Abstract
Sudan is a highly endemic area for hepatitis B virus (HBV), and >5% of blood donors are chronically infected. To examine potential strategies to improve HBV blood safety, 404 replacement donor samples previously screened for HBV surface antigen (HBsAg) were tested for antibody to HBV core (anti-HBc), anti-surface antigen (anti-HBs), and HBV DNA. Of 145 anti-HBc-containing samples (36%) identified, 16 retested were HBsAg positive (11%). Anti-HBs was detected in 43/77 (56%) anti-HBc-reactive samples. Six samples were HBsAg−/anti-HBc+/anti-HBs+ and contained HBV DNA, meeting the definition of occult HBV infection (OBI). OBIs had low HBV DNA loads (<10 IU/ml) and were genotype B (n = 1) or genotype D (n = 5). Pre-S/S and/or whole genome sequences were obtained from 47 randomly selected HBsAg-positive donors added to the previous 16. Genotype E was identified in 27 strains (57.5%), genotype D in 19 strains (40.5%), and genotype A2 in 1 strain (2%). Two outlier strains within genotype D ultimately were identified as recombinants of genotypes D and E with identical recombination points, suggesting circulating, infectious, recombinant strains. Anti-HBc screening does not appear to be a sustainable blood safety strategy because of the cost and the negative impact on the Sudanese blood supply, even when reduced by anti-HBs testing. Being at the junction between two main African HBV genotypes, genetic recombination occurred and became part of the molecular epidemiology of HBV in Sudan.
Hepatitis B virus (HBV) infection remains a major health problem causing considerable morbidity and mortality despite the availability of vaccine and antiviral treatments. The World Health Organization (WHO) estimates that more than one-third of the world population is or has been in contact with the virus, resulting in >350 million HBV chronic carriers worldwide, with >18% of them living in Africa. Sudan is classified among the African countries with high HBV endemicity. The reported prevalence of HBV chronic infection, characterized by the detectable level of HBV surface antigen (HBsAg), varied from region to region and ranged between 5 and 7% in the general population (1, 13, 28, 31) and 26% in hospital outpatients (25). The prevalence of adults having been in contact with HBV and identified by the presence of anti-core antibodies (anti-HBc) was high, ranging between 47.5 and 67% (25, 28). The introduction of vaccination and the screening of blood and blood products during the past 8 years is expected to reduce the rate of HBV infection and the carrier pool (28).
In sub-Saharan Africa, the risk of HBV transfusion-transmission is expected to be substantial because of the high prevalence of this infection, the frequent use of paid or replacement donors, and incomplete screening coverage (19). It has been estimated that the median overall risk of becoming infected with HBV from a blood transfusion in sub-Saharan Africa was 4.3 per 1,000 U (19). In Sudan, the screening of blood donations for HBV was introduced throughout the country in 2002, before which time screening was performed in only a few centers in Khartoum (28). Under these conditions, it is anticipated that HBV safety for recipients of blood transfusion might remain compromised even after routine blood testing for HBsAg. The residual risk of HBV transfusion transmission is related mainly to blood donations negative for HBsAg that have been collected during the preseroconversion window period (WP), defined as the time between infection and the detection of a viral antigen or antibody marker, during the late stages of infection, or during occult HBV infection or carriage (OBI) (2). Beyond sensitive HBsAg screening, two strategies have been considered to reduce the risk of HBV transfusion-transmission: anti-HBc screening, as implemented in areas in which HBV is endemic at low levels, or nucleic acid testing (NAT) either in pools of 6 to 24 individual donations or in individual donation samples (5, 12, 14, 24, 33). Precise cost-effectiveness studies have not been provided, but the cost of discarded blood units and the impact on the blood supply of countries in chronic shortage of blood are major considerations.
