Abstract
Due to a highly homogeneous genetic composition, the subtyping of Salmonella enterica serovar Enteritidis strains to an epidemiologically relevant level remains intangible for pulsed-field gel electrophoresis (PFGE). We reported previously on a highly discriminatory PFGE-based subtyping scheme for S. enterica serovar Enteritidis that relies on a single combined cluster analysis of multiple restriction enzymes. However, the ability of a subtyping method to correctly infer genetic relatedness among outbreak strains is also essential for effective molecular epidemiological traceback. In this study, genetic and phylogenetic analyses were performed to assess whether concatenated enzyme methods can cluster closely related salmonellae into epidemiologically relevant hierarchies. PFGE profiles were generated by use of six restriction enzymes (XbaI, BlnI, SpeI, SfiI, PacI, and NotI) for 74 strains each of S. enterica serovar Enteritidis and S. enterica serovar Typhimurium. Correlation analysis of Dice similarity coefficients for all pairwise strain comparisons underscored the importance of combining multiple enzymes for the accurate assignment of genetic relatedness among Salmonella strains. The mean correlation increased from 81% and 41% for single-enzyme PFGE up to 99% and 96% for five-enzyme combined PFGE for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains, respectively. Data regressions approached 100% correlation among Dice similarities for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains when a minimum of six enzymes were concatenated. Phylogenetic congruence measures singled out XbaI, BlnI, SfiI, and PacI as most concordant for S. enterica serovar Enteritidis, while XbaI, BlnI, and SpeI were most concordant among S. enterica serovar Typhimurium strains. Together, these data indicate that PFGE coupled with sufficient enzyme numbers and combinations is capable of discerning accurate genetic relationships among Salmonella serovars comprising highly homogeneous strain complexes.
The salmonellae comprise over 2,500 serovars, many of which are known to be intracellular pathogens of mammals, birds, and reptiles (33). Salmonella serovars remain some of the most common food-borne pathogens of humans, causing an estimated 1.4 million Salmonella cases, with 600 deaths, each year (25). The most notable of these include Salmonella enterica serovar Typhimurium and S. enterica serovar Enteritidis, both of which remain the most common etiologic agents of salmonellosis-induced gastroenteritis in humans (http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5714a2.htm). Outbreaks of nontyphoidal salmonellosis associated with the consumption of raw or undercooked foods are attributed regularly to S. enterica, including recent outbreaks from peanut paste, tree nuts, and fresh-cut produce (e.g., tomatoes and peppers).
Strain identification is essential for effective investigation of common-source outbreaks. Phage typing (PT) has been used widely to facilitate epidemiologic traceback of S. enterica serovar Enteritidis isolates. It has become clear, however, that most S. enterica serovar Enteritidis isolates are derived from a few endemic clones and belong to a limited number of PTs (35, 38). Pulsed-field gel electrophoresis (PFGE) remains a subtyping “gold standard” for public health investigation and has been shown to be highly effective for epidemiological investigation of Salmonella serovars (1, 16, 26). However, conventional PFGE has shown limited discriminatory power in subtyping certain highly clonal serotypes (e.g., S. enterica serovar Enteritidis and S. enterica serovar Hadar) (19, 24, 36). In the case of S. enterica serovar Enteritidis, in particular, both phage typing and single-enzyme (i.e., XbaI) PFGE often lack the discriminatory power to partition strains into epidemiologically meaningful clusters (3, 30).
Most recently, by combining several restriction enzyme data sets into a single analysis, we reported on a highly discriminatory PFGE-based subtyping scheme for S. enterica serovar Enteritidis to enhance the application of this method for differentiating highly homogeneous Salmonella strains (41). This scheme has also been applied successfully to other highly clonal serovars, including S. enterica serovar Saintpaul and S. enterica serovar Hadar, as well as to homogeneous strains of S. enterica serovar Heidelberg and S. enterica serovar Kentucky (40), reinforcing the potential application of this PFGE-based subtyping scheme for a variety of clonally derived serovars.
Although the discriminatory power of concatenated PFGE has compared favorably to that of traditional PFGE protocols in differentiating clonal complexes of S. enterica serovar Enteritidis (41), the genetic and epidemiologic accuracy of this method has not been evaluated stringently for Salmonella. Assignment of accurate genetic relationships among strains associated with food-borne outbreaks is critical for effective source tracking and traceback inference. The purpose of this study was to employ a series of genetic and phylogenetic measures to determine the suitability of the concatenated enzyme PFGE method to cluster closely related isolates of Salmonella into meaningful genetic hierarchies that reflect epidemiologically relevant groupings of strains.
MATERIALS AND METHODS
Bacterial strains.
Seventy-four strains each of S. enterica serovar Enteritidis and S. enterica serovar Typhimurium were included in this study and were obtained from the FDA's Center for Veterinary Medicine (CVM) and Center for Food Safety & Applied Nutrition (CFSAN) and the University of Georgia's Center for Food Safety. All strains were originally isolated from poultry and poultry-related sources.
Phage typing.
All S. enterica serovar Enteritidis strains were phage typed by previously described methods (38) at the National Microbiology Laboratory, Canadian Science Centre for Human and Animal Health, Winnipeg, Manitoba, Canada. Strains that reacted with phages but retained unrecognizable lytic patterns were atypical and were designated RDNC (reacts but does not conform).
PFGE.
