Abstract
A new isoform of the human estrogen receptor-alpha (hER-α) has been identified and characterized. This 46 kDa isoform (hERα46) lacks the N-terminal 173 amino acids present in the previously characterized 66 kDa isoform (hERα66). hERα46 is encoded by a new class of hER-α transcript that lacks the first coding exon (exon 1A) of the ER-α gene. We demonstrated that these Δ1A hER-α transcripts originate from the E and F hER-α promoters and are produced by the splicing of exon 1E directly to exon 2. Functional analysis of hERα46 showed that, in a cell context sensitive to the transactivation function AF-2, this receptor is an effective ligand-inducible transcription factor. In contrast, hERα46 is a powerful inhibitor of hERα66 in a cell context where the transactivating function of AF-1 predominates over AF-2. The mechanisms by which the AF-1 dominant-negative action is exerted may involve heterodimeri zation of the two receptor isoforms and/or direct competition for the ER-α DNA-binding site. hERα66/hERα46 ratios change with the cell growth status of the breast carcinoma cell line MCF7, suggesting a role of hERα46 in cellular proliferation.
Keywords: activation functions/estrogen receptor/gene regulation/isoforms/MCF7
Introduction
Estradiol (E2) controls a variety of physiological processes such as the establishment and maintenance of female sex differentiation patterns, reproductive cycle and pregnancy; liver, fat and bone cell metabolism; cardiovascular and neuronal activity; and embryonic and fetal development (Norman and Litwack, 1987; George and Wilson, 1988; Auchus and Fuqua, 1994). It is also well established that estrogens influence several pathological processes including breast, endometrium and ovarian cancers, osteoporosis and arteriosclerosis, and may also play a role in Alzheimer’s disease. E2 can have both desirable and harmful effects on these pathological processes (Norman and Litwack, 1987; Henderson et al., 1988; Auchus and Fuqua, 1994); however, the molecular mechanisms mediating these effects are poorly understood. These pleiotropic consequences result from the binding of E2 to specific intracellular receptors, the estrogen receptors (ERs). To date, two estrogen receptors (ER-α and ER-β), encoded by different genes, have been described (Green et al., 1986; Kuiper et al., 1996; Mosselman et al., 1996). These two receptors belong to the nuclear receptor superfamily of ligand-inducible transcription factors whose members, the steroid, thyroid hormone and retinoic acid receptors, regulate gene expression by interacting either in a protein–DNA manner with cognate DNA sequences called responsive elements (for reviews see Evans, 1988; Beato, 1989; Parker, 1991) or in a protein–protein manner with other transcriptional factors (Gaub et al., 1990; Paech et al., 1997). ERs are proteins with a modular structure that, on the basis of amino acid sequence homology with the other members of the family, can be subdivided into six distinct regions, A–F (Evans, 1988; Beato, 1989; Parker, 1991). Regions C and E are responsible for DNA and ligand binding, respectively. The A/B region contains a ligand-independent transactivation domain (AF-1) whereas a hormone-inducible transcription activating function (AF-2) is present in the hormone-binding domain. The relative contributions that both AF-1 and AF-2 exert on transcriptional control vary in a cell- and promoter-specific manner (Berry et al., 1990; Tzukerman et al., 1994).
One important route towards an understanding of how ER activation results in the pleiotropic effects of E2 is to study the molecular events involved in the differential and spatio-temporal expression of these receptors. Consequently, our laboratory has further characterized the human (h) and chicken (c) ER-α genes. We have shown that both hER-α and cER-α genes are complex genomic units exhibiting alternative splicing and promoter usage in a tissue-specific manner (Flouriot et al., 1998; Griffin et al., 1998). The six characterized hER-α mRNA isoforms (A–F) differ in their 5′ untranslated regions (5′ UTRs) as a consequence of alternative splicing of several upstream exons (1B–1F) to a common site 5′ to the translation initiation codon and, therefore, result in the generation of a common ER-α protein that is 66 kDa in size (Flouriot et al., 1998). Similarly, at least four (A1–D) ER-α mRNA 5′ UTR variants have been identified in chicken (Griffin et al., 1998). However, in contrast to the hER-α gene, the existence of a new class of cER-α mRNA (A2) encoding a novel 61 kDa cER-α protein (cERα61), which lacks the N-terminal 41 amino acids present in the previously characterized full-length cERα66, was also reported (Griffin et al., 1999). This cERα61was found to be expressed in oviparous species (chicken, Xenopus laevis and rainbow trout) but not in mammals. The cERα61 was shown to modulate, to a limited extent, estrogen-responsive promoter activity in an E2-independent manner (Griffin et al., 1999).
Therefore, further investigation into the genomic organization and expression of the hER-α gene was performed to determine whether the mammalian ER-α gene may also encode different ER-α isoforms. In this paper, we report the existence of a second hER-α protein, 46 kDa in size, referred to as hERα46. This isoform lacks the first 173 amino acids present at the N-terminus of the previously described hERα66 and consequently gives rise to a steroid receptor that does not possess an A/B region. The hERα46 acts as an AF-1 competitive inhibitor of hERα66 and is encoded by distinct hER-α mRNAs (E and F Δ1A hER-α mRNAs) that are generated by the alternative splicing of exon E (a non-coding exon 5′ to the initiating methionine) to exon 2 (a coding exon) of the hER-α gene.
Results
Evidence for the existence of hER-α mRNAs that lack exon 1A sequences: E–F Δ1A hER-α mRNAs
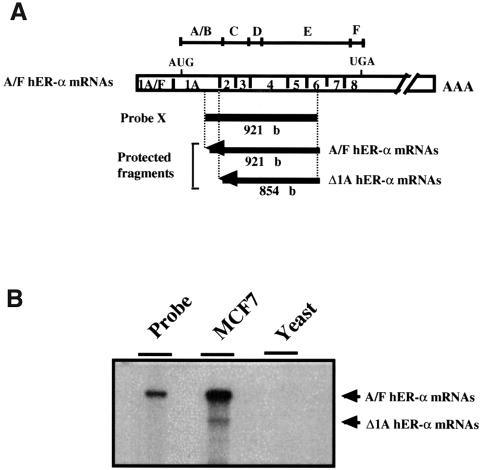
It is now well established that several genes that encode transcription factors belonging to the superfamily of nuclear receptors exhibit differential promoter usage and alternative splicing to generate receptor isoforms that differ at their N-terminus, resulting in different A or A/B regions (Kastner et al., 1990; Leroy et al., 1991; Zelent et al., 1991; Shi et al., 1992; Griffin et al., 1999). An S1 nuclease mapping analysis was performed using a probe (probe X) that encompassed the 3′ end of exon 1A through to exon 6, to evaluate whether hER-α transcripts that differ at their N-terminal region exist. This probe includes the region of hER-α mRNAs that encodes the C-terminal end of the B region, the DNA-binding domain and the beginning of the hormone-binding domain (Figure 1A). After hybridizing probe X with total RNA from the ER-α-positive breast carcinoma cell line MCF7, and S1 nuclease digestion, two major protected fragments of 921 and 854 nucleotides were detected (Figure 1B). As anticipated, the longest fragment, corresponding in size to the fully protected probe, resulted from hybridization of probe to the previously described A–F hER-α mRNA isoforms (Flouriot et al., 1998). The size of the second fragment was identical to that predicted for hER-α mRNAs that remained homologous to probe X until the junction between exon 1A and exon 2, and then diverged from probe X at their 5′ ends. Other experiments excluded the possibility that this band was due to a deletion of the 3′ end of the transcript. These results demonstrate the existence of hER-α transcripts lacking exon 1A sequences that are likely to arise from the splicing of exon(s) other than 1A to the acceptor splice site of exon 2.
Fig. 1. Evidence for an alternative splicing event at exon 2 acceptor splice site of the hER-α gene. (A) Experimental design for Δ1A hER-α mRNA detection, indicating the location and the size of the single-stranded probe X and each protected fragment obtained after S1 digestion of probe/hER-α mRNA hybrids. Probe X (from +617 to +1538) was specific for normal hER-α transcripts (A/F hER-α mRNAs) but was also able partially to protect Δ1A hER-α mRNA isoforms up to the splice acceptor site position of exon 2. Open boxes indicate the unique (1A–F) and common (1–8) exons encoding each normal hER-α mRNA isoform. The positions of the initiator methionine (AUG) and the termination codon (UGA) are indicated. The division of the hER-α protein into six regions, A–F, is shown directly above the cDNA. (B) Total RNA (30 µg) from MCF7 cells and 30 µg of yeast RNA used as a negative control were hybridized to the labeled S1 probe X, treated with S1 nuclease, and the resistant hybrids were separated on a sequencing gel as described in Materials and methods. The undigested probe is shown in a separate lane.
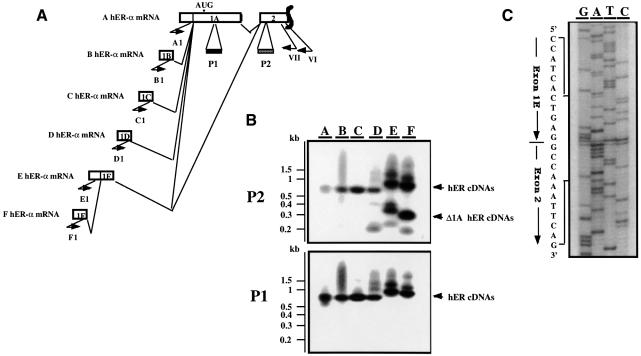
Reverse transcription PCR (RT–PCR) analysis was then performed in order to investigate whether one or several of the recently identified alternative upstream exons or leader sequences (1B–F), which are normally spliced alternatively to a common acceptor site 70 nucleotides upstream of the translation start site in exon 1A (Flouriot et al., 1998), can also splice directly to the acceptor site of exon 2, at position +685. Single strand cDNAs were synthesized from MCF7 total RNA using an hER-α gene specific primer (VI) chosen in exon 2. These hER-α cDNAs were then PCR amplified utilizing a common 3′ primer (VII) nested with primer VI in exon 2, in combination with 5′ primers specific for the different hER-α mRNA 5′ extremities (Figure 2A). Results from these experiments showed that, in addition to the expected amplified A–F hER-α cDNAs, shorter PCR products were present, and that these had E and F hER-α 5′ sequence. Southern blot analysis of these hER-α cDNA PCR products with two oligonucleotide probes (P1 and P2) specific for exon 1A and for exon 2, respectively, demonstrated that exon 1A sequences were not present in the shorter E–F hER-α cDNAs. Furthermore, sequencing analysis of these PCR products showed a direct splice junction between exon 1E and exon 2, as illustrated in Figure 2C. E and F hER-α transcripts share, in addition to the sequences from exon 1A to exon 8, the 3′ end of exon 1E (Figure 2A). Therefore, hER-α mRNAs transcribed from the E and F promoters may (E–F hER-α mRNAs) or may not [E–F Δ1A (exon 1A deleted) hER-α mRNAs] contain exon 1A sequences, as exon 1E can be alternatively spliced to either exon 1A or exon 2.