HBV is characterized by a high genetic variability, resulting in the recognition of eight well-established genotypes (A to H) based on >7.5% intergroup divergence in the complete genome and several subgenotypes within these genotypes (22). Subgenotypes are defined by more than 4% intragenotypic divergence but not greater than 7.5%. Recently, a ninth genotype, tentatively termed “I,” was proposed (35). Previous studies conducted in several African countries showed that HBV genotype E is the most prevalent genotype, spreading in western Africa from Senegal to Namibia, subgenotype A1 is dominant in southern and eastern Africa, subgenotype A3 is present in central and west Africa, and genotype D is dominant in northern Africa (20). However, to date no molecular epidemiology study on HBV infection in Sudan has been reported.
To assess the question of blood safety in Sudan, local evidence was collected on the prevalence of anti-HBc- and HBsAg-negative/HBV DNA-positive donations. This exercise was taken as a trigger to further explore the molecular epidemiology of HBV in asymptomatic blood donors from Khartoum.
MATERIALS AND METHODS
Samples.
Plasma samples were obtained from 404 replacement blood donors (403 males and 1 female) from Khartoum, Sudan. Blood donations were collected between February and August 2008 in Federal Hospitals in Khartoum state, Khartoum Teaching Hospital (KTH), and Radiation Isotopes Centre Khartoum (RICK), Federal Ministry of Health. Donors fulfilled the local donation requirements of normal blood pressure, aged between 18 and 50 years, a hemoglobin level of ≥12 g/dl, and the absence of a jaundice record. Plasma was obtained from 5 to 8 ml of citrated venous blood collected directly from the donated blood unit and stored at 4°C until mandatory serological testing for anti-HIV antibodies and p24 antigen (combo p24/anti-HIV-1&2; Enzygnost; Dade Behring, Marburg, Germany), HBsAg (Enzygnost HBsAg; 5.0 EIA), anti-Treponema antibodies (Enzygnost Syphilis), and anti-HCV antibodies (Innotest HCV Ab; Innogenetics, Belgium). Nonreactive samples were stored at −80°C in the Department of Virology, National Health Laboratory, Khartoum, until further tested. Antibodies to HBV core protein (HBc) were tested with MONOLISA anti-HBc plus (Bio-Rad, Marnes-la-Coquette, France) according to the manufacturer's instructions. Anti-HBc-reactive samples also were tested for quantitative antibodies to HBV surface antigen (anti-HBs) (MONOLISA anti-HBs plus; Bio-Rad).
Thirty-three anti-HBc-only-reactive samples, 65 anti-HBc-reactive/anti-HBs-reactive samples, and 50 HBsAg-reactive plasma samples were randomly selected for molecular analyses by the Division of Transfusion Medicine, Department of Haematology, University of Cambridge, United Kingdom.
The study was approved by the KTH general director, the laboratory supervisor, and the blood bank supervisors.
HBV DNA quantification.
Viral DNA was purified from 500 μl of plasma using the high pure viral nucleic acid kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions. HBV DNA was quantified with an in-house real-time quantitative PCR (qPCR) assay as described previously (4). An in-house reference was used and calibrated regularly against the WHO international standard for HBV DNA for nucleic acid testing (NAT) assays 97 and 746 (National Institute for Biological Standard and Controls, Potters Bar, United Kingdom).
Amplification and sequencing of the HBV genome.
The basic core promoter/precore (BCP/PC) region (∼300 bp), the Pre-S/S region (∼1,190 bp), 450 bp in the S region, including the major hydrophilic region (MHR), and the full-length HBV genome minus 50 bp were amplified using nested PCRs (8, 36).
HBV-specific amplified products were purified using the E.Z.N.A cycle-pure kit (Omega Bio-Tek Inc., Norcross, GA). The purified products were sequenced on an Applied Biosystems Prism 3730xl DNA analyzer (Applied Biosystems, Warrington, United Kingdom) as described previously (15, 36).
Cloning of PCR products.
Pre-S/S amplicons from suspected recombinant HBV strains were cloned with a dual-promoter TA cloning kit (Invitrogen Ltd., Paisley, United Kingdom) according to the manufacturer's instructions. For each strain investigated, plasmid DNA from eight clones was purified with a QIAprep spin miniprep kit (Qiagen GmbH, Hilden, Germany) and sequenced.
