative | |
| Staphylococcus aureus | Positive | ||
| BCB124 | Staphylococcus aureus | FN | Positive |
| Streptococcus oralis (viridans group) | Positive | ||
| Streptococcus sanguinis (viridans group) | Positive | ||
| BCB135 | Coagulase-negative staphylococcus sp. | FN | Positive |
| Streptococcus oralis (viridans group) | Positive | ||
| Streptococcus sanguinis (viridans group) | Positive | ||
| BCB138 | Viridans group streptococci | Positive | |
| Streptococcus pneumoniae | Positive | ||
| BCB144 | Staphylococcus hominis | Positive | |
| Staphylococcus carnosus | Positive | ||
| Staphylococcus auricularis | Positive | ||
| Staphylococcus piscifermentans | Positive | ||
| Staphylococcus aureus | Positive | ||
| BCB152 | Staphylococcus hominis | Positive | |
| Acinetobacter baumannii | Negative | ||
| BCB178 | Coagulase-negative staphylococcus sp. | Positive | |
| Staphylococcus aureus | Positive | ||
| BCB200 | Coagulase-negative staphylococcus sp. | FN | Positive |
| Beta-hemolytic streptococci | Positive | ||
| Enterococcus sp. | FN | Positive | |
| BCB206 | Klebsiella oxytoca | Negative | |
| Staphylococcus aureus | Positive | ||
| BCB217 | Klebsiella oxytoca | Negative | |
| Staphylococcus aureus | Positive | ||
| BCB218 | Streptococcus oralis (viridans group) | Positive | |
| Streptococcus sanguinis (viridans group) | Positive | ||
| BCB222 | Candida albicans | ||
| Acinetobacter baumannii | Negative | ||
| BCB228 | Staphylococcus epidermidis | Positive | |
| Enterococcus faecalis | FN | Positive | |
| BCB268 | Streptococcus pyogenes | Positive | |
| Staphylococcus aureus | Positive | ||
| BCB271 | Coagulase-negative staphylococcus sp. | FN | Positive |
| Streptococcus agalactiae | Positive | ||
| BCB284 | Enterobacter hormaechei | Negative | |
| Enterobacter hormaechei (second strain) | Negative | ||
| BCB293 | Acinetobacter lwoffii | Negative | |
| Acinetobacter baumannii | Negative | ||
| BCB296 | Streptococcus cristatus (viridans group) | Positive | |
| Streptococcus pneumoniae | Positive | ||
| BCB309 | Coagulase-negative staphylococcus sp. | Positive | |
| Enterococcus faecium | FN | Positive | |
| BCB338 | Acinetobacter baumannii | Negative | |
| Acinetobacter calcoaceticus | Negative | ||
| BCB347 | Stomatococcus mucilaginosus | Positive | |
| Streptococcus salivarius (viridans group) | Positive | ||
| BCB365 | Streptococcus sanguinis (viridans group) | Positive | |
| Streptococcus pneumoniae | Positive | ||
| BCB373 | Staphylococcus epidermidis | Positive | |
| Enterococcus faecalis | Positive | ||
| BCB374 | Staphylococcus epidermidis | Positive | |
| Candida albicans | |||
| BCB440 | Streptococcus oralis (viridans group) | Positive | |
| Streptococcus sanguinis (viridans group) | Positive |
If the organism was identified by standard reference methods but was not detected by PCR/ESI-MS, this is considered a false-negative result and is denoted FN.
Detection of Saccharomyces sp. in blood culture bottles.
In a significant proportion of samples (31.9%), a detection of Saccharomyces cerevisiae was made, in addition to the bacterial identification. It was determined, as discussed below, that this identification was a contamination from the blood culture medium itself and not a true bloodstream infection.
Negative blood culture bottle evaluation.