Fig. 2. Exon 1E is alternatively spliced to exon 1A or exon 2. (A) Schematic representation of the RT–PCR experiment designed to identify Δ1A hER-α mRNAs. Open boxes indicate the unique (1A–1F) and the two first common (1A, 2) exons encoding each hER-α mRNA variant. Approximate locations of primers are shown by short arrows. Primer VI, located in exon 2, was used to prime hER-α cDNA synthesis by reverse transcriptase. Primers A1–F1, which are specific for each hER-α cDNA 5′ region, were used in a round of PCR amplification with primer VII, which is nested to primer VI in exon 2. The oligonucleotide probes P1 and P2 from exon 1A and 2, respectively, were used to confirm the specificity of the PCR products as well as the exon 1A deletion for some hER-α transcripts. (B) The hER-α cDNA variants were amplified as described above, using total RNA from MCF7. PCR products were electrophoresed through an agarose gel and transferred by Southern blotting to a membrane, which was then hybridized with the oligonucleotide probes P1 and P2 as described in Materials and methods. Positions of migration of the molecular size markers are shown on the left side of the figure. (C) The sequence of the PCR products from lane E or F (B) that did not hybridize to the oligonucleotide probe P1 but hybridized to P2 probe revealed that they contain the donor site of exon 1E joined to the acceptor site of exon 2.
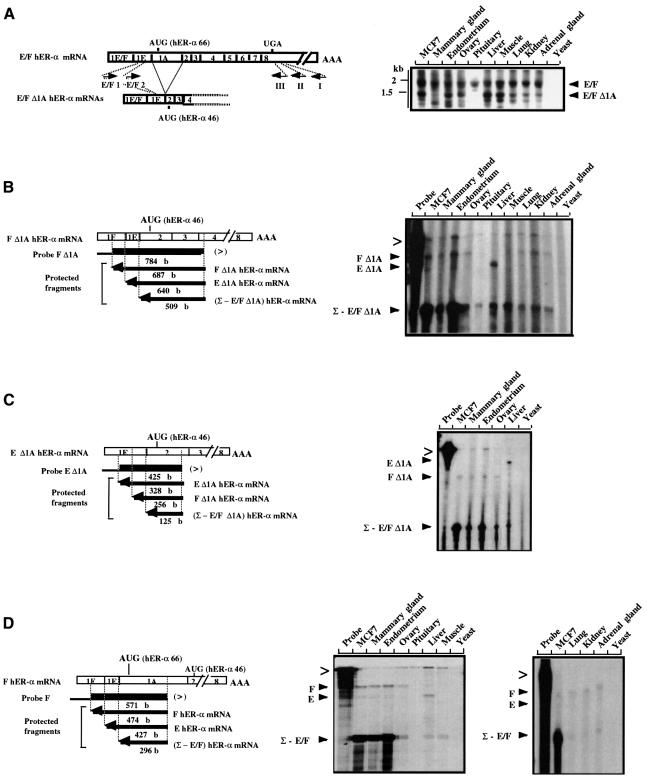
As the new E–F Δ1A hER-α mRNAs were described as a consequence of a direct splicing of exon 1E to exon 2, it was important to verify whether their full-length transcripts had hER-α sequences from exon 2 through to exon 8. To demonstrate this, PCR analysis was performed on single strand cDNAs synthesized from total RNA prepared from various sources, using an hER-α gene-specific primer (I) chosen from the hER-α mRNA 3′ UTR sequences (exon 8, Figure 3A). hER-α cDNAs were amplified by two rounds of PCR using the 3′ primer II and nested primer III located upstream from primer I in exon 8, in combination with the 5′ primer E/F1 and nested primer E/F2 specific for exon 1E part shared by E and F hER-α mRNAs. Two major cDNAs were amplified from almost all samples, the sizes of which were in agreement with those expected from the amplification of full-length and exon 1A-deleted E/F hER-α mRNAs (Figure 3A). These results were confirmed by Southern blotting and by hybridization of the PCR products with various oligonucleotide probes that recognized all eight coding exons of the hER-α gene (data not shown). This study also showed an amplification of E or F Δ1A hER-α cDNAs from all tissues analyzed, except the pituitary.
Fig. 3. E/F and E/F Δ1A hER-α mRNA variant distribution analysis. (A) RT–PCR analysis. Open boxes indicate the unique (1E or 1F) and common (part of 1E and 1A–8) exons encoding E/F hER-α mRNA isoforms. Approximate locations of primers are shown by short arrows. Primer I, located in the 3′ UTR of exon 8, was used to prime hER-α cDNA synthesis by reverse transcriptase, using total RNA from various sources as indicated at the top of each lane. Yeast total RNA was used as a negative control. Primer E/F1, which is specific for both E and F hER-α cDNA 5′ regions (in the common part of exon 1E), was then used in a first round of PCR amplification with primer II, which is nested to primer I in exon 8. A second round of PCR was performed with specific (E/F2) and common (III) nested primers. An oligonucleotide probe from exon 2 was used to confirm the specificity of the PCR products. Positions of migration of the molecular size markers are shown on the left side of the figure. (B–D) S1 nuclease mapping analysis. The S1 nuclease mapping assays of E/F and E/F Δ1A hER-α mRNA variants were performed as described in Materials and methods, with the single-stranded probes F (D), F Δ1A (B) and E Δ1A (C), and using 30 µg of total RNA from various sources as indicated at the top of each lane. Yeast total RNA was used as a negative control. The location and the size of each single-stranded probe (F, F Δ1A and E Δ1A) and each protected fragment obtained after S1 digestion of the probe/hER-α mRNA hybrids are indicated. Each probe was specific for one hER-α transcript (for example, F Δ1A hER mRNA) but was also able partially to protect the other hER-α mRNA isoforms [e.g. (Σ – E/F Δ1A) hER mRNA] up to the splice site positions. The probes were designed to contain vector sequence in their extremity (denoted by the thinner black line) in order to discriminate between undigested probes (>) and specific protected fragments.
S1 nuclease mapping experiments were then performed to estimate the abundance of E/F Δ1A and full-length hER-α mRNAs in the MCF7 cell line and in the different human tissues previously tested, using single strand DNA probes specific for each of the characterized hER-α transcripts (probes F Δ1A, E Δ1A and F, shown in Figure 3B–D). Each probe was able to measure the specific transcript and the residual expression resulting from the sum of the expression of other hER-α transcripts (for example Σ – E/F Δ1A in Figure 3C; see Figure 3B–D). As shown in Figure 3B and C, a protected fragment specific for F Δ1A hER-α mRNA was detected in MCF7, mammary gland, endometrium, ovary, lung, kidney and adrenal gland samples. E Δ1A hER-α mRNA was mainly expressed in the liver. Interestingly, in non-reproductive tissues E/F Δ1A hER-α transcripts were relatively abundant (20–50%) compared with the total hER-α mRNA, whereas they were less abundant in reproductive tissues (∼10%). It should be noted that, in agreement with the PCR analysis shown in Figure 3, pituitary did not express E or F Δ1A hER-α transcripts at a level detectable by S1 analysis. Finally, qualitative and quantitative comparison of E/F Δ1A hER-α mRNA expression in the various human tissues, with E/F hER-α mRNA expression pattern evaluated using probe F (Figure 3B and D), suggested that transcriptional activity from E and F promoters generates comparable amounts of E/F and E/F Δ1A hER-α mRNAs. This indicates that the splice donor of exon 1E has a similar probability of being spliced to either exon 1A or exon 2.
In conclusion, these data clearly demonstrate a significant level of expression of a new class of hER-α mRNAs that lack exon 1A (E/F hER-α Δ1A mRNAs) and are generated by alternative splicing and promoter usage.
E/F hER-α Δ1A mRNAs encode a novel hER-α protein: hERα46
Examination of the E/F Δ1A hER-α cDNA sequence showed that the first ATG codon in-frame with the remainder of the hER-α open reading frame (ORF) is at position +752/4 (methionine 174). Analysis of the sequence surrounding this ATG (5′-GAAGTATGG-3′) indicates a favorable ‘Kozak’ sequence for translation initiation (Kozak, 1989). Therefore, this ATG could function as a translation initiation codon for E/F Δ1A hER-α mRNAs to give rise to a 173 amino acid hER-α protein, devoid of the A/B domain, with a predicted size of 46 kDa. This protein is called hERα46, in contrast to the full-length receptor ER-α (hERα66) (Green et al., 1986) and the N-terminal 41 amino acid truncated cERα61 detected in oviparous species (Griffin et al., 1999).