Phylogenetic analysis.
The sequences of the full-length and BCP/PC regions were assembled with MacVector software version 7.2 and the CLUSTAL W alignment option. Phylogenetic analyses for genotyping and genomic distance calculations were performed using PAUP* version 1.0 b10 (34) and the neighbor-joining algorithm based on Kimura two-parameter distance estimation, ignoring all positions with gaps in pairwise comparisons. To confirm the reliability of phylogenetic tree topologies, bootstrap reconstruction was carried out 1,000 times and bootstrap values of >70% were considered significant. Genetic divergence was defined by the means of all pairwise genomic distances within or between a given (sub)genotype. Potential recombinant sequences were tested with the genotyping function of NCBI and verified with the SIMPLOT version 3.5.1 software (23). Window sizes of 300 and 200 bp and steps of 50 and 20 bp were used.
Nucleotide sequence accession numbers. All complete genome and Pre-S/S sequences were submitted to GenBank under the accession numbers HQ385227 to HQ385272.
RESULTS
HBV serological and molecular markers in HBsAg-negative Sudanese blood donors.
Plasma samples from 404 randomly selected donors initially tested HBsAg negative were investigated further for the presence of anti-HBc, anti-HBs, and HBV DNA. Anti-HBc was detected in 145 samples (36%). Anti-HBs was successfully quantified in 43/77 (56%) randomly selected anti-HBc-reactive samples (median titer, 192 IU/liter; range, 10 to >400 IU/liter).
All anti-HBc-reactive samples were first tested by qPCR, and 31 (21%) were positive, with an HBV DNA load ranging between <10 (18 cases) and 1.1 × 106 IU/ml (median of 99.5 IU/ml when including only quantifiable samples). To confirm these results, each HBV DNA-positive sample was retested serologically for HBsAg using an alternative assay to the initial routine screening assay and for HBV DNA with nested PCR assays. On the basis of results obtained from these additional assays, 16 HBV DNA-positive samples were reclassified HBsAg positive. The presence of HBV DNA was not confirmed by nested PCRs in nine samples that repeatedly tested as low positive by qPCR. However, the presence of HBV DNA in the absence of detectable levels of HBsAg defining occult HBV infection (OBI) was confirmed in 6 of 129 samples (4.6%). All six OBI samples carried anti-HBs and HBV DNA loads of <10 IU/ml (Table 1). Pre-S/S and/or S sequences were obtained, and phylogenetic analysis showed that one (S185) and five (S20, S51, S69, S152, and S206) OBI sequences were of HBV genotype B and D, respectively. The amino acid sequence of the S antigen major hydrophilic region (MHR) was deduced from the S sequences and compared to B and D consensus sequences previously obtained from asymptomatic HBsAg-positive blood donors. Genotype B S185 and genotype D S51 had a wild-type MHR. The four other genotype D OBIs showed 1 to 11 amino acid (aa) substitutions compared to the corresponding consensus sequence (S69, Q101H; S152, P127L and A168T; S206, P127L and E164G; S20, T115N, T118A, G119T, T123V, P127S, Y134H, P142L, S143L, G145R, P151L, and S167L). The intragroup average amino acid diversity within the MHR of genotype D OBIs was 9.8% (range, 2.8 to 19.4%).
TABLE 1.
Serological and virological characteristics of six Sudanese OBIs
| Samples | HBsAg | Anti-HBc | Anti-HBs | HBV DNA load (IU/ml) | HBV genotype |
|---|---|---|---|---|---|
| S20 | − | + | + | <10 | D |
| S51 | − | + | + | <10 | D |
| S69 | − | + | + | <10 | D |
| S152 | − | + | + | <10 | D |
| S185 | − | + | + | <10 | B |
| S206 | − | + | + | <10 | D |
Molecular characterization of HBV strains in HBsAg-positive Sudanese donors.