Six previously described (41) enzymes, including XbaI, BlnI, SpeI, SfiI, PacI, and NotI, were used in the PFGE analysis. The standard CDC PulseNet PFGE protocol for nontyphoidal Salmonella was performed as described previously (29). Individual run conditions were used as described in our previous report (41).
PCR screening for integrons and SGI1.
PCRs were performed by using chromosomal DNAs from S. enterica serovar Typhimurium and S. enterica serovar Enteritidis isolates included in this study as templates, with specific oligonucleotide primers for the amplification of class 1 integrons and Salmonella genomic island 1 (SGI1) (4, 32). The presence of class 1 integrons was probed using primer pair 5′CS (5′-GGCATCCAAGCAGCAAGC-3′) and 3′CS (5′-AAGCAGACTTGACCTGAT-3′), whereas identification of SGI1 and determination of its location were performed using primers U7-L12 (5′-ACACCTTGAGCAGGGCAAG-3′), LJ-R1 (5′-AGTTCTAAAGGTTCGTAGTCG-3′), and C9-L2 (5′-AGCAAGTGTGCGTAATTTGG-3′) or 104-RJ (5′-TGACGAGCTGAAGCGAATTG-3′) and 104-D (5′-ACCAGGGCAAAACTACACAG-3′), corresponding to left and right (with or without retron) junctions in the chromosome, respectively.
Data analyses.
Resultant PFGE gel images were analyzed using BioNumerics software package v.4.601 (Applied Maths, Sint-Martens-Latem, Belgium) and were converted to binary data format in such a way that all scorable bands were assigned to a specific bin or band class based on size variation. Band classes from S. enterica serovar Typhimurium and S. enterica serovar Enteritidis PFGE profiles were scored simultaneously in a single binary matrix. This ensured that all band bins from both serovars were represented in the resultant S. enterica serovar Typhimurium and S. enterica serovar Enteritidis data sets. Using these binary data matrices as input, genetic distance values among PFGE profiles for all enzymes and enzyme combinations (e.g., pairwise combinations) were calculated as uncorrected P distances, using MEGA v.3.0 (23), and were exported into Microsoft Excel to calculate the Pearson product moment correlation coefficient (r2) between all possible two-, three-, four-, and five-enzyme combinations.
Phylogenetic analysis.
Individual enzyme data sets were compiled into concatenated six-enzyme supermatrices for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains (15) and then subjected to phylogenetic analysis using a maximum likelihood approach available in PAUP* (phylogenetic analysis using parsimony) v.4.0b10 (37). The most likely trees were sought using heuristic search methods, with random addition of taxa (n = 20 iterations) and tree-bisection-reconnection (TBR) search methods in effect. Since the input data represented binary band scores for each strain, all frequencies were assumed to be equal, with equal distributions of rates at variable sites. Starting branch lengths were obtained using a Rogers-Swofford approximation.
Data robustness and accuracy were further evaluated using a series of parsimony-based phylogenetic measures for the six-enzyme S. enterica serovar Typhimurium and S. enterica serovar Enteritidis PFGE data sets, individually and concatenated as single six-enzyme character matrices. Mean pairwise diversity was calculated using MEGA v.3.0 (23). The number of equally most-parsimonious solutions, overall tree lengths, data skewedness values (17), retention indices (14), and consistency indices (CI) (14), reported here as reciprocals (i.e., % homoplasy), were calculated in PAUP*.
Congruence (i.e., phylogenetic concordance) between enzymes was assessed using the incongruence length difference (ILD) test. The version of the ILD test employed here is available in PAUP* v.4.0b10. ILD testing was used to measure statistical significance in the observed discordance between data sets (e.g., concatenated fingerprint alignments). Enzyme petitions were assembled as nexus files in interleaved format, using the PAUP* text editor, such that each enzyme data set represented a single interleaved portion of the combined data matrix. ILD tests were performed with 1,000 data partitions, using simple heuristic searches with 20 random taxon additions and TBR branch swapping in effect. Concordance among independent enzyme data matrices was explored further by compatibility analyses. Overall compatibility of sites was measured for each enzyme data set by using COMPATDNA (R. Stones, 2008), a Windows-based program that uses the compatibility algorithm (RETICULATE) of Jakobsen and Eastal (20), whereby two sites are deemed compatible if the character changes at these sites can be accounted for only once in a phylogeny. Incompatible sites may be the result of convergent evolution or redundancy among restriction sites between two strains. In this study, binary sites only—informative sites containing exactly two distinct bins—were included in the analysis.
RESULTS
Genetic and phylogenetic diversity of PFGE patterns resolved by individual restriction enzymes for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium.
Using the binary data matrices generated from PFGE profiles for all enzymes, the mean pairwise diversity of all enzyme combinations was calculated for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains (Table 1). Mean distances for the 15 two-enzyme combination analyses of S. enterica serovar Enteritidis strains ranged from 9.7% to 20%, with an average two-enzyme diversity for the six enzymes of 14.8%. All of these values, regardless of enzyme choice, fell well short of the two-enzyme diversity value for S. enterica serovar Typhimurium strains—combined XbaI/BlnI analysis of S. enterica serovar Typhimurium yielded a diversity value of 47.9%. Furthermore, these low diversity values underscore the genetically depauperate condition of S. enterica serovar Enteritidis strains. Interestingly, mean diversity readings do appear to serve as reliable correlates for discerning the relative discriminatory power of particular enzymes or enzyme combinations (Fig. 1). The SfiI/PacI enzyme pair, for example, yielded the lowest pairwise diversity reading (9.7%) and also yielded the fewest parsimony-informative sites. Conversely, the SpeI/NotI combination had a high level of diversity relative to that of other enzyme pairs and yielded the largest number of informative and parsimony-informative sites. Thus, simple genetic distances among enzymes and enzyme pairs may represent an accurate surrogate for identifying taxonomically informative enzyme combinations for PFGE.