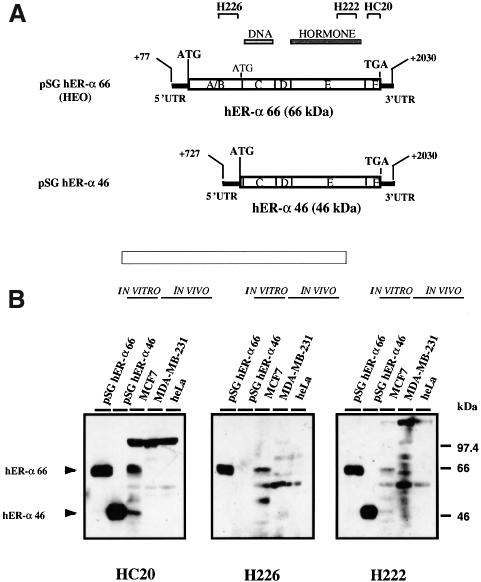
To demonstrate that E/F Δ1A hER-α transcripts encode a novel hER-α protein and that this protein is produced in vivo, western blot analysis was performed with both in vitro translated hER-α proteins and with whole cell extracts from ER-α-positive (MCF7) and ER-α-negative (MDA-MB-231 and HeLa) cell lines. In order to test whether hERα46 could be produced in vitro, a PCR product containing hER-α cDNA sequences from +727 to +2030 was inserted in pSG5, and then transcribed and translated in vitro by the rabbit reticulocyte lysate system. The expression vector HEO (pSG hERα66) was used to translate hERα66 (Figure 4A) (Green et al., 1988).
Fig. 4. E/F Δ1A hER-α mRNA isoforms encode a 46 kDa protein, called hERα46, which lacks the A/B domain present in the 66 kDa hER-α. (A) Schematic representation of the cDNAs inserted within the expression vector pSG5, which gave rise to pSG hERα66 (HEO) and pSG hERα46. The position of the initiator methionine for hERα66, the initiator methionine for hERα46 and the common termination codon (TGA) are indicated. The division of the hER-α protein (66 kDa) into six regions, A–F, together with the DNA- (region C) and hormone- (region E) binding domains, is shown directly above the cDNAs. Also shown are the epitopes recognized by the anti-hER antibodies, HC20, H226 and H222, used in (B). HC20 is a polyclonal antibody, and H226 and H222 are monoclonal antibodies. (B) pSG hERα66 and pSG hERα46 plasmids were in vitro transcribed and translated in rabbit reticulocyte lysate. Two microliters of the obtained translation products as well as 20 µg of whole cell extracts from MCF7 (ER-α-positive breast cancer cell line), MDA-MB-231 (ER-α-negative breast cancer cell line) and HeLa (ER-α-negative cell line) were resolved on a 10% SDS–polyacrylamide gel and then subjected to immunoblotting with the HC20, H226 and H222 antibodies. Immunoreactive bands 66 and 46 kDa in size were visualized by ECL.
Rabbit reticulocyte lysates and whole cell extracts were subjected to SDS–PAGE, transferred onto a nitrocellulose membrane and immunoblotted with the polyclonal antibody HC20 directed against the C-terminus of hER-α, with the monoclonal antibody H226 directed against the B domain of the hER-α protein and with the monoclonal antibody H222 raised against the ligand-binding domain (Greene et al., 1984). Analysis of the in vitro translation product from pSG hERα46 showed a 46 kDa hER-α protein recognized by HC20 and H222 antibodies (Figure 4B). The size of this protein and its failure to react with the antibody H226, to which hER-α form I cross-reacted specifically, correlated to an hER-α form lacking the N-terminus of hERα66 and thus giving rise to a receptor devoid of the A/B domain. These data demonstrated that the in-frame ATG codon at position +752/4 could initiate transcription by a rabbit reticulocyte lysate. Western blot analysis also showed that, in addition to hERα66, hERα46 was present in MCF7 whole cell extract (Figure 4B). As expected, MDA-MB-231 and HeLa did not express detectable levels of either hER-α isoform.
hERα46 heterodimerizes with hERα66
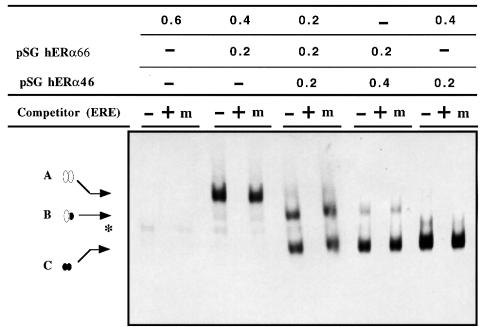
The capacity of both hER-α isoforms to form homo- and heterodimers that are able to bind to an estrogen response element (ERE) was tested in vitro by electrophoretic mobility shift assays. Extracts containing various hERα66/hERα46 ratios were produced using the rabbit reticulocyte lysate system. As shown in Figure 5, these extracts were able to form DNA–protein complexes with a radiolabeled consensus ERE from the chicken apoVLDL II promoter (Van Het Schip et al., 1983). The specificity of these complexes was confirmed by competition experiments. A suppression of the signal was observed with a 10-fold excess of unlabeled consensus apoVLDLII-ERE, whereas a 10-fold excess of a mutated ERE had no effect. Interestingly, depending on the extract used, up to three complexes with different mobilities were observed (Figure 5). The slower migrating complex (A) was obtained from extracts producing hERα66 whereas the faster one (C) was found in extracts containing hERα46. As ER binds to its cognate ERE as a dimer, A and C complexes presumably corresponded to homodimers of hERα66 and hERα46, respectively. An additional intermediate mobility complex (B) was generated when both receptor forms were synthesized simultaneously, demonstrating the formation of a heterodimer between the two hER-α forms. The presence of the intermediate complex at a low level in the reticulocyte lysate, where only HEO was transcribed and translated, indicated that hERα46 can also be translated from normal A–F hER-α transcripts in vitro as a consequence of leaky ribosome scanning (Kozak, 1989). Interestingly, the absence of a visible hERα46 homodimer in the same HEO-translated extract suggested that this hER-α protein forms heterodimers preferentially with hERα66. Finally, it should be noted that a reduction in the hERα66/hERα46 input ratios resulted in a progressive reduction of the initial levels of first the hERα66 homodimer followed by the hERα66/46 heterodimer in favor of the formation of hERα46 homodimer. This would be in keeping with, and may be a consequence of, an increased affinity of hERα46 homodimer for this ERE, in comparison with the hERα66 homodimer.
Fig. 5. hERα46 binds specifically in vitro to an ERE as a homodimer or a heterodimer with hERα66. Plasmid samples (0.6 µg) containing different combinations of pSG5, pSG hERα66 and pSG hERα46 vectors, as indicated at the top of each lane (expressed in µg), were in vitro transcribed and translated in rabbit reticulocyte lysate. Four microliters of in vitro translated products were incubated with 60 000 c.p.m. of labeled apoVLDLII-ERE. Specificity was determined in the absence (–) or presence (+) of a 10-fold excess of unlabeled apoVLDLII-ERE competitor, or a 10-fold amount of unlabeled mutant ERE (m) as a non-specific competitor. The positions of the three specific hER-α–DNA complexes (A–C) are indicated by arrows. A corresponds to hERα66 homodimer–ERE complex; B represents hERα66–46 heterodimer–ERE complex; and C represents hERα46 homodimer–ERE complex. An asterisk indicates a non-specific complex.
hERα46 is a competitive inhibitor of hERα66 for the activation function AF-1
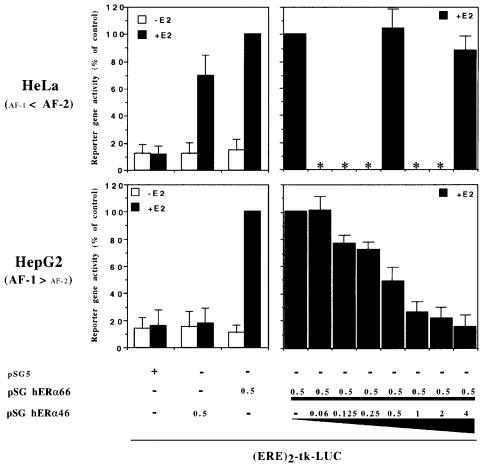
The full-length ER-α contains two major transactivation functions, one located in the A/B domain (AF-1), the other in the C-terminal part of the hormone-binding domain (AF-2) (Berry et al., 1990; Tzukerman et al., 1994). As hERα46 is devoid of an A/B domain, it was expected that transactivation by hERα46 would be effective in a cell context sensitive to AF-2 but inefficient in a cell context predominately mediated through AF-1 activation. To confirm this assumption, both hER-α receptors were assayed in transient mammalian cell transfection experiments using a luciferase-expressing reporter construct that contained two EREs (sequences from –331 to –289 of the Vitellogenin A2 gene) placed upstream of the thymidine kinase promoter [(ERE)2-tk-LUC] (Paech et al., 1997). The two cell lines selected for this study were HeLa and the liver cell line HepG2, as it has been reported previously that AF-2 is the dominant hER-α transactivation function in HeLa cells whereas HepG2 cells mediate ER-α signaling through the AF-1 hER-α transactivation function (Berry et al., 1990; Tzukerman et al., 1994; Norris et al., 1997). As shown in Figure 6, in the presence of estradiol hERα46 was able to activate reporter gene expression in HeLa cells. However, neither E2-dependent nor -independent transactivation resulting from hERα46 expression was observed in HepG2 cells.
Fig. 6. hERα46 transcriptional properties differ in accordance with the cell sensitivity to ER-α transactivation functions, AF-1 and AF-2. HeLa and HepG2 cell lines are known to present different sensitivity to the two transactivation functions of ER-α, AF-1 and AF-2, as indicated on the left side of the graph (Berry et al., 1990; Tzukerman et al., 1994; Norris et al., 1997). Therefore, these two cell lines were transiently transfected with 5 µg of the reporter plasmid (ERE)2-tk-LUC together with 0.5 µg of the expression vector pSG5, 0.5 µg of pSG hERα46 or 0.5 µg of pSG hERα66 (HEO) alone, or with increasing concentration of pSG hERα46 (0–4 µg). Cells were treated with or without estradiol (10–8 M) for 48 h before being assayed for luciferase activity. Results are expressed as a percentage of the reporter gene activity measured in the presence of the expression vector pSG hERα66 alone and E2. Luciferase activities were normalized using the internal reference control EF-1α–CAT. Values correspond to the average ± standard deviation (SD) of at least three separate transfection experiments. Values not determined are indicated by an asterisk.