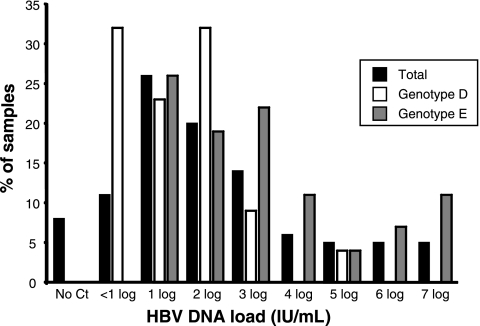
Plasma samples containing HBsAg were obtained from 48 deferred asymptomatic Sudanese blood donors. The 16 HBsAg-positive samples retrospectively identified above were included in the analysis for a total of 64 samples. HBV DNA was quantified in 59 samples (92.2%), and the median viral load was 297 IU/ml (range, <10 to 7 × 107 IU/ml), including samples giving a signal of <10 IU/ml but confirmed to contain at least 1 IU/ml. The majority of samples (75%) had a viral load below 104 IU/ml (Fig. 1). Four samples that were only slightly positive by qPCR (<10 IU/ml) were not confirmed by nested PCR, and five HBsAg-positive samples showed no detectable viral DNA. When the samples were classified according to HBV genotype (see below), the median HBV DNA load of 17 genotype D-infected samples was 103 IU/ml (range, <10 × 105 to 8.5 × 105 IU/ml). Most genotype D samples (95.5%) carried viral loads of less than 104 IU/ml. Viral load could not be reliably quantified (<10 IU/ml) in 32% of samples. The median viral load was significantly higher in 27 genotype E-infected samples (median, 3.1 × 103 IU/ml; range, 19 to 7 × 107 IU/ml; P < 0.0001). A biphasic distribution was observed, with 67 and 18.5% of samples having HBV DNA loads below 104 and above 106 IU/ml, respectively (Fig. 1).
FIG. 1.
HBV viral load distribution of 64 Sudanese HBsAg-positive blood donor samples. Black bars characterize the total population of donors, white bars characterize donors infected with HBV genotype D, and gray bars characterize donors infected with HBV genotype E.
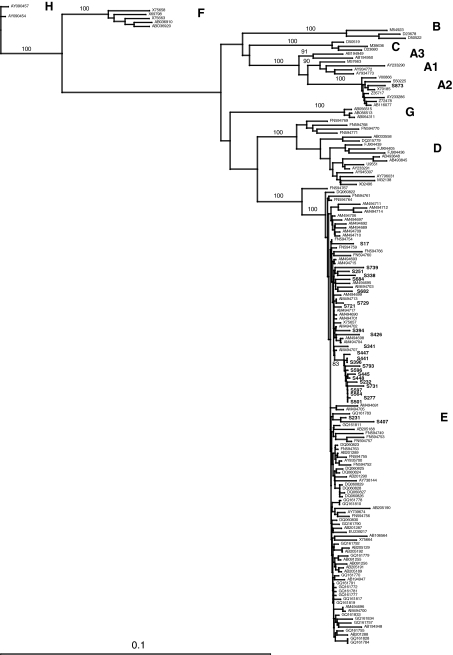
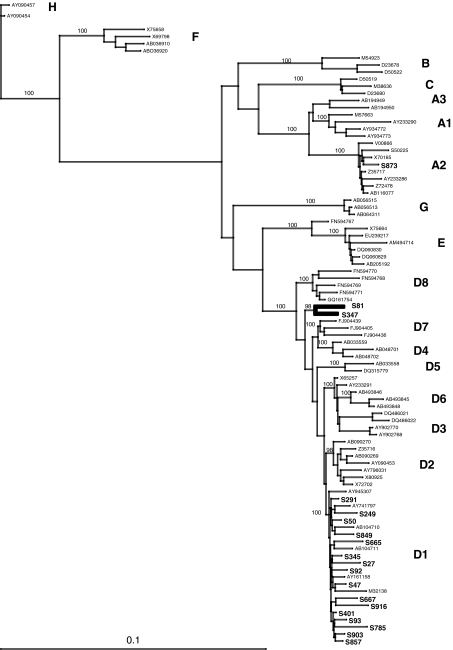
The Pre-S/S region and the whole HBV genome (minus 50 bp in the BCP region) were successfully amplified and sequenced from 47 and 22 samples, respectively. HBV genotyping was performed by phylogenetic analysis using both sets of sequences. Results showed that one sequence (2%) clustered with HBV genotype A2 (HBV/A2), 19 (40.5%) with genotype D (HBV/D), and 27 (57.5%) with genotype E (HBV/E) (Fig. 2).