An additional experimental set was investigated to determine how PCR/ESI-MS handles negative blood culture bottles that should contain no microorganism. For this evaluation, 45 blood culture bottles, deemed negative after extended growth and continuous monitoring, were evaluated by PCR/ESI-MS using the same protocol used for positive blood culture bottles. All 45 negative blood culture specimens were identified as negative by PCR-ESI/MS, resulting in 100% concordance. An artifact Saccharomyces sp. signal was detected in 39 of the 45 negative bottles, and an additional signal from Bacillus coagulans was detected. These are spurious false-positive results determined to be DNA contamination within the bottle, as evidenced by a lack of growth after 5 days determined by reference standard methods, as well as the fact that the results were found in a single batch of bottles and the same base composition was demonstrated.
DISCUSSION
The critical nature of bloodstream infections and sepsis creates a clear and imposing unmet need for rapid and accurate diagnostic methods to identify and characterize bloodstream pathogens. Under these premises, the overall significance of rapid results is clear because they have the potential to alter the course of diagnosis and treatment of bloodstream infections and sepsis.
Identification of additional organisms.
The most important finding in this experiment was from the samples in which additional organisms that were not previously characterized by standard clinical testing methods were identified. Because the PCR/ESI-MS technology uses broad-range PCR amplification, it is capable of amplifying and detecting very low levels of bacterial DNA in a sample. This factor is highly desirable in a clinical setting, enabling physicians to obtain information about infections and coinfections much sooner in the patient's disease progression.
One example of how the PCR/ESI-MS technique was able to identify additional organisms was for samples containing both Streptococcus pneumoniae and viridans group streptococci, which were represented in 6 of the 234 samples. In each of these samples, the standard clinical testing identified viridans group streptococci as the only organism present in the blood culture. However, PCR/ESI-MS results demonstrated the presence of a coinfection with Streptococcus pneumoniae, which was confirmed by additional enrichment techniques and subculturing with an extended incubation time (44). Viridans group streptococci are potentially nonpathogenic organisms, whereas Streptococcus pneumoniae is pathogenic and can cause bacterial meningitis and pneumonia. We found that for bottles in which a mixed infection was found, more than 24 h of incubation was sometimes required before the typical glistening colony morphology or central colony depressions of S. pneumoniae (45) could be readily identified among the colonies on a lawn of other alpha-hemolytic colonies. Misidentification of S. pneumoniae and viridans group streptococci has also previously been known to occur when traditional phenotypic methods (20, 43) or MALDI-TOF approaches (10) are used for identification of alpha-hemolytic streptococci. Overall, the reference standard clinical microbiological methods were incorrect in six cases when S. pneumoniae was identified by PCR/ESI-MS.
Mixtures.
The major drawback to the current clinical methods for identification of bloodstream infections is the requirement for subculture to isolate organisms prior to evaluation. The PCR/ESI-MS technique is advantageous due to its ability to characterize mixtures of organisms directly from raw blood culture broth without the need for separation of colonies by subculture. Additionally, because the PCR/ESI-MS is able to provide semiquantitative data, it is possible to approximate microbial burden of infections. A clear example of the power of the PCR/ESI-MS mixture resolution can be found for sample BCB115, the results for which are illustrated in Fig. 3. This sample contained a mixture of Bacteroides thetaiotaomicron and methicillin-resistant Staphylococcus aureus detected by PCR/ESI-MS, allowing the detection of a Gram-positive aerobic organism and an anaerobic Gram-negative organism in one signal assay.
A second example of PCR/ESI-MS distinguishing mixtures of organisms was demonstrated with BCB222, from which both a Gram-negative organism, Acinetobacter baumannii, and a yeast, Candida albicans, were identified from a single specimen. In this specific example, it is important to note that the original clinical evaluation was not able to detect the presence of C. albicans, when reference standard methods were used in a routine clinical setting. The presence of C. albicans was confirmed by additional enrichment subculture and biochemical testing.