TABLE 1.
Pairwise diversity values of combined two-enzyme PFGE profiles for Salmonella enterica serovar Enteritidis and Salmonella enterica serovar Typhimurium
| Enzyme | % diversitya when combined with enzyme |
|||||
|---|---|---|---|---|---|---|
| XbaI | BlnI | SpeI | SfiI | PacI | NotI | |
| XbaI | 47.9 ± 3.0 | 36.1 ± 2.3 | 32.2 ± 2.3 | 37.5 ± 2.5 | 38.3 ± 2.6 | |
| BlnI | 15.3 ± 2.0 | 45.2 ± 3.0 | 39.2 ± 2.9 | 45.9 ± 2.8 | 47.5 ± 3.1 | |
| SpeI | 17.4 ± 1.9 | 19.8 ± 2.5 | 30.8 ± 2.0 | 35.8 ± 2.2 | 36.2 ± 2.3 | |
| SfiI | 10.8 ± 1.2 | 12.4 ± 1.6 | 14.2 ± 1.7 | 32.0 ± 2.2 | 33.0 ± 2.3 | |
| PacI | 11.7 ± 1.3 | 13.4 ± 1.8 | 15.4 ± 1.9 | 9.7 ± 1.1 | 38.1 ± 2.3 | |
| NotI | 15.8 ± 1.7 | 18.2 ± 2.1 | 20.0 ± 2.1 | 13.4 ± 1.4 | 14.3 ± 1.6 | |
Diversity values were calculated as mean P distances for 74 Salmonella enterica serovar Enteritidis strains (lower diagonal) and 74 Salmonella enterica serovar Typhimurium strains (upper diagonal), with standard errors, using MEGAv.3.0 (23).
FIG. 1.
Relative levels of informative PFGE bands among six different restriction enzymes for PFGE of Salmonella enterica serovar Typhimurium (ST) and S. enterica serovar Enteritidis (SE) strains. The bars in the graph note the percentages of informative and parsimony-informative bands among the total number of bands for the specific enzyme and serovar indicated. An informative band is defined here as any polymorphic band that varies by its presence or absence in at least one strain. Parsimony-informative bands are defined as polymorphic bands represented in two or more species. Informative and parsimony-informative sites are represented by black and light grey bars, respectively, for S. enterica serovar Enteritidis, and by dark grey and open bars, respectively, for S. enterica serovar Typhimurium.
Phylogenetic robustness measures for six-enzyme data sets from S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains revealed a stark contrast in the levels of consistency, homoplasy, data skewedness, and overall tree length (Table 2). These data suggest that S. enterica serovar Typhimurium enzyme matrices contain substantially more convergence (i.e., homoplasy) than corresponding S. enterica serovar Enteritidis data sets. Surprisingly, however, S. enterica serovar Enteritidis enzyme matrices yielded fewer equally parsimonious solutions when analyzed individually versus S. enterica serovar Enteritidis data sets for the same enzyme. Regardless, these data buttress the notion that single-enzyme PFGE analysis, which is largely incapable of reconstructing accurate relationships for Salmonella strains, is particularly intangible for S. enterica serovar Typhimurium strains, since an astonishing 80 or 90% of PFGE characters from any given single-enzyme data set displayed homoplasy (10, 41).
TABLE 2.
Phylogenetic robustness and taxonomic confidence measures among various enzymes
| S. enterica serovar and enzyme(s) used for analysis | Mean % diversitya | No. of maximum parsimony treesb | Tree lengthb | Tree skewednessc (G1) | Retention indexb | % homoplasyd |
|---|---|---|---|---|---|---|
| Enteritidis | ||||||
| XbaI | 10.2 | 8 | 84 | −0.54 | 0.79 | 46 |
| BlnI | 13.7 | 4 | 68 | −0.22 | 0.91 | 40 |
| SpeI | 16.2 | 2 | 140 | −0.25 | 0.71 | 64 |
| SfiI | 7.3 | 23 | 90 | −2.20 | 0.71 | 46 |
| PacI | 8.9 | 13 | 99 | −0.94 | 0.68 | 54 |
| NotI | 14.4 | 5 | 169 | −0.31 | 0.69 | 66 |
| Six enzymes | 9.8 | |||||
| Four enzymes (PAe) | 11.6 | |||||
| Typhimurium | ||||||
| XbaI | 27.1 | 1 | 322 | −0.22 | 0.56 | 85 |
| BlnI | 37.4 | 3 | 433 | −0.11 | 0.54 | 90 |
| SpeI | 25.0 | 1 | 332 | −0.08 | 0.49 | 85 |
| SfiI | 21.5 | 1 | 321 | −0.28 | 0.57 | 81 |
| PacI | 26.8 | 1 | 411 | −0.13 | 0.47 | 87 |
| NotI | 28.0 | 1 | 445 | −0.26 | 0.50 | 87 |
| Six enzymes | 27.2 | |||||
| Three enzymes (PA) | 29.7 |
Diversity values were calculated as mean P distances, using MEGAv.3.0 (23).