FIG. 2.
Phylogenetic tree of Sudanese HBV strains based on Pre-S/S sequences. Phylogenetic analysis was performed with the neighbor-joining algorithm based on the Kimura two-parameter distance estimation method. Only bootstrap values of ≥75% are shown (1,000 replicates). Sudanese sequences are indicated in boldface, and HBV reference sequences of genotypes/subgenotypes A1 to A3, B, C, D1 to D8, E, F, G, and H are identified by their GenBank accession numbers (HQ385227 to HQ385272). For a more comprehensive analysis, genotype E sequences (a) and genotype D sequences (b) are shown in two distinct trees. Suspected recombinant strains are indicated by thick bars.
Within the genotype E clade, 13 (48%) Pre-S/S Sudanese sequences formed a distinct subcluster, supported by 83% bootstrapping for 1,000 replicates (Fig. 2a). This distinct branching was confirmed with the full-genome sequences available for four strains (100% bootstrap) (not shown). The average intragroup genetic variability of the Sudanese HBV/E Pre-S/S and full-genome sequences was 1.2% (range, 0 to 3.0%) and 1.4% (range, 0.1 to 2.3%), respectively. Diversity between genotype E Sudanese sequences and 89 genotype E references used for comparison was 1.3% (range, 0.3 to 3.0%) in the Pre-S/S region and 1.9% (range, 0.9 to 3.4%) for the full genome. The average overall amino acid diversity within the Pre-S/S region of 27 HBV/E was 2.1% (range, 0 to 6.8%). In the Pre-S2 region, two strains presented a 4-aa deletion (positions ps136 to ps139), two strains had a 5-aa deletion (positions ps135 to ps139 and ps136 to ps140), and one had a 7-aa deletion (positions ps133 to ps139). One strain showed a 1-aa deletion at the N-terminal region of the S antigen (position 10). The MHR was remarkably conserved (1.4% average diversity; range, 0 to 7%). One strain (S232) showed the substitution M133T, which previously was reported to be associated with vaccine escape. The complete sequence of the core and polymerase genes was obtained for eight and nine genotype E Sudanese strains, respectively. The average amino acid diversity within the core was 3.7% (range, 0 to 11%) and was 1.7% (range, 0.5 to 2.6%) within the polymerase. Complete X gene sequences were available for four strains. Two strains had the xK130M/xV131I and xV131I substitutions, respectively, which have been associated with an increased risk of developing hepatocellular carcinoma.
As shown in Fig. 2b, 17 Sudanese sequences clustered with subgenotype D1 (HBV/D1) within the HBV/D clade (100 and 67% bootstrap with full genome and Pre-S/S, respectively). The average intragroup nucleotide diversity was 2.6% (range, 0.4 to 10.6%) and 2.1% (range, 1.3 to 3.0%) for the Pre-S/S region and full genome, respectively. Diversity between subgenotype D1 Sudanese sequences and 91 previously characterized HBV/D1 sequences from Egypt (n = 29), Tunisia (n = 15), and Iran (n = 47) (16), which were used as references, was 3.1% (range, 1.1 to 11.0%) for the complete genome minus the BCP/PC region. The average amino acid diversity within the Pre-S/S region of Sudanese HBV/D1 strains was 1.9% (range, 0 to 3.8%). All Sudanese strains had a wild-type MHR. Polymerase and core sequences were analyzed for 13 strains, and the average amino acid diversity was 2.4% (range, 0.9 to 4.4%) and 5.9% (1.1 to 11.5%), respectively. In the core, the genotype-specific insertion of aspartic acid (position 152) and arginine (position 153) was present in the genotype A2 strain. Despite the lack of BCP/PC sequence, the sequence of the X gene was obtained for 12 HBV/D1 strains. Five strains presented the xK130M and xV131I substitutions, one had the xV131I substitution alone, and two had the xV131L substitution. The average amino acid diversity was 3.3% (range 0 to 6.5%) in the Sudanese HBV/D1 X protein.