The ability of the technique to resolve mixtures is not limited to genetically dissimilar organisms, as discussed above, but the technique can also distinguish a mixture of several different species of organisms, as demonstrated by BCB028. This sample contains a mixture of CoNS, which would not ordinarily be distinguished at the species level by routine clinical testing. The PCR/ESI-MS technology was able to identify three different species of staphylococci: Staphylococcus capitis, S. epidermidis, and S. warneri. This information provides a more accurate representation of the blood culture than that which occurs when only the predominant organism (CoNS) is identified in a two-bottle positive set of blood cultures. This example demonstrates the PCR/ESI-MS resolving capability to distinguish even genetically similar organisms and will aid in tracing the epidemiology of infection. This level of detail has not been the norm for historic laboratory methods and can, in some instances, provide more information than what is necessary for diagnosis. Proper training would be required for qualitative assessments made with complex samples to allow standardization. In some cases, a polyphasic approach (genotypic and phenotypic approaches) will need to be combined with clinical assessment of the patient to resolve the clinical relevance of mixed infections.
Although the PCR/ESI-MS technology has the ability to resolve mixtures, there are known limitations to this capability, as demonstrated by the relatively high false-negative error rate of 24%. Of the 29 samples that contained mixtures, 7 of these samples were identified to be false negative and the secondary organism was undetected by PCR/ESI-MS, as illustrated in Table 4, but was found to be present by traditional bacterial culture and biochemical testing. Four of these seven false-negative samples included beta-hemolytic streptococci suppressing the detection of staphylococci, and three of these seven included a Staphylococcus sp. suppressing the signal of enterococci, resulting in preferential amplification. One possible explanation for this result is dependent on the relative amount of DNA for each organism present in the sample. However, another cause may be more plausible. Although not thoroughly investigated to date, the primers used in the PCR/ESI-MS assay are able to amplify genes only when the DNA concentrations of each species were of a similar order of magnitude. The primers will quickly be saturated with DNA from the most abundant organism, leaving microorganisms present in lower titers undetected. This effect is mitigated if the organisms are identified by nonoverlapping pairs of primers, eliminating the competition of each target for the primers. This is of additional concern when samples are analyzed after some period of growth, such as with blood culture bottles, where one organism may grow more rapidly than another. This may result in drastically different titers of organisms present in the blood culture specimen at the time of positivity. However, this is not the case with staphylococci and streptococci, which have similar times to positivity in blood culture bottles (38).
One potential resolution to this issue would be to perform a controlled experiment, comparing the results produced when DNA is extracted and amplified from suspensions of bacterial mixtures that were prepared by varying each of the microbial densities to mimic common bacterial mixtures that may be encountered in blood culture bottles. This experiment was beyond the scope and purpose of this study, yet resolution of issue will be important in future experiments with pure cultures and mixed infections.
Discordance.
In both genus and species analyses, discordance was evaluated and characterized. A false-negative result for mixtures is discussed above as likely being due to preferential amplification. False-positive results for mixtures rarely occurred (1.71% for genus identification, 2.99% for species identification), and in all instances, the primary organism identified by reference standard methods was correctly identified by PCR/ESI-MS. In these false-positive identifications, the secondary organism identified was a low-confidence identification, as rated by the Ibis T5000 software. This is likely due to a deviation of the clinical strain from the reference standard strain, resulting in an additional false identification that would require additional testing in a clinical situation. Theoretically, contamination from environmental organisms might happen on this open platform, but this is highly unlikely due to the rigorous cleaning of instrument components between assays with different samples. Finally, the discordant results listed in Table 3 for genus identification (0.43%) and species identification (2.56%) cannot be resolved. In all cases, the discrepant result is very genetically similar to the clinical identification. For example, for 4/6 discordant species results, the clinical identification is Enterobacter cloacae, while PCR/ESI-MS identified E. hormaechei. Because the PCR/ESI-MS technique uses genetic information, which is more variable between species than the phenotypic profile, it is likely that the PCR/ESI-MS identification is correct, and without sequence analysis, this problem cannot be resolved.