Derived from maximum parsimony analysis in PAUP* v.4.0b10 (37).
Derived from evaluation of 10,000 iterations of a Random Trees analysis, available in PAUP* v.4.0b10.
Defined as the percentage of convergent characters that map to the tree more than once and calculated as (1 − CI)100, where CI is the consistency index for the most parsimonious tree in PAUP* v.4.0b10.
PA, prior-agreement concordance based on simultaneous ILD analysis of 6 enzymes.
Combined six-enzyme analyses.
In order to ascertain the minimum number of concatenated enzyme matrices necessary to bring the genetic relatedness coefficient for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium to an epidemiologically relevant level, the mean correlation coefficients and r2 (coefficient of determination) values for all enzyme combinations for one-, two-, three-, four-, and five-enzyme concatenated data sets were determined. A logarithmic function computed from these data allowed for projection of the six-enzyme correlation as well. In the case of S. enterica serovar Enteritidis, the correlation for genetic relatedness among strains increased from 81% for one enzyme to 99% for five enzymes (Fig. 2, top panel). This placed the six-enzyme data set at 100% correlation (r2 = 1.0) for S. enterica serovar Enteritidis strains, indicating that a single and highly informative genetic signal should be attainable using a concatenated data set comprised of six combined PFGE-based enzyme matrices. For S. enterica serovar Typhimurium strains, this was even more remarkable. Single-enzyme genetic correlates were poor, at 41%. However, as more enzymes were concatenated, the r2 value increased arithmetically, until an r2 value of 0.96 was achieved for the concatenated five-enzyme supermatrix (Fig. 2, bottom panel). Similar to the S. enterica serovar Enteritidis analysis, this placed the six-enzyme data set at 100% correlation (r2 = 1.0) for S. enterica serovar Typhimurium strains, indicating that a single genetic signal could be attained for S. enterica serovar Typhimurium strains by use of a data concatenation approach.
FIG. 2.
Effect of concatenated number of enzymes on correlation coefficiency (r2) of genetic similarities among Salmonella strains, determined by the P distance. The graphs shown represent regression plots of correlation coefficients versus enzyme numbers for S. enterica serovar Enteritidis (top) and S. enterica serovar Typhimurium (bottom). In both graphs, the ordinate is defined by the concatenated number of enzymes (1 to 5), and the abscissa is denoted by corresponding r2 values. The shaded diamonds represent pairwise r2 values resulting from pairwise comparisons of all possible enzyme combinations at the concatenated enzyme number indicated. The circular data points represent the six-enzyme analysis.
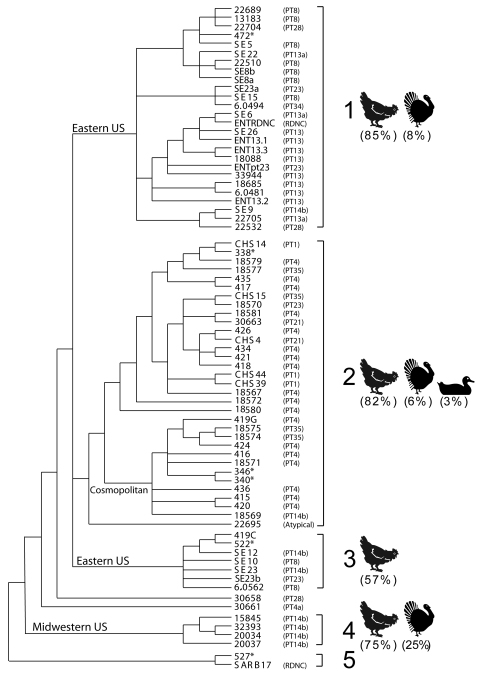
Maximum parsimony analysis of the six-enzyme concatenated data sets for the poultry-derived S. enterica serovar Enteritidis strains included in this study revealed the presence of six distinct clades (Fig. 3). Three of these strain clusters were composed of seven or more S. enterica serovar Enteritidis strains and retained strains derived from both turkey and chicken sources. These clades may represent epidemic clones that evolved independently during the radiation of S. enterica serovar Enteritidis strains. Surprisingly, phylogeographic components could be discerned from the tree, based on clade assignment of various strains. As an example, clade 2 retained international strains from Mexico, Scotland, and China almost in their entirety, while clade 1 represented a domestic cluster of S. enterica serovar Enteritidis strains from six different states in the United States coalescing into a single group.
FIG. 3.
Combined six-enzyme phylogenetic tree of S. enterica serovar Enteritidis strains. The tree depicts the enhanced epidemiological congruence resulting from the concatenated molecular phylogenetic analysis of PFGE data obtained with six enzymes (XbaI, BlnI, SpeI, SfiI, PacI, and NotI). Brackets to the right of the tree define the five major lineages (1 to 5) of S. enterica serovar Enteritidis strains. The host species (i.e., chicken, turkey, or mallard) and prevalence of S. enterica serovar Enteritidis strains within each clade are noted. Major geographical regions, where noted, are listed on the basal branches of several clades. Phage types are presented to the right of the tree (in parentheses). The tree was the most probable tree derived from a maximum likelihood analysis available in PAUP* 4.0b v.10.0 (37) and was constructed with empirical frequencies and equal rates for all sites. The tree shown was midpoint rooted.