Two Pre-S/S Sudanese sequences (S81 and S347) were outliers that clustered together on a branch distinct from subgenotypes D1 to D8 supported by 82% bootstrap (Fig. 2b). The sequence of the entire genome of strain S81 was obtained, and phylogenetic analysis confirmed the Pre-S/S analysis. The full-length sequence of S81 was compared to previously characterized HBV/D subgenotypes, including the recently proposed HBV/D6-8. As shown in Table 2, the average intersubgenotype nucleotide diversity ranged from 3.9% with HBV/D8 (a recently characterized HBV D/E recombinant form) and 5.9% with HBV/D5. The Pre-S/S genetic diversity between S81 and S347 was 2.3%. In addition, while closely related to HBV D subgenotypes, these two strains did not present the specific 33-nucleotide deletion in the Pre-S1 region carried by reference genotype D strains. Therefore, the presence of genetic recombination within these two HBV strains was suspected.
TABLE 2.
Genetic divergence of Sudanese recombinant HBV-D/E strain S81 compared to described HBV-D subgenotypes
| D subgenotype | N | Divergencea (%) |
|
|---|---|---|---|
| Mean | Range | ||
| D1 | 104 | 5.4 | 4.6-11.0 |
| D2 | 37 | 5.1 | 4.7-5.5 |
| D3 | 29 | 5.4 | 4.6-6.4 |
| D4 | 4 | 4.6 | 4.2-5.5 |
| D5 | 2 | 5.9 | 5.8-6.0 |
| D6 | 3 | 5.2 | 5.0-5.4 |
| D7 | 29 | 4.1 | 3.4-4.8 |
| D8 | 5 | 3.9 | 3.8-4.1 |
Genetic divergence was defined as the mean value for pairwise distance between sequences, calculated as the number of nucleotide differences between two individual sequences, corrected for sequence length.
Characterization of recombination events in two Sudanese HBV strains.
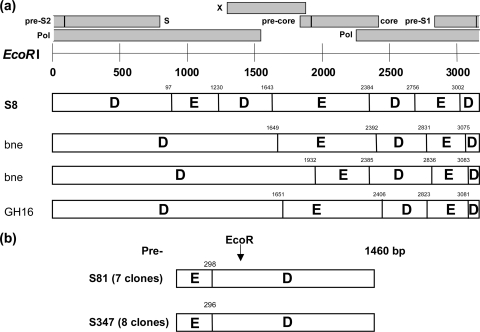
To investigate further the presence of recombination events, the complete HBV sequence of S81 was analyzed with SIMPLOT software. Whole HBV genome amplification was not obtained from sample S347 due to plasma volume limitation and low viral DNA load (3.1 ×102 IU/ml in S347 and 1.3 × 106 IU/ml for S81). As summarized in Fig. 3 a, S81 appeared as a complex D/E recombinant form containing six putative recombination points between genotype D and genotype E. Breakpoints were identified approximately at positions 978, 1230 (pol gene), 1643 (X gene), 2384 (core/pol overlapping region), 2756 (pol gene), and 3000 (Pre-S1/pol overlapping region).
FIG. 3.
Schematic representation of Sudanese HBV D/E recombinant strains. (a) Schematic representation of recombination points in Sudanese strain S81 and in three recombinant HBV-D/E (D8) strains from Niger (bne281 and bne367; GenBank accession numbers FN594769 and FN294770) and Ghana (GH16; GenBank accession number GQ161754), which were included for comparison. Numbers above the bars represent the exact genome positions (EcoRI cut) of the recombination points as indicated by SIMPLOT in an alignment of 3,257 nucleotides that includes all genotype-specific gaps. A representation of the three open reading frames of the HBV genome is shown above the figure. (b) Schematic representation of the clones isolated from the Sudanese S81 and S347 plasma samples in the Pre-S/S region. Numbers indicate the nucleotide positions of recombination as indicated by SIMPLOT.