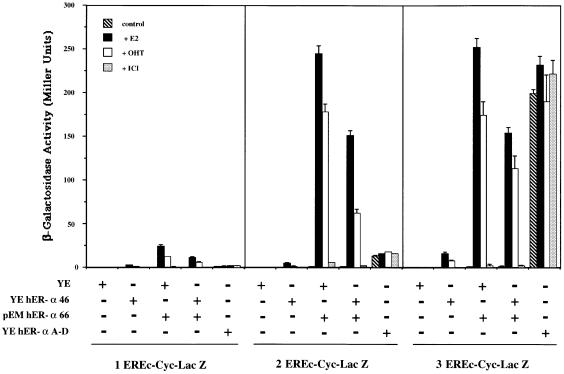
The consequence of hERα46 on estrogen-dependent transcriptional activation by hERα66 in a cell context sensitive to AF-1 was then evaluated. A series of transfections with various pSG hERα66/46 ratios was performed in HepG2. The results obtained demonstrated that hERα46 is a potent competitive inhibitor of hERα66 in these cells, at all DNA input ratios tested (Figure 6). A complete suppression of hERα66 activity was achieved with an input DNA ratio of hERα46 to hERα66 of 4/1. However, a similar experiment in HeLa cells in which the main hER-α transactivation activity is mediated through AF-2 did not show inhibition of hERα66 transactivation by hERα46 at either 1/1 or 1/8 input DNA ratios. These data indicate that hERα46 is an effective competitive inhibitor of hERα66 where transactivation is mediated through the AF-1 domain. This analysis was further strengthened by a functional study of hERα46 in the yeast Saccharomyces cerevisiae, which previous work has also reported to exhibit a predominant AF-1-dependent transactivation activity of hERα66 (Metzger et al., 1992, 1995; Pham et al., 1992). cDNAs for hERα46, hERα66 and an hER-α A–D deletion mutant retaining only AF-1 function (deletion of the hormone-binding domain and AF-2 region) were subcloned in the YEpucG (Wrenn and Katzenellenbogen, 1993) or pEMBL (Banroques et al., 1986) yeast expression vectors. These plasmids were then cotransformed in the BJ2168 yeast host strain with the pLG Δ178 reporter genes containing 1, 2 or 3 consensus EREs (1–3 EREc-Cyc-Lac Z) (Guarente and Masson, 1983; Petit et al., 1999). The results of this study are illustrated in Figure 7. AF-1 was confirmed to be the dominant hER-α transactivation function in yeast: first, the hER-α A–D mutant had a constitutive activity that reached 85% of the maximum induction obtained with the hER-α form I activating the triple EREs, and secondly, 4-hydroxytamoxifen (OHT) functioned as a potent agonist of hERα66 (Berry et al., 1990). In this cell context, hERα46 was characterized by a low ability (2–10% of hERα66 transactivation) to transactivate reporter gene expression from an ERE. Finally, as expected from the data obtained in HepG2 cells, coexpression of hERα66 and hERα46, at an input ratio of 1/1 in yeast, resulted in a 40–50% inhibition of hERα66 transactivation activity. Therefore, hERα46 again behaved as a competitive inhibitor of hERα66 in the AF-1-dependent context in yeast.
Fig. 7. hERα46 acts as an inhibitor of hERα66 in yeast. Yeast cells transformed with the reporter genes 1, 2 or 3 EREc-Cyc-Lac Z and a combination of the expression vectors YEpucG (YE), YE hERα46, pEM hERα66 and YE hER-α A–D (as indicated at the bottom of the graph) were grown in the presence or absence of 1 µM estradiol (E2), 10 µM 4-hydroxytamoxifen (OHT) or 10 µM ICI 164,384 (ICI). β-galactosidase activity was assayed and expressed in Miller units. Values correspond to the average ± SD of at least four separate experiments.
hER-α form 66/46 ratios change with cell growth status in the breast carcinoma cell line MCF7
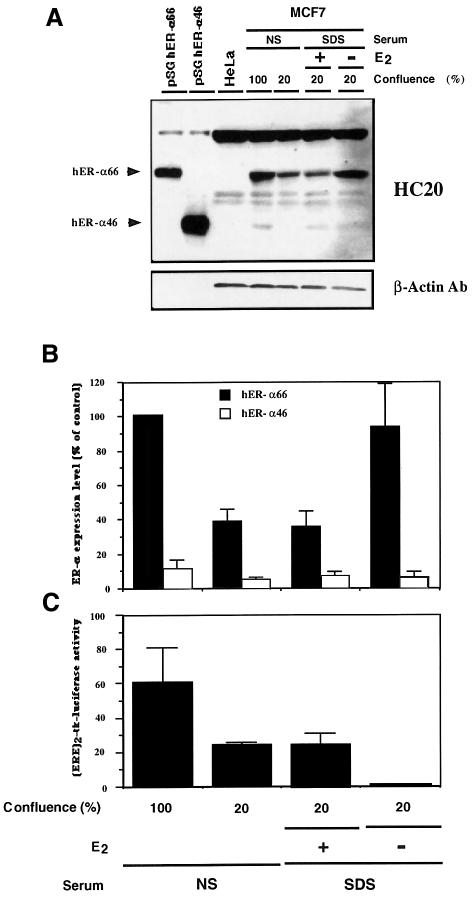
In the light of the above observations that the ability of hER-α to transactivate target genes via AF-1 may be modulated by the relative proportion of hERα66 and hERα46, an examination of physiological situations in which different hERα66/46 ratios may arise was carried out. Estrogen is essential for the growth of normal human mammary gland (Topper and Freedman, 1980; Norman and Litwack, 1987; George and Wilson, 1988; Auchus and Fuqua, 1994) as well as for the proliferation of ER-positive carcinomas in vivo and in vitro (Dickson and Lippman, 1988; Henderson et al., 1988; Auchus and Fuqua, 1994). Therefore, the relative levels of hERα66 and hERα46 present in the cell line MCF7 were analyzed in relation to growth status. In addition to estrogenic treatment, the rate of proliferation of MCF7 cells can also be altered by plating the cells at different densities (Jakesz et al., 1984). Consequently, MCF7 cell extracts from confluent and non-confluent (∼20% confluence) cells growing in normal serum (NS) (steroids present in the serum), as well as from estrogen-treated and non-treated cells cultivated in the presence of charcoal-treated calf serum (SDS for steroid-deprived serum), were evaluated for hER-α protein content by western blot analysis using the polyclonal antibody HC20. Results showed that, whereas the expression level of hERα46 did not really change with the cell growth conditions, the expression level of hERα66 was significantly reduced in proliferating cells [cells at 20% of confluence and growing in the presence of estrogen (lanes 5 and 6 in Figure 8A)], reaching 30–40% of the level detected in confluent cells or in cells cultivated in the absence of estrogen (Figure 8A and B). Densitometric analysis of hER-α signals in slowly or non-proliferating MCF7 cells indicated an ∼10-fold excess of the hERα66 compared with hERα46 (Figure 8B). Therefore, these data suggest an inverse relationship between the proliferation rate of MCF7 cells and hERα66/hERα46 ratios in these cells. Finally, the alteration in ER cellular content observed between confluent and non-confluent MCF7 cells resulted in a change in the estrogen-dependent transcriptional activation of the (ERE)2-tk-LUC reporter gene after transient transfection of these cells (Figure 8C). Similar results were also obtained after assay of the endogenous estrogen-regulated creatine kinase activity (data not shown) (Spatz et al., 1992).
Fig. 8. hERα66/46 ratios in the MCF7 cell line differ in confluent and non-confluent cells as well as in estradiol-treated and untreated cells. For the study of confluent and non-confluent cells, MCF7 cells were grown to confluency (100%) or non-confluency (20%) in normal DMEM containing 10% calf serum (NS for normal serum); medium was then changed and cells were kept for an additional 3 days under those conditions before harvesting. For the study of estradiol-treated and untreated cells, MCF7 cells were first grown under normal conditions to non-confluency (20%) and, after a PBS wash, were kept for 3 days in phenolred-free medium supplemented with 2.5% charcoal-treated calf serum (SDS for steroid-deprived serum) with (+) or without (–) 10 nM estradiol, before harvesting. Whole cell extracts were prepared as described in Materials and methods. The obtained protein extract (20 µg), as well as 20 µg of HeLa protein extract (negative control) and 2 µl of pSG hERα66 and pSG hERα46 in vitro translated products in rabbit reticulocyte lysate (positive control) were resolved on a 10% SDS–polyacrylamide gel and then subjected to immunoblotting with the HC20 antibody and a β-actin antibody as a control. Immunoreactive proteins were visualized by ECL (A). hERα66 and hERα46 signals were quantified by densitometry and results were expressed as a percentage of the hERα66 level detected in confluent cells (B). Values correspond to the average ± SD of three independent experiments. (C) MCF7 cells, grown in the conditions as previously described, were transiently transfected with 5 µg of the reporter plasmid (ERE)2-tk-LUC. Two days later, cells were assayed for luciferase activity. The luciferase activities were normalized using the internal reference control EF-1α–CAT. Values correspond to the average ± SD of three separate transfection experiments.
Discussion
A novel isoform of ER-α has been identified and characterized in this study. This receptor is referred to as hERα46 to distinguish it from the first characterized ER-α protein, hERα66 (Green et al., 1986).