Parsimony analysis of S. enterica serovar Typhimurium strains revealed several interesting observations (Fig. 4). First, seven distinct clades of S. enterica serovar Typhimurium strains were described that assorted largely along host source. That is, five clades retained S. enterica serovar Typhimurium strains almost entirely from chicken sources, save for two strains from turkey sources in clade 1. Clade 2 was found to comprise turkey strains solely, while another clade (i.e., clade 7) appeared to be composed of S. enterica serovar Typhimurium strains obtained equally from chickens and turkeys. Second, several notable phylogeographic partitions were evident in the tree. Several subclades within larger clades in the tree were identified that reiterated geographic similarity among strains, including a Midwest strain cluster, a Mexican cluster, and two southeastern U.S. lineages, each of which may represent the radiation of a single epidemic clone of S. enterica serovar Typhimurium among poultry sources. Finally, it was found that several strains known to retain the multidrug resistance element SGI1 were in fact clustered together tightly at the top of the tree, in clade 1. The only exception was a single Mexican S. enterica serovar Typhimurium strain (18563), which resided in clade 4. Taken together, these data illustrate the predictive value of six-enzyme concatenated analysis to reiterate the source type and geographic locale of closely related Salmonella strains.
FIG. 4.
Combined six-enzyme phylogenetic tree of S. enterica serovar Typhimurium strains. The tree depicts enhanced epidemiological congruence resulting from concatenated molecular phylogenetic analysis of six-enzyme PFGE data (XbaI, BlnI, SpeI, SfiI, PacI, and NotI). Brackets to the right of the tree define seven distinct lineages (1 to 7) of S. enterica serovar Typhimurium strains. The host species (i.e., chicken or turkey) and prevalence of S. enterica serovar Typhimurium strains within each clade are noted. Major geographical regions, where noted, are listed on the basal branches of several clades. The presence of SGI1 and the O5− (Copenhagen) serotype is listed to the right of the tree (in parentheses). The tree was the most probable derived tree from a maximum likelihood analysis available in PAUP* 4.0b v.10.0 (37) and was constructed with empirical frequencies and equal rates for all sites. The tree shown was midpoint rooted.
Congruence and compatibility of combined enzymes.
Phylogenetic concordance between enzymes for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains was examined further by using the ILD test (12), which evaluates the goodness of fit of phylogenetic signals between two independent data matrices (Fig. 5). For S. enterica serovar Enteritidis, ILD testing of all six enzymes partitioned separately led to the conclusion of incongruence. In order to isolate the enzymes responsible for incongruence (i.e., data discordance), each of the six data sets was partitioned against a combined matrix consisting of the remaining five data sets. While XbaI, BlnI, SfiI, and PacI were congruent with the combined enzyme matrix, SpeI and NotI were each highly discordant in their signals. For S. enterica serovar Typhimurium, ILD testing of all six enzymes partitioned separately also led to the conclusion of incongruence. In this case, XbaI, BlnI, and SpeI were congruent, while SfiI, PacI, and NotI were each discordant.
FIG. 5.
ILD analysis of the six enzymes used for concatenated analysis of S. enterica serovar Enteritidis (SE) and S. enterica serovar Typhimurium (ST) strains. Each individual cell represents the ILD test conducted between the enzyme indicated and the remaining five enzymes. White cells denote an ILD score of >0.10 (congruence), gray cells denote an ILD score between 0.10 and 0.05 (borderline incongruence), and black cells denote an ILD score of ≤0.05 (incongruence). Scores reflect the results of 1,000 independent ILD iterations.
These findings were largely supported by an independent analysis of compatibility of sites (Fig. 6). Similar to the case for ILD testing, XbaI, BlnI, SfiI, and PacI were found to retain the highest intraenzyme compatibilities for S. enterica serovar Enteritidis strains. Similarly, among S. enterica serovar Typhimurium strains, XbaI and SpeI retained the highest intraenzyme compatibility scores. It is also notable that overall compatibility among S. enterica serovar Enteritidis strains was markedly higher than that for S. enterica serovar Typhimurium strains, a finding consistent with the homoplasy levels uncovered for S. enterica serovar Typhimurium analysis (Table 2).
FIG. 6.
Intraenzyme compatibility of informative binary sites. Enzyme names are provided under their respective compatibility matrices. Compatibility data for S. enterica serovar Enteritidis (SE) strains are presented in the lower diagonal of each plot, in grayscale, while compatibility results for S. enterica serovar Typhimurium (ST) are provided in the upper right diagonal of each matrix. Compatibility scores (percentage of compatible binary sites within each enzyme) are reported in the lower left and upper right corners of each matrix for S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains, respectively. The positions of all informative binary sites are provided along the x and y axes of each matrix for each enzyme. Open cells in the matrices denote compatibility between two sites, while shaded cells represent an incompatible comparison.
Prevalence and distribution of S. enterica serovar Typhimurium and S. enterica serovar Enteritidis phenotypes.
The prevalence and distribution of several important phenotypes retained by the two populations of S. enterica serovar Typhimurium and S. enterica serovar Enteritidis strains, including the SGI1 multidrug resistance cassette and the O5 antigen complex in S. enterica serovar Typhimurium and the phage type for S. enterica serovar Enteritidis, were examined. Additionally, strain groupings were examined in light of their geographic origin and are noted on both trees (Fig. 3 and 4).