To confirm further the recombination event, the successfully amplified Pre-S/S region (1460 bp) of S81 and S347 strains that contained the putative recombination point at position 3000 was cloned and sequenced (seven to eight clones per strain). No evidence of dual infection was observed, and all clone sequences belonged to a monophyletic cluster within the genotype D clade that included the corresponding consensus sequence obtained from the direct sequencing of the original PCR product (100 and 96% bootstrap support for S81 and S347 clones, respectively). In both cases, the quasispecies observed consisted of closely related variants. However, sample S81 contained two major variants in similar proportions that differed from the other species by 3- and 9-nucleotide deletions in the Pre-S2 region. The quasispecies genetic diversity was 1.1% (range, 0.6 to 1.2%) and 1.4% (range, 0.4 to 2.4%) in sample S81 (excluding the deletions) and S347, respectively. Despite this intrasample genetic variability, SIMPLOT analysis showed that the recombination point around position 3000 remained present in each clone and at the same location in individual variants constitutive of the quasispecies (Fig. 3b).
DISCUSSION
The first objective of this study was to determine the prevalence of anti-HBc among Sudanese blood donors and to explore the potential benefits of implementing such a screening test in addition to HBsAg in blood donor screening to improve HBV blood safety. Since the prevalence of HBsAg in the initial group of blood donors screened was not known, we could not determine the overall HBsAg prevalence. However, HBsAg prevalence of 5 to 7% was previously reported in the general healthy population from different regions of Sudan (13, 28, 31). A higher prevalence of 26% was reported in hospital outpatients from southern Sudan (25). The initial screening of 404 randomly selected HBsAg-negative plasma samples indicated a high prevalence of anti-HBc among Sudanese blood donors (145/404). However, further investigation revealed the presence of 16 confirmed HBsAg-positive samples (4%) that were not detected at the time of the initial screening. This unexpected finding emphasized the need for a sensitive and well-controlled HBsAg screening process. The confirmed yield of anti-HBc in HBsAg-negative blood donors was 33% (129/388). This anti-HBc prevalence is lower than the 47.5 and 67% prevalence reported in previous studies in Sudan (25, 28). The difference might relate to the fact that high prevalence was observed in patients attending clinics and/or with a history of jaundice as opposed to apparently healthy blood donors. Among these anti-HBc-positive donors, 56% were anti-HBs positive, indicating that 18.5% of Sudanese donors had been in contact with the virus, recovered, and maintained detectable neutralizing antibody levels after natural infection. Should anti-HBc screening be introduced in the blood donation testing algorithm, it will cause a significant shortage in the blood supply. However, anti-HBs of >100 IU/liter in anti-HBc-positive donations generally is considered safe for transfusion, and anti-HBs quantification would identify approximately 15% donations with anti-HBs of >100 IU/liter, reducing the deferral of collected blood bags to 20.9%. To select the best strategy for HBV blood safety in Sudan, the implementation of anti-HBc would considerably worsen the chronic shortage of blood. Even with retaining the high-titer anti-HBs-positive units, anti-HBc screening would remain unaffordable from a blood supply standpoint. The alternative strategy of HBV DNA screening would only defer infectious units either from the window period or from the less-infectious occult HBV carriage, avoiding the loss of noninfectious units. A cost comparison between the anti-HBc and nucleic acid testing strategy should take into consideration not only the cost of testing and of discarded and destroyed units but also the cost of replacing these units with others necessary to maintain the blood supply.