Detection of Saccharomyces sp. in blood culture bottles.
The identification of a Saccharomyces sp. contaminant was made in several blood culture bottles. An evaluation of blood culture bottles confirmed that DNA was present in the broth, likely due to DNA from yeast extracts used in the broth formulation. In all situations, the number of genomes per well of the yeast contamination was reported to be very low and never competed with the identification of the predominant organism present. Although Saccharomyces cerevisiae is known to cause empyema and fungemia in some rare cases (3, 19, 27), the resulting DNA amplicon detected was from the same strain in all cases, having the same base composition and being present at a low signal strength, barely breaking the cutoff signal requirements of the assay. Therefore, for this sample set, low Saccharomyces cerevisiae signals were considered to be an artifact of the assay. For fungal testing, manufacturers of blood culture bottles and suppliers of broad-range amplification methods will need to work toward the removal of background fungal DNA from broth used for PCR-based testing.
Comparison to other methods.
Overall, the results obtained by PCR/ESI-MS are able to achieve high levels of concordance with those of existing reference standard techniques currently being used in the clinical setting. Results from PCR/ESI-MS can be achieved in 5 to 6 h, whereas culture and biochemical characterization techniques typically require 1 to 5 days to confirm microbial identification. PCR/ESI-MS has the potential to overcome a major bottleneck in diagnostic microbiology and could guide medical practitioners toward a more rapid deescalation from the use of broad-spectrum empirical antimicrobials to the administration of appropriate targeted antibiotics.
Despite the potential advantages of the PCR/ESI-MS technique, there are several drawbacks. This work was performed on the Abbott Molecular T5000 PCR/ESI-MS, the first model available to laboratories for research-use-only (RUO) testing. The T5000 was prone to a number of significant mechanical malfunctions during the course of this study. To overcome this limitation, Abbott Molecular has designed a newer model of this instrument, called the PLEX-ID, which is designed to be more robust and provide a streamlined platform and software interface more suitable for use in the clinical laboratory. An additional limitation is the T5000's current requirement for batching of samples in sets of six, preventing the random access that would be optimal in a clinical setting. While this issue has been partly addressed by the override function of the next-generation instrument, which allows STAT samples to be preemptively analyzed, the bulk of the system still requires batch processing and limits the practical routine use of the instrument under low-volume testing conditions. Finally, the costs of the instrument and reagents remain high and represent a significant disadvantage to the routine use of the technology in clinical laboratories.
A comparison to the MALDI-TOF/MS technique is relevant to the discussion (10, 46). MALDI-TOF/MS measures the m/z values of proteins expressed by a microorganism and correlates the spectra back to the spectra in databases comprised of experimental spectra collected from culture collection strains. This type of analysis is very rapid, from the moment of sample preparation to the time of identification of microorganisms, and achieves high levels of concordance with standard methods, comparable to those reported in this paper and others (10, 30, 39, 40, 46). The cost per analysis for MALDI-TOF/MS methods is very low. The instrument allows random access, is highly automated, requires little training, and has the potential to be integrated into clinical labs. However, MALDI-TOF/MS analysis may in some instances require a bacterial isolation step prior to analysis, which slows the time to results. In addition, the instrument currently does not have the capacity to identify mixed samples or the analytical sensitivity to detect less than 104 to 105 CFU. As discussed above, the PCR/ESI-MS technique does not have a requirement for subculture and can therefore provide faster, unambiguous identifications of microorganisms present in blood culture bottles. Furthermore, at this time MALDI-TOF/MS does not have the ability to identify antimicrobial resistance genes like the PCR/ESI-MS technology does. Although not discussed at length in this report, PCR/ESI-MS has the ability to evaluate samples for the antibiotic resistance genes contained in the genomes of particular organisms of interest, particularly β-lactam resistance-encoding genes for Staphylococcus spp. (mecA), the vancomycin resistance gene for Enterococcus spp. (vanA/vanB), and the carbapenem resistance gene for Klebsiella spp. (blaKPC) The ability to detect antibiotic resistance genes is indicated in Fig. 3, where the detection of the mecA gene was seen for sample BCB115. The ability to do so will further increase the ability to administer targeted antibiotics and avoid the use of unnecessary ineffective medication. Since there are no highly abundant proteins routinely measured that are directly associated with these antibiotic resistance genes to date, MALDI-TOF/MS methods are not capable of identifying these potential antibiotic-resistant organisms.