Analysis of the SGI1 element revealed a limited distribution among poultry-derived Salmonella strains. This sequence was detected in only four (5%) S. enterica serovar Typhimurium strains and in no S. enterica serovar Enteritidis strains. An examination of the O5 antigen complex among S. enterica serovar Typhimurium strains revealed that the majority (57%; n = 42) of this population lacked the O5 antigen (i.e., O5− or Copenhagen phenotype).
The prevalence and distribution of PTs across the 74 poultry-derived strains of S. enterica serovar Enteritidis are noted in Fig. 3. Eleven different PTs were represented, along with several strains that were untypable using current reagents. The most common PTs were 4 (23%), 8 (12%), 13 (14%), and 14b (12%). It is noteworthy that phage type distributions partitioned largely with the major strain groupings present in the six-enzyme S. enterica serovar Enteritidis tree (Fig. 3). For example, clade 2 comprised largely S. enterica serovar Enteritidis strains with PT 4, while clade 4 retained strains solely from PT 14b.
Host source and geographic information was available for most of the Salmonella strains included in our study. S. enterica serovar Typhimurium strains partitioned largely along host source, with five of seven clades comprising strains from a single host species, four from chicken and one from turkey. The only exceptions were clades 1 and 7. It is notable, however, that clade 1 was primarily chicken in origin, with only three (14%) strains originating in turkeys. Surprisingly, the S. enterica serovar Enteritidis tree was more mosaic in terms of host partitions. That is, of the four clades for which the host source was known, only one comprised a single poultry host. The remaining three clades retained S. enterica serovar Enteritidis strains from multiple chicken, turkey, or duck sources. However, when geography was mapped across the S. enterica serovar Typhimurium and S. enterica serovar Enteritidis six-enzyme strain clusters, several interesting phylogeographic trends emerged (Fig. 3 and 4). First, S. enterica serovar Typhimurium clades 5 and 6 contained strains solely from the southeastern United States, suggesting that these lineages represent two independent epidemic clones from the same geographic locale. Second, sublineages within clades 2 and 4 were found to represent source-specific clusters from the midwestern United States and Mexico, respectively. Third, four clades from the S. enterica serovar Enteritidis tree also revealed phylogeographic associations. Clades 1 and 3 could be traced back almost in their entirety to the eastern United States, while clade 4 could be linked entirely to strains originating in the midwestern United States. Surprisingly, clade 2, designated a “cosmopolitan” clade in this study, consisted of strains from several widely disparate geographic sources, including Mexico, China, and Scotland. Taken together, these data highlight the epidemiological relevance of the six-enzyme PFGE method and underscore its predictive power in discerning host source (i.e., S. enterica serovar Typhimurium strains) and geographical origin (i.e., S. enterica serovar Typhimurium and Enteritidis strains) for these two Salmonella serovars.
DISCUSSION
S. enterica serovar Enteritidis remains a significant clinical and food-borne Salmonella pathogen. However, it is one of the most genetically homogeneous serotypes of Salmonella and is poorly differentiated by the most commonly used subtyping methods (18, 27). In a previous report where simultaneous analyses of multiple restriction enzymes were performed, we were able to easily resolve S. enterica serovar Typhimurium and S. enterica serovar Enteritidis strains along serological partitions as well as to genetically resolve a collection of tightly clustered S. enterica serovar Enteritidis strains (41). Through the combining of data for additional typing methods and/or subsequent restriction enzymes, several studies have already shown vast improvements in both discriminatory power and epidemiological congruence for Escherichia coli O157:H7 (10), Salmonella enterica (40, 41), and other human pathogens (21). While certain technical parameters (e.g., PFGE run conditions) associated with this six-enzyme concatenated data approach may preclude its application during an actual outbreak, the analytical scheme presented here can provide important retrospective information regarding the epidemiological relatedness of outbreak and potential source strains of Salmonella (41). In addition, the method may yield novel insight related to the source attribution of outbreak strains of Salmonella (13, 39).
A recent empirical evaluation of the effects of data concatenation on phylogenetic accuracy found that, in general, concatenation approaches outperformed consensus tree approaches for discerning evolutionary relationships (15). In the present study, a six-enzyme concatenated data approach yielded substantial increases in the accuracy of genetic relatedness among strains and subsequently allowed for discerning of molecular epidemiological relationships of closely related S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains. Empirical assessments of the accuracy of genetic relatedness demonstrated improved outcomes for both serovars. It is notable that while the combining of restriction enzyme data sets into a single supermatrix improved genetic accuracy for both serovars, S. enterica serovar Typhimurium strains enjoyed an increase in correlations of relatedness of about 60%, increasing from 40% for a single-enzyme analysis to nearly 100% for the combined six-enzyme approach. Although the six-enzyme analysis was unnecessary for differentiation of S. enterica serovar Typhimurium strains, the concatenation of separate enzyme data sets greatly enhanced the genetic and epidemiologic accuracy of PFGE for S. enterica serovar Typhimurium strains as well.