In humans, ERα66 is the translation product of at least six ER-α transcripts (A–F hER-α mRNA variants) that differ in their 5′ UTRs as a consequence of alternative splicing of several upstream exons (1B–F) to a common site 70 nucleotides upstream of the translation initiation codon in exon 1A (Flouriot et al., 1998). Unlike hERα66, hERα46 is encoded by a new class of hER-α transcripts that result from direct splicing to exon 2 of the hER-α gene. As demonstrated by RT–PCR and S1 nuclease mapping experiments, these Δ1A hER-α transcripts originate from the E and F hER-α promoters. The resulting mRNA transcripts are identical from exon 2–8 to the previously described mRNAs that generate hERα66 (Green et al., 1986). The E–F Δ1A hER-α mRNA sequence has an ORF starting at position +752/4 (methionine 174) [numbering from hERα66 mRNA A (Green et al., 1986; Flouriot et al., 1998)] in exon 2 that is in-frame with the hERα66 ORF. The protein encoded by this transcript has a predicted size of 46 kDa and is devoid of the first 173 amino acids of 66 kDa hER-α. This ER-α isoform was shown to be present in several different tissues. In vitro translation of mRNAs encoding hERα66 generated, in addition to hERα66, a low level of hERα46. This indicates that truncated hER-α forms may also be translated from A–F hER-α mRNAs as a consequence of leaky ribosome scanning. Previous investigations with bicistronic vectors in transient transfection experiments indicated that translational initiation at ATG codon 174 of hER-α cDNA may occur by internal ribosome entry (Barraille et al., 1999).
Analysis of the pattern of expression of A–F and E–F Δ1A hER-α transcripts revealed that their relative levels vary in the different human tissues and cell lines evaluated. A and C hER-α transcripts have previously been shown to be the main mRNA variants detected in tissues associated with reproduction, such as mammary gland and endometrium, where the ER-α mRNAs are expressed at a high level (Flouriot et al., 1998). The expression of A and C hER-α mRNA was considerably reduced in non-reproductive tissues (Flouriot et al., 1998), where the mRNAs encoding hERα66 were predominantly due to E or F hER-α promoter activity. The expression level and tissue distribution of E/F Δ1A hER-α transcripts paralleled those of E/F hER-α mRNAs. Both E and F promoters produce a transcript that is spliced to a common exon, known as exon 1E (Flouriot et al., 1998). The data presented in this paper indicate that exon 1E presents a splice donor that has an apparently equal chance of being spliced either to the acceptor site of exon 1A or to exon 2, thereby generating both E/F or E/F Δ1A hER-α mRNAs. Analysis of this splicing process should provide more information about potential mechanisms that may be involved in regulating the ratio of E/F or E/F Δ1A production.
In contrast to tissues that utilize only E and F hER-α promoters and that contain both hER-α isoforms at a similar level, cells expressing mainly A, B and/or C hER-α mRNAs show predominant expression of hERα66. Western blot analysis of the breast carcinoma cell line MCF7 supports this hypothesis, with hERα66 accounting for ∼90% of the total ER-α immunoreactivity in confluent cells and hERα46 accounting for the remainder of this activity. In contrast, analysis of hER-α gene expression in human osteoblast primary cultures has shown that the principal hER-α transcription is from the F promoter. This resulted in the production of a similar amount of both F and F Δ1A hER-α mRNAs. As predicted from the above hypothesis, both hER-α protein isoforms were present at similar levels (S.Denger, G.Flouriot, M.Kos, D.Parsch, G.Reid, H.Brand, V.Sontag-Buck and F.Gannon, submitted).
The hERα46 isoform described here is identical to hERα66 apart from a deletion of the first 173 amino acids. This isoform is therefore devoid of the domain previously mapped as having AF-1 function. However, the transactivation function AF-2, and the DNA and ligand- binding activities are not abolished by the deletion of this region, thereby potentially allowing hERα46 to act as a ligand-inducible transcription factor in some cells in a promoter-specific manner. Indeed, analysis of hERα46 transactivation efficiency demonstrated that, in a cell context mainly sensitive to AF-2, hERα46 effectively induced transcriptional activity in a ligand-dependent manner by interacting with an ERE-derived reporter gene construct. In contrast, this N-terminal truncated form of hER-α was unable to transactivate the same reporter gene constructs in a cellular context, such as in the HepG2 cell line (Tzukerman et al., 1994; Norris et al., 1997) or in yeast (Metzger et al., 1992, 1995; Pham et al., 1992) where AF-1 has been shown to be predominantly involved in the hER-α transactivation mechanism. Moreover, when both hER-α forms are coexpressed (as seems to be the most frequent situation in vivo) hERα46 is a powerful competitor that can efficiently suppress the AF-1 activity of hERα66 in a cell-specific context.
Several studies have reported that receptor deletion mutants devoid of the N-terminal A/B region are characterized by an increased affinity for their corresponding hormone responsive element (Palvimo et al., 1993; Xing et al., 1995). For instance, using a promoter interference assay, Xing et al. (1995) found that the Xenopus (x)ER-α mutant 160/586 exhibited almost a 2-fold increase in affinity for an ERE in comparison with the wild-type xER-α. Therefore, the ability of hERα46 to behave as an effective AF-1 negative competitor may also be due, in part, to its ability to out-compete hERα66 for binding to an ERE. An indication that this may occur was seen in gel mobility shift experiments where the binding of hERα66 homodimer to a constant amount of radiolabeled ERE was first reduced and then eliminated in the presence of increasing quantities of hERα46. It is also possible that hERα46 has different binding affinities for coactivators or corepressors compared with hERα66, and that this could also have a role in the interplay between these two isoforms and their interaction with different promoters. Further investigations are required to determine the relative contribution of these different mechanisms to the AF-1 dominant-negative action of hERα46.
The existence and the potential activities of hERα46, a protein that had previously been ignored or considered to be a degradation product when detected in western blots (Abbondanza et al., 1993), suggests that some data in the area of ER-α function should be re-evaluated. For instance, a mutant mouse line, termed αERKO, with an insertional disruption of the ER-α gene has been created and assessed for estrogen responsiveness (Lubahn et al., 1993). This disruption proved not to be lethal, but rather αERKO mice were found to develop normally and demonstrate no gross external phenotype, except for complete infertility (for review see Couse and Korach, 1999). As the disruptive insertion was performed in the first coding exon of the mouse (m)ER-α gene (the exon that is skipped in the generation of the transcripts encoding ERα46 in human), it is possible that the production of any mouse equivalent of ER-α46 is not affected by the disruption of the ER-α gene. In this regard, residual [3H]E2 binding with high affinity (Kd of 0.2 nM) was detected in some tissues from ER-α knock-out mice, representing ∼3–10% of the levels measured in the wild type (Couse et al., 1995). Sucrose gradient analysis indicated that this residual [3H]E2 binding was probably ER-α specific since the H222 antibody, which recognizes ER-α but not ER-β, was able to shift the E2 binding peak observed in αERKO extracts (Couse et al., 1995). The authors attributed this to a splicing variant generated by the insertion of the disruptive sequence and resulting in the production of a smaller mutant ER-α protein that could be the source of residual E2 binding (Couse et al., 1995). Recent experiments performed in our laboratory demonstrated that the mouse ER-α gene generates transcripts equivalent to the human E–F Δ1A ER-α mRNAs, characterized by a deletion of the first coding exon and containing the ER-α46 ORF (Kos et al., 2000). Therefore, residual [3H]E2 binding in some αERKO mouse tissues may be explained retrospectively by the production of ER-α46, with the αERKO more correctly viewed as being an αER66 knock-out.
Given the potential of hER-α46 to modulate hERα66 action, it is interesting that the hERα66/46 ratios change with the cell growth status in the breast carcinoma cell line MCF7 (Figure 8A and B). Using an approach where the growth rate of MCF7 cells is modified by plating the cells either at low density (20% confluence) in normal calf serum (rapidly proliferating cells), or at high density until confluence (slowly to non-proliferating cells) (Jakesz et al., 1984), we observed that the level of hERα66 was 3- to 4-fold lower in rapidly dividing cells compared with slowly to non-proliferating cells. This result was confirmed using both an exogenous reporter, which was transiently transfected into MCF7 (Figure 8C), and the endogenous ER-inducible marker creatine kinase (data not shown). Likewise, estradiol, which has a mitotic effect on MCF7 cells (Dickson and Lippman, 1988), was shown to downregulate the level of hERα66 in non-confluent cells, confirming previous studies (Saceda et al., 1988; Read et al., 1989). Corroborating these observations, it has been reported that there is a correlation between ER-α expression and the different phases of the cell cycle. ER-α is predominantly expressed in the G1 phase (Jakesz et al., 1984; Dong et al., 1991). Furthermore, recent studies investigating the proliferative status of ER-α-positive and ER-α-negative cells in normal human breast, by in situ immunohistochemical staining for ER-α and proliferation markers, demonstrated that it is primarily ER-α-negative cells and not ER-α-positive cells that proliferate (Clarke et al., 1997; Russo et al., 1999). Finally, it is known that tumor cells with an ER-α phenotype are more differentiated and have lower metastatic potential than ER-α-negative tumors (McGuire, 1986). In light of these data, it is conceivable that in estrogen-sensitive breast carcinoma cells the presence of high levels of hER66α is able to prevent proliferation. Conversely therefore, cell growth may require a low hER-α 66/46 ratio, which is obtained through a reduction of hERα66 expression or stability. In support of this hypothesis, several studies report that estradiol treatment results in growth inhibition of ER-negative cell lines that had been stably transfected with the ER-α cDNA and that express high amounts of hERα66; this contrasts with the fact that E2 stimulates proliferation in ER-α-positive breast carcinomas (for review see Levenson and Jordan, 1994). Taken in isolation, these results are surprising given the accepted mitogenic role of estradiol and its receptor. They may simply reflect differences in biological systems. However, as described above, the actions of hERα46 may also provide an explanation for this apparent paradox. As hERα46 is shown here to be an effective inhibitor of hER-α AF-1 activity, it can also be speculated that the transactivation function AF-1, which was suggested to be the dominant transcriptional activation function of ER-α (Tzukerman et al., 1994; Tremblay et al., 1999), must be reduced or inactivated for cell proliferation to occur. In this regard, the partial ER-α agonist/antagonist OHT has been shown to inhibit cell growth by an ER-dependent mechanism, in addition to its anti-estrogenic effect (Vignon et al., 1987). In a manner similar to estradiol, OHT may repress the growth of cell lines that express high amounts of hERα66 following transfection (Levenson and Jordan, 1994). The inhibition of growth factors and/or the induction of inhibitory growth factors are probably involved in this mechanism (for reviews see Dickson and Lippman, 1988; Parker, 1991). For instance, it was recently reported that estrogens as well as tamoxifen, which has an estrogenic effect on bone resorption, promote TGF-β-mediated apoptosis of murine osteoclasts (Hughes et al., 1996). Since the mixed agonist/antagonist effect of the tamoxifen has been explained by its ability to activate the AF-1 function of ER-α but not AF-2 (Berry et al., 1990), these data may suggest that growth inhibition is mediated by the AF-1 function of ER-α. Further studies would be informative to identify the exact function of both ER-α protein isoforms in the control of estrogen target cell proliferation. In all of these clinically important situations, the potential role of hERα46 must be integrated into the models that describe the observed functions of estrogen analogs.