Conclusion.
This study was designed to evaluate the feasibility of the use of PCR/ESI-MS to identify microorganisms directly from blood culture bottles in the clinical microbiology laboratory. The high concordance of the results of this technique with those of standard methods, particularly at the genus level, demonstrates that the PCR/ESI-MS technique is capable of rapidly evaluating clinically complex specimens, providing information as to the selection and administration of targeted antibiotics. The use of this technique in a clinical setting has the potential of lowering the turnaround time for microbial identification. Additionally, the method provides a potential mechanism for intervention with faster, targeted antibiotic therapy, which in turn may provide a decrease in the mortality rate and reduce the amount of financial resources spent by providing medical personnel with accurate information about bloodstream infections. If adapted for diagnostic purposes, the method could provide an innovation that may improve the speed and accuracy of pathogen identification. Full evaluation with fresh blood culture samples and clinical utility studies that include cost-benefit analysis are needed before the full impact of the technology can be ascertained. Maximum impact could occur if the technology was adapted for use with a high-volume blood extraction technology with whole-blood samples, which represents the focus of our future research. Although it should be noted that currently the error rate for identification of mixtures in this study was relatively high, at 24%, the PCR/ESI-MS method remains at the forefront of microbiological identification methods for single pathogen detection directly from blood culture bottles.
This study was designed to demonstrate the performance of the BAC detection assay by challenging it with a positive sample set handpicked for diversity of species. The study was not designed to be a full assessment of the BAC assay's sensitivity and specificity, which, of course, must follow prior to clinical use. On the basis of assay performance with this sample set, this novel technique has the potential for rapid and accurate analysis of a wide range of microbes, alone or present as coinfections.
Acknowledgments
This study was supported by research grant funds provided by Abbott Molecular to D.M.W. and NIH grant U54-AI065359 (Pacific Southwest Regional Center for Excellence) to V.H.W. and by a NIH Chemistry-Biology Interface training grant T32GM008804 to E.J.K.
The researchers acknowledge the contributions of Elizabeth Serbi of the University of Arizona Genetics Core for daily operation of the T5000 instrument; Lorraine Dominguez, Ellen Tuttle, Joe Marano, and Erica Isaacs for their support in banking blood culture specimens; and Elizabeth Ingram for providing assistance with banking, microbiological culture, and organism identification. Finally, we thank the technical support team at Ibis Biosciences and Abbott Molecular.
Footnotes
Published ahead of print on 3 November 2010.
REFERENCES
- 1.Angus, D. C., W. T. Linde-Zwirble, J. Lidicker, G. Clermont, J. Carcillo, and M. R. Pinsky. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29:1303-1310. [DOI] [PubMed] [Google Scholar]
- 2.ASM Press. 2010. Manual of clinical microbiology, 10th ed. ASM Press, Washington, DC.