It is noteworthy that several individual enzymes were highly discordant by ILD testing when evaluated against the concatenated supermatrices for S. enterica serovar Typhimurium and S. enterica serovar Enteritidis, despite the fact that concordance among genetic similarities rose in parallel with the number of enzymes added. That is, correlation coefficients and congruence values revealed starkly contrasting signals across various concatenated enzyme combinations (data not shown). While r2 values (i.e., correlation) rose consistently as the concatenated data matrix grew larger, ILD scores (i.e., congruence) did not. In fact, mean ILD values for S. enterica serovar Enteritidis actually decreased substantially between one and three concatenated enzymes. This difference may be accounted for by the fact that correlation among genetic similarities increases sharply as additional shared and derived PFGE band characters (i.e., synapomorphies) are added to the overall data matrix. Additionally, since a large proportion of characters were likely to display homoplasy for each additional enzyme added, the ILD score never improved substantially, as it reflects the congruence of the overall data set. That is, even though some informative characters were added with each additional enzyme, the more sensitive ILD test was most likely clouded by additional homoplasies that poured into the data set alongside useful characters. Previous studies have demonstrated the effects of combining even a few incongruent characters on resultant ILD test scores (5, 7, 9).
Although geographic partitions of S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains were fairly informative, the delineation of clear host partitions was more circumspect among the poultry-derived isolates studied here. For example, the S. enterica serovar Enteritidis strain tree yielded only one example of host monophyly. An explanation for observed differences in host specificity among S. enterica serovar Enteritidis and S. enterica serovar Typhimurium strains is not easily discerned, particularly given the broad host-adaptive ranges of both serovars (2, 22). However, unlike S. enterica serovar Typhimurium strains, S. enterica serovar Enteritidis strains appear more panmictic in poultry, revealing no evidence of host specificity among distinct poultry-derived hosts. This distinction likely stems from the more recent introduction of a few highly clonal lineages of S. enterica serovar Enteritidis into poultry and related environmental niches (31).
The unusual genetic homogeneity observed among S. enterica serovar Enteritidis strains remains intriguing. Recent population genetic studies suggested that most S. enterica serovar Enteritidis strains belong to a single multilocus genotype (31). A subpopulation of this clone was shown to associate more frequently with egg-related salmonellosis and clinical illness. Thus, specific requirements for colonization and survival may select for only a few genotypes of S. enterica serovar Enteritidis in poultry environments. Alternatively, horizontal gene transfer is now widely accepted as a significant factor in driving genome composition among enteric bacteria (8, 28), and both diversifying and homogenizing genome effects have been noted as outcomes of the lateral transfer of DNA among closely related strains (6, 11). Akin to the case of polymerase genes in E. coli (6, 34), the repeated horizontal transfer and subsequent recombination of a few preferred alleles may have a significant effect on homogenizing the S. enterica serovar Enteritidis genome. Whatever explanation accounts for the depauperate genetic condition of S. enterica serovar Enteritidis, our findings denote an additional scheme for genetically partitioning the often clonally related strains common to this serovar. Moreover, these findings highlight PFGE as a continued essential and informative subtyping tool for the molecular epidemiological investigation of this and other group I Salmonella pathogens.
Acknowledgments
We dedicate this work to the memory of David Derse.
Footnotes
Published ahead of print on 27 October 2010.
REFERENCES
- 1.Amavisit, P., P. F. Markham, D. Lightfoot, K. G. Whithear, and G. F. Browning. 2001. Molecular epidemiology of Salmonella Heidelberg in an equine hospital. Vet. Microbiol. 80:85-98. [DOI] [PubMed] [Google Scholar]
- 2.Baumler, A. J., R. M. Tsolis, T. A. Ficht, and L. G. Adams. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boxrud, D., K. Pederson-Gulrud, J. Wotton, C. Medus, E. Lyszkowicz, J. Besser, and J. M. Bartkus. 2007. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J. Clin. Microbiol. 45:536-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E. W., M. L. Kotewicz, and T. A. Cebula. 2002. Detection of recombination among Salmonella enterica strains using the incongruence length difference test. Mol. Phylogenet. Evol. 24:102-120. [DOI] [PubMed] [Google Scholar]
- 6.Brown, E. W., J. E. LeClerc, M. L. Kotewicz, and T. A. Cebula. 2001. Three R's of bacterial evolution: how replication, repair, and recombination frame the origin of species. Environ. Mol. Mutagen. 38:248-260. [DOI] [PubMed] [Google Scholar]
- 7.Brown, E. W., J. E. LeClerc, B. Li, W. L. Payne, and T. A. Cebula. 2001. Phylogenetic evidence for horizontal transfer of mutS alleles among naturally occurring Escherichia coli strains. J. Bacteriol. 183:1631-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, E. W., M. K. Mammel, J. E. LeClerc, and T. A. Cebula. 2003. Limited boundaries for extensive horizontal gene transfer among Salmonella pathogens. Proc. Natl. Acad. Sci. U. S. A. 100:15676-15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bull, J. J., J. P. Huelsenbeck, C. W. Cunningham, D. L. Swofford, and P. J. Wadell. 1993. Partitioning and combining data in phylogenetic analysis. Syst. Biol. 42:384-397. [Google Scholar]
- 10.Davis, M. A., D. D. Hancock, T. E. Besser, and D. R. Call. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173:7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farris, J. S., M. Kallersjo, A. G. Kluge, and C. Bult. 1995. Testing significance of incongruence. Cladistics 10:315-319.
- 13.Foley, S. L., D. G. White, P. F. McDermott, R. D. Walker, B. Rhodes, P. J. Fedorka-Cray, S. Simjee, and S. Zhao. 2006. Comparison of subtyping methods for differentiating Salmonella enterica serovar Typhimurium isolates obtained from food animal sources. J. Clin. Microbiol. 44:3569-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forey, P. L., C. J. Humphries, I. Kitching, R. W. Scotland, D. J. Siebert, and D. Williams. 1992. Cladistics, a practical course in systematics. Clarendon Press, Oxford, United Kingdom.