In conclusion, the identification of a protein isoform of hER-α, produced by alternative splicing and promoter usage, that is able to modulate ER-α-mediated transactivation reveals a previously unknown mechanism that contributes towards understanding how the pleiotropic effects of estrogen and its analogs are integrated into a wide range of physiological and pathological processes.
Materials and methods
RNA isolation
Total RNA from MCF7 cell line and tissues was extracted with TRIzol (Gibco-BRL) as described by the manufacturer. Total RNAs from human mammary gland, endometrium, liver, skeletal muscle, lung, kidney and adrenal gland were purchased from Clontech. Human pituitary RNA was kindly provided by Professor J.Duval (Université de Rennes, France). Ovarian tissues were provided from patients undergoing ovariectomy (Dr R.Lepin, University of Heidelberg, Germany).
RT–PCR analysis
cDNAs were synthesized from 1 µg of total RNA following reverse transcription with 50 U of expand™ reverse transcriptase (Boehringer Mannheim) under the conditions recommended by the supplier, using either the oligonucleotide primer VI [5′-CTCACAGGACCAGACTCCATAATGGA, from exon 2 (see Figure 2A)] or primer I [5′-TTGGCTAAAGTGGTGCATGATGAGG, from the 3′ UTR (exon 8)] of hER-α mRNAs (see Figure 3A). An aliquot of the reverse transcriptase reaction (2.5 µl) resulting from primer VI was then amplified by exon-specific primers by 30 cycles of PCR amplification. The 5′ primers for A, B, C, D, E and F hER-α cDNA amplification were: A1 (5′-CTCGCGTGTCGGCGGGACAT), B1 (5′-CTGGCCGTGAAACTCAGCCT), C1 (5′-TCTCTCGGCCCTTGACTTC), D1 (5′-CACATTCAACGGAGGAGCCA), E1 (5′-AGCCTCAAATATCTCCAAAATCT) and F1 (5′-TTCTATAGCATAAGAAGACAG), respectively (see Figure 2A). The 3′ primer VII (5′-AGCATAGTCATTGCACACTGC) was from exon 2, immediately upstream of the primer used for reverse transcription. cDNAs reverse transcribed from primer I were amplified by PCR in two rounds of 30 cycles using the 5′ primer E/F1 (5′-AAGGAGTAAGCACAAAGATCTC) and the nested primer E/F2 (5′-CAGCACTTCTTCAAAAAGGATGTAGA) with the 3′ primer II (5′-ATTATCTGAACCGTGTGGGAG) and the nested primer III (5′-CTCTCAGACTGAGGCAGGGAAACC), which were located in exon 8 (see Figure 3A). Both rounds of amplification were performed using the expand™ long template PCR system (Boehringer Mannheim) as recommended by the manufacturer. Samples (5 µl) from each reaction were analyzed on 1% agarose gels and transferred to nylon membranes (Hybond N+, Amersham) with 20× SSC as transfer solution. The membranes were incubated in a pre-hybridization buffer containing 6× SSC, 5× Denhart’s solution, 0.05% sodium pyrophosphate, 100 µg/ml salmon sperm DNA and 0.5% SDS, at 37°C for 1 h. The membranes were then hybridized in 6× SSC, 1× Denhart’s solution, 0.05% sodium pyrophosphate and 100 µg/ml yeast tRNA with the oligonucleotide probe P1 (5′-TCTGACCGTAGACCTGCG) (from exon 1A) or P2 (5′-CCCTGGCGTCGATTATCTGAAT) (from exon 2, see Figure 2A), which had been end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mmol).
Modified S1 nuclease mapping
Biotinylated single-stranded DNA templates were used to prepare highly labeled single-stranded DNA probes by extension from a specific primer with T7 DNA polymerase in the presence of [α-32P]dCTP (3000 Ci/mmol) (Flouriot et al., 1996). The origin of probe X (see Figure 1A) template was a PCR product obtained by amplification from pHEO (pSG5 expression vector containing hER-α form I cDNA) (Green et al., 1988) using the upstream 5′ biotinylated primer X1 (5′-CCTACTACCTGGAGAACGAG, located in exon 1A) with the downstream primer III located in exon 8. In order to prepare the template used to make probes E and F (see Figure 3D), RT–PCR reactions were performed with the 5′ primer E1 or F1 and the common 3′ primer VIII (5′-CTGGCCGTGGGGCTGCAGGAAA, located in exon 1A). The RT–PCR products were subcloned downstream of T7 in the TA cloning vector pCR2.1 (Invitrogen) giving rise to pCR-E and pCRM-F, respectively. Then, PCR was performed using a biotinylated T7 primer with primer VIII. Finally, probe F Δ1A and E Δ1A (see Figure 3B and C) templates were prepared by PCR using the biotinylated T7 primer with primer IV (5′-GAACCGAGATGATGTAGCCAG, located in exon 6) and, for each reaction, two partially overlapping templates in order to link directly exon 1E/F sequences to exon 2. The partially overlapping templates were obtained from the TA cloning vector pCRTM-E or pCRTM-F (see above) and an RT–PCR product obtained utilizing the upstream primer E2 (5′-TCTGAACTTTGAACCATCACTGAGGCCAAATTCAGATAATCGACGCCA) with the downstream primer III.
All biotinylated PCR products were bound to streptavidin-coated magnetic beads (Dynal) as recommended by the manufacturer, and the non-biotinylated DNA strands were removed by denaturation with 0.1 M NaOH. X, F, F Δ1A and E Δ1A S1 single-stranded DNA probes were obtained by extending the respective IV (in exon 6), IX (5′-TCTGACCGTAGACCTGCG, in exon 1A), V (5′-CCAACAAGGCACTGACCATC, in exon 4) and VI (in exon 2) primers annealed to the corresponding biotinylated single-stranded template. After elution of the single-stranded DNA probes by alkaline treatment and magnetic separation, the probe was purified on a sequencing gel. The probe (105 c.p.m.) was coprecipitated with 30 µg of total RNA and then dissolved in 20 µl of hybridization buffer (80% formamide, 40 mM PIPES pH 6.4, 400 mM NaCl, 1 mM EDTA pH 8), denatured at 70°C for 10 min and hybridized overnight at 55°C. S1 digestions were then carried out as previously described (Ausubel et al., 1989) and the samples electrophoresed through denaturing polyacrylamide/urea gels. The relative amounts of mRNAs encoding hERα66 and hERα46 were determined from the densitometric scanning of the protected fragments obtained after the S1 nuclease mapping analysis. This was possible due to the fact that F Δ1A and E Δ1A probes were able to measure the total expression of hER-α mRNAs [F Δ1A hER mRNA + E Δ1A hER mRNA + (Σ – E/F Δ1A) hER mRNA].
Expression vectors
Expression vectors pSG hERα46 and pYE hERα46 were prepared by cloning the hER-α coding region from +727 to +2030 into the BamHI site of the parental expression vectors pSG5 (Green et al., 1988) and pYEpucG (Wrenn and Katzenellenbogen, 1993). This region was previously amplified using primers designed to introduce a BamHI restriction site at the 5′ and 3′ ends of the PCR product. pSG hER-α form I (HEO) (Green et al., 1988), pEMBL (Banroques et al., 1986), pYEpucG and pYE hER-α form I (pYEpER) (Wrenn and Katzenellenbogen, 1993) were gifts from P.Chambon, J.H.Camonis and B.S.Katzenellenbogen, respectively. pYE hER-α A–D was constructed as previously described (Petit et al., 1999). pEM hERα66 was made by inserting the hER-α66 ORF BamHI restricted fragment from pYE hERα66 into the BglII site of pEMBL.
In vitro transcription and translation
In vitro transcription and translation was accomplished with the TNT-coupled reticulocyte lysate system from Promega Biotech (Madison, WI) following the manufacturer’s directions. pSG5 recombinant expression vectors, pSG ER-α66 and pSG ER-α46, were used as templates for transcription with T7 RNA polymerase followed by translation to generate hERα66 and hERα46 proteins. Translation efficiency was checked by incorporating [35S]methionine. Cold methionine was used in the in vitro transcription and translation of proteins for electromobility shift assays and for western blot analysis.
Whole cell extracts
Whole cell extracts from MCF7, MDA-MB-231 and HeLa cell lines were prepared using RIPA-Lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) as previously described (Harlow and Lane, 1988). Protein concentrations were determined using the Bradford protein assay obtained from Bio-Rad (Richmond, CA).
Western blot analysis
Twenty micrograms of whole cell extracts and 2 µl of in vitro transcription and translation mix were subjected to SDS–PAGE. Proteins were denatured at 95°C for 15 min and resolved on a 10% SDS–polyacrylamide gel next to pre-stained Rainbow marker (Amersham Pharmacia, Freiburg, Germany) and electrotransferred onto Immobilon membrane (Millipore, Bedford, MA). The membrane was blocked in phosphate-buffered saline (PBS) containing 0.05% Tween and 6% (wt/vol) non-fat dry milk powder. The membrane was then incubated with different primary anti-ER-α antibodies—the monoclonal antibodies H222 or H226 (0.5 µg/ml) kindly provided by Dr G.L.Greene (Greene et al., 1984) and the polyclonal antibody HC20 (Fa. Santa Cruz) (0.3 µg/ml)—in PBS containing 0.05% Tween and 3% non-fat milk powder for 1.5 h at room temperature (RT). Incubation with peroxidase-coupled goat anti-rat (for H222 and H226) or anti-rabbit (for HC20) antibodies was then performed. ER-α proteins were visualized by chemiluminesence using the ECL system from Amersham according to the manufacturer’s instructions. Signals were quantified by densitometry.