- 3.Aucott, J. N., J. Fayen, H. Grossnicklas, A. Morrissey, M. M. Lederman, and R. A. Salata. 1990. Invasive infection with Saccharomyces cerevisiae: report of three cases and review. Rev. Infect. Dis. 12:406-411. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, C. D., G. B. Howe, R. Sampath, L. B. Blyn, H. Matthews, V. Harpin, T. A. Hall, J. J. Drader, S. A. Hofstadler, M. W. Eshoo, K. Rudnick, K. Studarus, D. Moore, S. Abbott, J. M. Janda, and C. A. Whitehouse. 2009. Usefulness of multilocus polymerase chain reaction followed by electrospray ionization mass spectrometry to identify a diverse panel of bacterial isolates. Diagn. Microbiol. Infect. Dis. 63:403-408. [DOI] [PubMed] [Google Scholar]
- 5.Balk, R. A. 2000. Severe sepsis and septic shock. Definitions, epidemiology, and clinical manifestations. Crit. Care Clin. 16:179-192. [DOI] [PubMed] [Google Scholar]
- 6.Barenfanger, J., C. Drake, and G. Kacich. 1999. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 37:1415-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barenfanger, J., M. A. Short, and A. A. Groesch. 2001. Improved antimicrobial interventions have benefits. J. Clin. Microbiol. 39:2823-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control. 1990. Increase in national hospital discharge survey rates for septicemia—United States. 1979-1987. MMWR Morb. Mortal. Wkly. Rep. 49:31-34. [PubMed] [Google Scholar]
- 9.Chen, J. R., S. Y. Lee, B. H. Yang, and J. J. Lu. 2008. Rapid identification and susceptibility testing using the VITEK 2 system using culture fluids from positive BacT/ALERT blood cultures. J. Microbiol. Immunol. Infect. 41:259-264. [PubMed] [Google Scholar]
- 10.Cherkaoui, A., J. Hibbs, S. Emonet, M. Tangomo, M. Girard, P. Francois, and J. Schrenzel. 2010. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J. Clin. Microbiol. 48:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed., vol. 29, no. 2. CLSI document M07-A8. Approved guidelines. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 10th ed. CLSI document M02-A10, vol. 26, no. 2. Approved guidelines. Clinical and Laboratory Standards Institute, Wayne, PA.
- 13.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial suspectibility testing, 20th informational supplement. CLSI document M100-S20, vol. 30, no. 1. Approved guidelines. Clinical and Laboratory Standards Institute, Wayne, PA.
- 14.de Cueto, M., E. Ceballos, L. Martinez-Martinez, E. J. Perea, and A. Pascual. 2004. Use of positive blood cultures for direct identification and susceptibility testing with the Vitek 2 system. J. Clin. Microbiol. 42:3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doern, G. V., R. Vautour, M. Gaudet, and B. Levy. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ecker, D. J., C. Massire, L. B. Blyn, S. A. Hofstadler, J. C. Hannis, M. W. Eshoo, T. A. Hall, and R. Sampath. 2009. Molecular genotyping of microbes by multilocus PCR and mass spectrometry: a new tool for hospital infection control and public health surveillance. Methods Mol. Biol. 551:71-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ecker, D. J., R. Sampath, C. Massire, L. B. Blyn, T. A. Hall, M. W. Eshoo, and S. A. Hofstadler. 2008. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6:553-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ecker, D. J., R. Sampath, P. Willett, J. R. Wyatt, V. Samant, C. Massire, T. A. Hall, K. Hari, J. A. McNeil, C. Buchen-Osmond, and B. Budowle. 2005. The Microbial Rosetta Stone Database: a compilation of global and emerging infectious microorganisms and bioterrorist threat agents. BMC Microbiol. 5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eschete, M. L., and B. C. West. 1980. Saccharomyces cerevisiae septicemia. Arch. Intern. Med. 140:1539. [PubMed] [Google Scholar]