- 15.Gadagkar, S. R., M. S. Rosenberg, and S. Kumar. 2005. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J. Exp. Zool. B Mol. Dev. Evol. 304:64-74. [DOI] [PubMed] [Google Scholar]
- 16.Guerra, B., P. Schrors, and M. C. Mendoza. 2000. Application of PFGE performed with XbaI to an epidemiological and phylogenetic study of Salmonella serotype Typhimurium. Relations between genetic types and phage types. New Microbiol. 23:11-20. [PubMed] [Google Scholar]
- 17.Hillis, D. M., and J. P. Huelsenbeck. 1992. Signal, noise, and reliability in molecular phylogenetic analyses. J. Hered. 83:189-195. [DOI] [PubMed] [Google Scholar]
- 18.Hudson, C. R., M. Garcia, R. K. Gast, and J. J. Maurer. 2001. Determination of close genetic relatedness of the major Salmonella Enteritidis phage types by pulsed-field gel electrophoresis and DNA sequence analysis of several Salmonella virulence genes. Avian Dis. 45:875-886. [PubMed] [Google Scholar]
- 19.Hyytia-Trees, E., S. C. Smole, P. A. Fields, B. Swaminathan, and E. M. Ribot. 2006. Second generation subtyping: a proposed PulseNet protocol for multiple-locus variable-number tandem repeat analysis of Shiga toxin-producing Escherichia coli O157 (STEC O157). Foodborne Pathog. Dis. 3:118-131. [DOI] [PubMed] [Google Scholar]
- 20.Jakobsen, I. B., and S. A. Eastal. 1996. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput. Appl. Biosci. (Cambridge) 12:291-295. [DOI] [PubMed] [Google Scholar]
- 21.Keim, P., M. N. Van Ert, T. Pearson, A. J. Vogler, L. Y. Huynh, and D. M. Wagner. 2004. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect. Genet. Evol. 4:205-213. [DOI] [PubMed] [Google Scholar]
- 22.Kingsley, R. A., and A. J. Baumler. 2000. Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Mol. Microbiol. 36:1006-1014. [DOI] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 24.Lukinmaa, S., U. M. Nakari, M. Eklund, and A. Siitonen. 2004. Application of molecular genetic methods in diagnostics and epidemiology of food-borne bacterial pathogens. APMIS 112:908-929. [DOI] [PubMed] [Google Scholar]
- 25.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nesse, L. L., K. Nordby, E. Heir, B. Bergsjoe, T. Vardund, H. Nygaard, and G. Holstad. 2003. Molecular analyses of Salmonella enterica isolates from fish feed factories and fish feed ingredients. Appl. Environ. Microbiol. 69:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen, J. E., M. N. Skov, E. J. Threlfall, and D. J. Brown. 1994. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J. Med. Microbiol. 40:15-22. [DOI] [PubMed] [Google Scholar]
- 28.Porwollik, S., and M. McClelland. 2003. Lateral gene transfer in Salmonella. Microbes Infect. 5:977-989. [DOI] [PubMed] [Google Scholar]
- 29.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, and T. J. Barrett. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]
- 30.Ridley, A. M., E. J. Threlfall, and B. Rowe. 1998. Genotypic characterization of Salmonella Enteritidis phage types by plasmid analysis, ribotyping, and pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeed, A. M., S. T. Walk, M. Arshad, and T. S. Whittam. 2006. Clonal structure and variation in virulence of Salmonella Enteritidis isolated from mice, chickens, and humans. J. AOAC Int. 89:504-511. [PubMed] [Google Scholar]
- 32.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 33.Selander, R. K., J. Li, and K. Nelson. 1996. Evolutionary genetics of Salmonella enterica, p. 2691-2707. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 34.Shinkai, A., P. H. Patel, and L. A. Loeb. 2001. The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J. Biol. Chem. 276:18836-18842. [DOI] [PubMed] [Google Scholar]
- 35.Stanley, J., M. Goldsworthy, and E. J. Threlfall. 1992. Molecular phylogenetic typing of pandemic isolates of Salmonella enteritidis. FEMS Microbiol. Lett. 69:153-160. [DOI] [PubMed] [Google Scholar]
- 36.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford, D. L. 1999. Phylogenetic analysis using parsimony (PAUP* v. 4.03b) program and documentation. The Smithsomian Institution, Washington, DC.
- 38.Ward, L. R., J. D. de Sa, and B. Rowe. 1987. A phage-typing scheme for Salmonella Enteritidis. Epidemiol. Infect. 99:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittam, T. S., and T. M. Bergholz. 2007. Molecular subtyping, source tracking, and food safety, p. 93-136. In J. W. Santo Domingo and M. J. Sadowsky (ed.), Microbial source tracking. American Society for Microbiology, Washington, DC.
- 40.Xi, M., J. Zheng, S. Zhao, E. W. Brown, and J. Meng. 2008. An enhanced discriminatory pulsed-field gel electrophoresis scheme for subtyping Salmonella serotypes Heidelberg, Kentucky, SaintPaul, and Hadar. J. Food Prot. 71:2067-2072. [DOI] [PubMed] [Google Scholar]
- 41.Zheng, J., C. E. Keys, S. Zhao, J. Meng, and E. W. Brown. 2007. Enhanced subtyping scheme for Salmonella Enteritidis. Emerg. Infect. Dis. 13:1932-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]