Electrophoretic mobility shift assay
ER-α proteins were prepared by in vitro transcription and translation as described above. In vitro translated product (4 µl) was pre-incubated in GSA buffer [10 mM Tris–HCl pH 7.5, 1 mM dithiothreitol, 100 mM KCl, 10% glycerol, 100 µg/ml bovine serum albumin, 5 µg/ml of each protease inhibitor (aprotinin, leupeptin and pepstatin A) and 1 mM PMSF] with 1 µg of poly(dI/dC) for 15 min at RT. The samples were then incubated for 15 min at RT with 1 ng of radioactive oligonucleotide probe (6 × 104 c.p.m.) end-labeled with [γ-32P] ATP (3000 Ci/mM) using T4 polynucleotide kinase (Roche). Protein–DNA complexes were separated from free probe by non-denaturing electrophoresis on 5% polyacrylamide gels in 0.5× TBE. The gels were pre-run at 4°C for 30 min followed by 2–3 h running at 200 V. After electrophoresis, the gels were dried and exposed to Kodak Biomax film. The sequence of the consensus ERE 30 base pair oligonucleotide was derived from the 5′ flanking region of chicken apoVLDL II gene (–186 to –156) (Van Het Schip et al., 1983). The nucleotide sequence was 5′-ctgtgctcaGGTCAgacTGACCttccatta-3′ with the wild-type consensus ERE sequence shown in capitals. The sequence of a mutant version of this oligonucleotide (m, mismatches underlined) was 5′-ctgtgctcaGGACAgacTGTACttccatta-3′. Both oligonucleotides were used as double-stranded DNA for the electrophoretic mobility shift assay. In competition assays, extracts were incubated with a 10-fold molar excess of unlabeled double-stranded oligomer during the pre-incubation step.
Human cell transfections
The MCF7, HepG2 and HeLa cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (Gibco-BRL), penicillin (100 U/ml) and streptomycin (100 µg/ml) at 37°C in a 5% CO2 incubator. One day prior to transfection, the medium was replaced with phenolred-free DMEM containing 2.5% charcoal-stripped calf serum. Cells were then transiently transfected using the DNA/calcium phosphate coprecipitation method (Graham and Van der Eb, 1973). Briefly, 6 cm dishes were seeded with 0.5 × 105 cells, propagated for 4 days and were then transfected with a total of 10 µg of DNA per dish [5 µg of reporter plasmid (ERE)2-tk-LUC (Paech et al., 1997), 0.5 µg of expression vector, 0.25 µg of internal control (EF-1α–CAT) (Mizushima and Naguta, 1990), and carrier DNA to 10 µg (pBluescript)]. Medium was changed 6 h before transfection. After 16 h incubation with the DNA/calcium phosphate precipitate, the medium was aspirated and cells washed twice with PBS, and fresh serum-stripped phenolred-free medium was added. Transfected cells were cultured for 48 h in the absence or presence of 10–8 M 17β-estradiol before harvesting for luciferase and CAT assays. Luciferase assays were performed on 2% of the lysate, as described by Brasier and Ron (1992). CAT activity was determined with the ELISA kit from Boehringer Mannheim using 2% of the lysate. Reporter gene activity results were normalized for transfection efficiency according to the activity of the co-transfected reference control (EF-1α–CAT).
Yeast cell transformations
The yeast strain BJ2168 (Yeast Genetic Stock Center, Berkeley, CA) was used in this study. Yeast cells were transformed using a lithium acetate method (Ausubel et al., 1989) and BJ2168 transformants were selected by growth on complete minimal medium [0.13% dropout powder lacking uracil and tryptophan, 0.67% yeast nitrogen base, 0.5% (NH4)2SO4 and 1% dextrose]. Liquid assays for LacZ activity were performed as described previously (Petit et al., 1999) in the presence of either ethanol carrier alone, estradiol (10–6 M), OHT (10–5 M) or ICI 164,384 (10–5 M). β-galactosidase activity was measured using o-nitrophenyl β-d-galactopyranoside substrate and quantified at 420 nm with a spectrophotometer. The activity was expressed in Miller units (Miller, 1972).
Acknowledgments
Acknowledgements
We thank G.L.Greene for the gift of H226 and H222 antibodies; P.Webb for the reporter gene (ERE)2-tk-LUC; P.Chambon for the expression vectors pSG 5 and HEO; J.H.Camonis for pEMBL vector; and B.S.Katzenellenbogen for the gift of YEpucG and YEpER vectors. This work was supported by Dompe S.P.A. (S.D.), an EMBO long-term fellowship (G.F.) and the Ligue contre le Cancer. The work forms part of the European network program GENOSPORA (QLK6-1999-02108).
References
- Abbondanza C., De Falco,A., Nigro,V., Medici,N., Armetta,I., Molinari,A.M., Moncharmont,B. and Puca,G.A. (1993) Characterization and epitope mapping of a new panel of monoclonal antibodies to estradiol receptor. Steroids, 58, 4–12. [DOI] [PubMed] [Google Scholar]
- Auchus R.J. and Fuqua,S.A.W. (1994) The oestrogen receptor. In Bailliere’s Clinical Endocrinology and Metabolism: Hormones, Enzymes and Receptors, Vol. 8. Baillière Tindall, London, UK, pp. 433–449. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. Wiley-Interscience, New York, NY. [Google Scholar]
- Banroques J., Delahodde,A. and Jacq,C. (1986) A mitochondrial RNA maturase gene transferred to the yeast nucleus can control mitochondrial mRNA splicing. Cell, 46, 837–844. [DOI] [PubMed] [Google Scholar]
- Barraille P., Chinestra,P., Bayard,F. and Faye,J.C. (1999) Alternative initiation of translation accounts for a 67/45 kDa dimorphism of the human estrogen receptor ERα. Biochem. Biophys. Res. Commun., 257, 84–88. [DOI] [PubMed] [Google Scholar]
- Beato M. (1989) Gene regulation by steroid hormones. Cell, 56, 335–344. [DOI] [PubMed] [Google Scholar]
- Berry M., Metzger,D. and Chambon,P. (1990) Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J., 9, 2811–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier A.R. and Ron,D. (1992) Luciferase reporter assay in mammalian cells. Methods Enzymol., 216, 386–397. [DOI] [PubMed] [Google Scholar]
- Clarke R.B., Howell,A., Potten,C.S. and Anderson,E. (1997) Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res., 57, 4987–4991. [PubMed] [Google Scholar]
- Couse J.F. and Korach,K.S. (1999) Estrogen receptor null mice: what have we learned and where will they lead us? Endocrinol. Rev., 20, 358–417. [DOI] [PubMed] [Google Scholar]
- Couse J.F., Curtis,S.W., Washburn,T.F., Lindzey,J., Golding,T.S., Lubahn,D.B., Smithies,O. and Korach,K.S. (1995) Analysis of transcription and estrogen insensitivity in the female mouse after targeted disruption of the estrogen receptor gene. Mol. Endocrinol., 9, 1441–1454. [DOI] [PubMed] [Google Scholar]
- Dickson R. and Lippman,M. (1988) Control of human breast cancer by estrogen, growth factors and oncogenes. In Lippman,M. and Dickson,R. (eds), Breast Cancer: Cellular and Molecular Biology. Kluwer, Boston, MA, pp. 119–165. [DOI] [PubMed] [Google Scholar]
- Dong X.F., Berthois,Y., Colomb,E. and Martin,P.M. (1991) Cell cycle phase dependence of estrogen and epidermal growth factor (EGF) receptor expression in MCF-7 cells: implications in antiestrogen and EGF cell responsiveness. Endocrinology, 129, 2719–2728. [DOI] [PubMed] [Google Scholar]
- Evans R.M. (1988) The steroid and thyroid hormone receptor superfamily. Science, 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G., Nestor,P., Kennealy,M.R., Pope,C. and Gannon,F. (1996) An S1 nuclease mapping method for detection of low abundance transcripts. Anal. Biochem., 237, 159–161. [DOI] [PubMed] [Google Scholar]
- Flouriot G., Griffin,C., Kennealy,M.R., Sonntag-Buck,V. and Gannon,F. (1998) Differentially expressed messenger RNA isoforms of the human estrogen receptor-α gene are generated by alternative splicing and promoter usage. Mol. Endocrinol., 12, 1939–1954. [DOI] [PubMed] [Google Scholar]
- Gaub M.P., Bellard,M., Scheuer,I., Chambon,P. and Sassone-Corsi,P. (1990) Activation of the ovalbumin gene by the estrogen receptor involves the fos–jun complex. Cell, 63, 1267–1276. [DOI] [PubMed] [Google Scholar]
- George F.W. and Wilson,J.D. (1988) Sex determination and sex differentiation. In Knobil,E., Neil,J.D., Ewing,L.L., Greenwald,G.S., Market,C.L. and Pfaff,D.W. (eds), The Physiology of Reproduction. Raven Press, New York, NY, pp. 3–26. [Google Scholar]
- Graham F.L. and Van Der Eb,A.J. (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology, 52, 456–467. [DOI] [PubMed] [Google Scholar]
- Green S., Walter,P., Kumar,V., Krust,A., Bornert,J.M., Argos,P. and Chambon,P. (1986) Human estrogen receptor cDNA: sequence, expression and homology to v-erbA. Nature, 320, 134–139. [DOI] [PubMed] [Google Scholar]
- Green S., Issemann,I. and Sheer,E. (1988) A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res., 16, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene G.L., Sobel,N.B., King,W.J. and Jensen,E.V. (1984) Immunochemical studies of estrogen receptors. J. Steroid Biochem., 20, 51–56. [DOI] [PubMed] [Google Scholar]