- 20.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrest, G. N., K. Mankes, M. A. Jabra-Rizk, E. Weekes, J. K. Johnson, D. P. Lincalis, and R. A. Venezia. 2006. Peptide nucleic acid fluorescence in situ hybridization-based identification of Candida albicans and its impact on mortality and antifungal therapy costs. J. Clin. Microbiol. 44:3381-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrest, G. N., S. Mehta, E. Weekes, D. P. Lincalis, J. K. Johnson, and R. A. Venezia. 2006. Impact of rapid in situ hybridization testing on coagulase-negative staphylococci positive blood cultures. J. Antimicrob. Chemother. 58:154-158. [DOI] [PubMed] [Google Scholar]
- 23.Forrest, G. N., M. C. Roghmann, L. S. Toombs, J. K. Johnson, E. Weekes, D. P. Lincalis, and R. A. Venezia. 2008. Peptide nucleic acid fluorescent in situ hybridization for hospital-acquired enterococcal bacteremia: delivering earlier effective antimicrobial therapy. Antimicrob. Agents Chemother. 52:3558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin, P. J., J. R. Warren, A. A. Obias, S. M. Collins, and L. R. Peterson. 2002. Evaluation of the Vitek 2 system for rapid identification of clinical isolates of gram-negative bacilli and members of the family Streptococcaceae. Eur. J. Clin. Microbiol. Infect. Dis. 21:869-874. [DOI] [PubMed] [Google Scholar]
- 25.Grace, C. J., J. Lieberman, K. Pierce, and B. Littenberg. 2001. Usefulness of blood culture for hospitalized patients who are receiving antibiotic therapy. Clin. Infect. Dis. 32:1651-1655. [DOI] [PubMed] [Google Scholar]
- 26.Hall, T. A., R. Sampath, L. B. Blyn, R. Ranken, C. Ivy, R. Melton, H. Matthews, N. White, F. Li, V. Harpin, D. J. Ecker, L. K. McDougal, B. Limbago, T. Ross, D. M. Wolk, V. Wysocki, and K. C. Carroll. 2009. Rapid molecular genotyping and clonal complex assignment of Staphylococcus aureus isolates by PCR coupled to electrospray ionization-mass spectrometry. J. Clin. Microbiol. 47:1733-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazen, K. C. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heron, M., D. L. Hoyert, S. L. Murphy, J. Xu, K. D. Kochanek, and B. Tejada-Vera. 2006. Deaths: final data for 2006. Natl. Vital Stat. Rep. 57:1-134. [PubMed] [Google Scholar]
- 29.Hofstadler, S. A., R. Sampath, L. B. Blyn, M. W. Eshoo, T. A. Hall, Y. Jiang, J. J. Drader, C. Hannis, K. A. Sannes-Lwery, L. L. Cummins, B. Libby, D. J. D. Knize, D. Robbins, K. Rudnik, A. Desai, E. Moradi, and D. J. Ecker. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 242:23-41. [Google Scholar]
- 30.Ilina, E. N., A. D. Borovskaya, M. M. Malakhova, V. A. Vereshchagin, A. A. Kubanova, A. N. Kruglov, T. S. Svistunova, A. O. Gazarian, T. Maier, M. Kostrzewa, and V. M. Govorun. 2009. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J. Mol. Diagn. 11:75-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan, J. A., A. R. Butchko, and M. B. Durso. 2005. Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. J. Mol. Diagn. 7:105-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jordan, J. A., J. Jones-Laughner, and M. B. Durso. 2009. Utility of pyrosequencing in identifying bacteria directly from positive blood culture bottles. J. Clin. Microbiol. 47:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karakousis, A., L. Tan, D. Ellis, H. Alexiou, and P. J. Wormald. 2006. An assessment of the efficiency of fungal DNA extraction methods for maximizing the detection of medically important fungi using PCR. J. Microbiol. Methods 65:38-48. [DOI] [PubMed] [Google Scholar]
- 34.Kumar, A., D. Roberts, K. E. Wood, B. Light, J. E. Parrillo, S. Sharma, R. Suppes, D. Feinstein, S. Zanotti, L. Taiberg, D. Gurka, A. Kumar, and M. Cheang. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34:1589-1596. [DOI] [PubMed] [Google Scholar]
- 35.Ling, T. K., Z. K. Liu, and A. F. Cheng. 2003. Evaluation of the VITEK 2 system for rapid direct identification and susceptibility testing of gram-negative bacilli from positive blood cultures. J. Clin. Microbiol. 41:4705-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling, T. K., P. C. Tam, Z. K. Liu, and A. F. Cheng. 2001. Evaluation of VITEK 2 rapid identification and susceptibility testing system against gram-negative clinical isolates. J. Clin. Microbiol. 39:2964-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ly, T., J. Gulia, V. Pyrgos, M. Waga, and S. Shoham. 2008. Impact upon clinical outcomes of translation of PNA FISH-generated laboratory data from the clinical microbiology bench to bedside in real time. Ther. Clin. Risk Manag. 4:637-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez, J. A., L. Pozo, M. Almela, F. Marco, A. Soriano, F. Lopez, V. Balasso, J. Aguilar, and J. Mensa. 2007. Microbial and clinical determinants of time-to-positivity in patients with bacteraemia. Clin. Microbiol. Infect. 13:709-716. [DOI] [PubMed] [Google Scholar]
- 39.Mellmann, A., F. Bimet, C. Bizet, A. D. Borovskaya, R. R. Drake, U. Eigner, A. M. Fahr, Y. He, E. N. Ilina, M. Kostrzewa, T. Maier, L. Mancinelli, W. Moussaoui, G. Prevost, L. Putignani, C. L. Seachord, Y. W. Tang, and D. Harmsen. 2009. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 47:3732-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellmann, A., J. Cloud, T. Maier, U. Keckevoet, I. Ramminger, P. Iwen, J. Dunn, G. Hall, D. Wilson, P. Lasala, M. Kostrzewa, and D. Harmsen. 2008. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munson, E. L., D. J. Diekema, S. E. Beekmann, K. C. Chapin, and G. V. Doern. 2003. Detection and treatment of bloodstream infection: laboratory reporting and antimicrobial management. J. Clin. Microbiol. 41:495-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prod'hom, G., A. Bizzini, C. Durussel, J. Bille, and G. Greub. 2010. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J. Clin. Microbiol. 48:1481-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richter, S. S., K. P. Heilmann, C. L. Dohrn, F. Riahi, S. E. Beekmann, and G. V. Doern. 2008. Accuracy of phenotypic methods for identification of Streptococcus pneumoniae isolates included in surveillance programs. J. Clin. Microbiol. 46:2184-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seifert, H., C. von Eiff, and G. Fatkenheuer. 1999. Fatal case due to methicillin-resistant Staphylococcus aureus small colony variants in an AIDS patient. Emerg. Infect. Dis. 5:450-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spellerberg, B., and C. Brandt. 2007. Streptococcus, p. 412-429. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 46.Stevenson, L. G., S. K. Drake, and P. R. Murray. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:444-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolk, D. M., L. B. Blyn, T. A. Hall, R. Sampath, R. Ranken, C. Ivy, R. Melton, H. Matthews, N. White, F. Li, V. Harpin, D. J. Ecker, B. Limbago, L. K. McDougal, V. H. Wysocki, M. Cai, and K. C. Carroll. 2009. Pathogen profiling: rapid molecular characterization of Staphylococcus aureus by PCR/electrospray ionization-mass spectrometry and correlation with phenotype. J. Clin. Microbiol. 47:3129-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolk, D. M., M. J. Struelens, P. Pancholi, T. Davis, P. Della-Latta, D. Fuller, E. Picton, R. Dickenson, O. Denis, D. Johnson, and K. Chapin. 2009. Rapid detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in wound specimens and blood cultures: multicenter preclinical evaluation of the Cepheid Xpert MRSA/SA skin and soft tissue and blood culture assays. J. Clin. Microbiol. 47:823-826. [DOI] [PMC free article] [PubMed] [Google Scholar]