- Griffin C., Flouriot,G., Sonntag-Buck,V., Nestor,P. and Gannon,F. (1998) Identification of novel chicken estrogen receptor-α messenger ribonucleic acid isoforms generated by alternative splicing and promoter usage. Endocrinology, 139, 4614–4625. [DOI] [PubMed] [Google Scholar]
- Griffin C., Flouriot,G., Sonntag-Buck,V. and Gannon,F. (1999) Two functionally different protein isoforms are produced from the chicken estrogen receptor-α gene. Mol. Endocrinol., 13, 1571–1587. [DOI] [PubMed] [Google Scholar]
- Guarente L. and Masson,T. (1983) Heme regulates transcription of the CYC1 gene of S.cerevisiae via an upstream activation site. Cell, 32, 1279–1286. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies, A Laboratory Manual, 1st edn. Cold Spring Harbor Laboratory Press, Cold Sping Harbor, NY, p. 447. [Google Scholar]
- Henderson B.E., Ross,R. and Bernstein,L. (1988) Estrogens as a cause of human cancer: the Richard and Hinda Rosenthal Foundation award lecture. Cancer Res., 48, 246–253. [PubMed] [Google Scholar]
- Hughes D.E., Dai,A., Tiffee,J.C., Li,H.H., Mundy,G.R. and Boyce,B.F. (1996) Estrogen promotes apoptosis of murine osteoclasts mediated by TGF-β. Nature Med., 2, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Jakesz R., Smith,C.A., Aitken,S., Huff,K., Schuette,W., Shackney,S. and Lippman,M. (1984) Influence of cell proliferation and cell cycle phase on expression of estrogen receptor in MCF-7 breast cancer cells. Cancer Res., 44, 619–625. [PubMed] [Google Scholar]
- Kastner P., Krust,A., Mendelsohn,C., Garnier,J.M., Zelent,A., Leroy,P., Staub,A. and Chambon,P. (1990) Murine isoforms of retinoic acid receptor γ with specific patterns of expression. Proc. Natl Acad. Sci. USA, 87, 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos M., O'brien,S., Flouriot,G. and Gannon, F. (2000) Tissue-specific expression of multiple mRNA variants of the mouse estrogen receptor α gene. FEBS Lett., 477, 15–20. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1989) The scanning model for translation: an update. J. Cell Biol., 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper G.J.M., Enmark,E., Pelto-Huikko,M., Nilsson,S. and Gustafsson,J.A. (1996) Cloning of a novel estrogen receptor expressed in rat prostate and ovary. Proc. Natl Acad. Sci. USA, 93, 5925–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy P., Krust,A., Zelent,A., Mendelsohn,C., Garnier,J.M., Kastner,P., Dierich,A. and Chambon,P. (1991) Multiple isoforms of the mouse retinoic acid receptor α are generated by alternative splicing and differential induction by retinoic acid. EMBO J., 10, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson A.S. and Jordan,V.C. (1994) Transfection of human estrogen receptor (ER) cDNA into ER-negative mammalian cell lines. J. Steroid Biochem. Mol. Biol., 51, 229–239. [DOI] [PubMed] [Google Scholar]
- Lubahn D.B., Moyer,J.S., Golding,T.S., Couse,J.F., Korach,K.S. and Smithies,O. (1993) Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc. Natl Acad. Sci. USA, 90, 11162–11166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire W.L. (1986) Prognostic factors in primary breast cancer. Cancer Surv., 5, 527–536. [PubMed] [Google Scholar]
- Metzger D., Losson,R., Bornert,J.M., Lemoine,Y. and Chambon,P. (1992) Promoter specificity of the two transcriptional activation functions of the human oestrogen receptor in yeast. Nucleic Acids Res., 20, 2813–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D., Ali,S., Bornert,T.J.C. and Chambon,P. (1995) Characterization of the amino-terminal transcriptional activation function of the human estrogen receptor in animal and yeast cells. J. Biol. Chem., 270, 9535–9542. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Mizushima S. and Naguta,S. (1990) pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res., 18, 5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosselman S., Polman,J. and Dijkema,R. (1996) ERβ: identification and characterization of a novel human receptor. FEBS Lett., 392, 49–53. [DOI] [PubMed] [Google Scholar]
- Norman A.W. and Litwack,G. (1987) Estrogens and progestins. In Litwack,G. (ed.), Hormones. Academic Press, London, UK, pp. 550–560. [Google Scholar]
- Norris J.D., Fan,D., Kerner,S.A. and McDonnell,D.P. (1997) Identification of a third autonomous activation domain within the human estrogen receptor. Mol. Endocrinol., 11, 747–754. [DOI] [PubMed] [Google Scholar]
- Paech K., Webb,P., Kuiper,G.G., Nilsson,S., Gustafsson,J.A., Kushner,P.J. and Scanlan,T.S. (1997) Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science, 277, 1508–1510. [DOI] [PubMed] [Google Scholar]
- Palvimo J.J., Kallio,P.J., Ikonen,T., Mehto,M. and Janne,O.A. (1993) Dominant-negative regulation of trans-activation by the rat androgen receptor: roles of the N-terminal domain and heterodimer formation. Mol. Endocrinol., 7, 1399–1407. [DOI] [PubMed] [Google Scholar]
- Parker M.G. (1991) Nuclear Hormone Receptors. Molecular Mechanisms, Cellular Functions, Clinical Abnormalities. Academic Press, London, UK. [Google Scholar]
- Petit F.G., Metivier,R., Valotaire,Y. and Pakdel,F. (1999) Synergism between a half-site and an imperfect estrogen-responsive element and cooperation with COUP-TFI are required for estrogen receptor (ER) to achieve a maximal estrogen-stimulation of rainbow trout ER gene. Eur. J. Biochem., 259, 385–395. [DOI] [PubMed] [Google Scholar]
- Pham T.A., Hwung,Y.P., Santiso-Mere,D., McDonnell,D.P. and O’Malley,B.W. (1992) Ligand-dependent and -independent function of the transactivation regions of the human estrogen receptor in yeast. Mol. Endocrinol., 6, 1043–1050. [DOI] [PubMed] [Google Scholar]
- Read L.D., Greene,G.L. and Katzenellenbogen,B.S. (1989) Regulation of estrogen receptor messenger ribonucleic acid and protein levels in human breast cancer cell lines by sex steroid hormones, their antagonists and growth factors. Mol. Endocrinol., 3, 295–304. [DOI] [PubMed] [Google Scholar]
- Russo J., Ao,X., Grill,C. and Russo,I.H. (1999) Pattern of distribution of cells positive for estrogen receptor α and progesterone receptor in relation to proliferating cells in the mammary gland. Breast Cancer Res. Treat., 53, 217–227. [DOI] [PubMed] [Google Scholar]
- Saceda M., Lippman,M.E., Chambon,P., Lindsey,R.L., Ponglikitmongkol,M., Puente,M. and Martin,M.B. (1988) Regulation of the estrogen receptor in MCF-7 cells by estradiol. Mol. Endocrinol., 2, 1157–1162. [DOI] [PubMed] [Google Scholar]
- Shi Y.B., Yaoita,Y. and Brown,D.D. (1992) Genomic organization and alternative promoter usage of two thyroid hormone receptor genes in Xenopus laevis. J. Biol. Chem., 267, 733–738. [PubMed] [Google Scholar]
- Spatz M., Waisman,A. and Kaye,A.M. (1992) Responsiveness of the 5′-flanking region of the brain type isozyme of creatine kinase to estrogens and antiestrogen. J. Steroid Biochem. Mol. Biol., 41, 711–714. [DOI] [PubMed] [Google Scholar]
- Topper J. and Freedman,C. (1980) Multiple hormone interactions in the developmental biology of the mammary gland. Physiol. Rev., 60, 1049–1060. [DOI] [PubMed] [Google Scholar]
- Tremblay G.B., Tremblay,A., Labrie,F. and Giguere,V. (1999) Dominant activity of activation function 1 (AF-1) and differential stoichiometric requirements for AF-1 and -2 in the estrogen receptor α–β heterodimeric complex. Mol. Cell. Biol., 19, 1919–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzukerman M.T., Esty,A., Santiso-Mere,D., Danielian,P., Parker,M.G., Stein,R.B., Pike,W.J. and McDonnell,D.P. (1994) Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol. Endocrinol., 8, 21–30. [DOI] [PubMed] [Google Scholar]
- Van Het Schip A.D., Meijlink,F.C., Strijker,R., Gruber,M., Van Vliet, A.J., Van De Klundert,J.A. and Ab,G. (1983) The nucleotide sequence of the chicken apo very low density lipoprotein II gene. Nucleic Acids Res., 11, 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignon F., Bouton,M.M. and Rochefort,H. (1987) Anti-estrogens inhibit the mitogenic effect of growth factors on breast cancer cells in the total absence of estrogens. Biochem. Biophys. Res. Commun., 146, 1502–1508. [DOI] [PubMed] [Google Scholar]
- Wrenn C.K. and Katzenellenbogen,B.S. (1993) Structure–function analysis of the hormone-binding domain of the human estrogen receptor by region-specific mutagenesis and phenotypic screening in yeast. J. Biol. Chem., 268, 24089–24098. [PubMed] [Google Scholar]
- Xing H., Mattick,S., Lew,D. and Shapiro,D.J. (1995) An N-terminal deletion mutant of estrogen receptor exhibits increased synergism with upstream activators and enhanced binding to the estrogen response element. Biochemistry, 34, 3956–3963. [DOI] [PubMed] [Google Scholar]
- Zelent A., Mendelsohn,C., Kastner,P., Krust,A., Garnier,J.M., Ruffenach,F., Leroy,P. and Chambon,P. (1991) Differentially expressed isoforms of mouse retinoic acid receptor β are generated by usage of two promoters and alternative splicing. EMBO J., 10, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]