There is no previously published data reporting the presence of occult HBV infection (OBI) in Sudan, defined as HBV DNA positive with low viral load (<10 IU/ml) but HBsAg negative. In the present study, the prevalence of occult HBV carriage was 4.6% in anti-HBc-positive Sudanese donors, leading to a 1.5% estimate in the total blood donor population. This frequency was similar to data collected in Ghana, in west Africa (32). All Sudanese OBI samples were anti-HBs positive, suggesting that these donors had recovered from past infection but were unable to fully control lowly replicating virus (9). The high anti-HBs prevalence in Sudanese OBIs differed from the ∼50% prevalence reported for OBI donors from western Europe (HBV genotypes A2 and D) and South Africa (HBV genotype A1) and the 37.5% prevalence observed in OBIs genotype E from Ghana (3, 9, 36). Compared to HBsAg-positive samples, HBV genotype D was dominant in OBIs (five of six), and none was genotype E (Table 1). This is consistent with other studies describing a high frequency of genotype D OBI relative to its prevalence in a population with mixed genotypes, such as Poland (A2 and D) (7, 9). In the case of Sudan, genotype E is dominant, but no genotype E OBI was identified. The average amino acid diversity within the MHR of the Sudanese genotype D OBIs was lower than the diversity reported in European genotype D and genotype A2 OBIs (9.8% versus 16 and 11%, respectively), but it was higher than that observed in South African genotype A1 (3%) and west African genotype E OBI strains (<1%) (Table 2) (3, 9, 36).
Despite a limited number of large-scale epidemiological studies, a trend in the distribution of HBV genotypes in Africa has emerged (20). HBV genotype A is dominant in southern and eastern Africa and parts of central Africa. Most African HBV/A strains characterized to date belong to subgenotype A1. Subgenotype A3 initially was identified in central Africa and was sporadically reported from west Africa (15, 30). HBV genotype E predominates throughout a vast crescent spanning from Senegal to Namibia, one-third of the continent, and extending to the Central African Republic in the east and to Mali and Niger in the north (6, 11, 18, 29). A few HBV/E strains also were identified as minority genotypes further east in Mozambique and Madagascar or further north in Tunisia (21). Finally, HBV genotype D is dominant in northern Africa, with two subgenotypes recently identified in Tunisia (D7) and Niger (D8) (11, 26). Sudan is a large country at the presumed geographical junction of the distribution of these three HBV (sub)genotypes. However, no clear data regarding the molecular epidemiology of HBV in Sudan was available. Among the 53 samples sequenced and analyzed, phylogenetic analysis showed that genotype E was dominant in Sudan (51%), followed by genotype D (41.5%) (Fig. 2). Strains of other genotypes, such as the European A2 or the Asian B genotypes, were detected occasionally but probably were not autochthonous. Genotype A1 was not found in the present study. This could be due to the small size of the cohort studied and to the northern location of Khartoum, where samples were collected.
Sudanese donors infected with genotype D strains tended to carry lower levels of HBV DNA than those infected with genotype E strains (103 versus 3,100 IU/ml; P < 0.0001). In addition, when examining the distribution of HBV DNA load, genotype D had a unimodal distribution with a large number of outliers from the median with a very low DNA load (Fig. 1). A similar viral load distribution was reported for genotype D-infected blood donors from Poland, Tunisia, and Iran (16, 17, 26). In contrast, a bimodal distribution was observed in Sudanese genotype E-infected donors, as previously reported for a similar blood donor population from west Africa (4, 10) or for donors from southeast Asia infected with genotypes B and C (27). However, a smaller proportion of genotype E-infected donors had a viral load above 104 IU/ml compared to that of donors infected with genotypes B and C.
Two strains (38%) were recombinant between genotypes D and E, the two dominant genotypes in Sudan. The whole genome (minus 50 bp, including the BCP/PC region) of one strain could be analyzed and was identified as a mosaic of six sections alternating genotypes D and E (Fig. 3). D/E breakpoints located between nucleotides 1500 and 3200 were similar to those reported in subgenotype D8 strains from Niger (11) and in a D/E recombinant strain from Ghana (15). Only the Pre-S/S region of the second Sudanese strain could be cloned and analyzed. It contained only one recombination site from genotype D to genotype E around nucleotide 3000 from the EcoRI start. All clones from both strains included this recombination site, suggesting that these individuals had been infected with a similar, if not identical, HBV-D/E circulating recombinant form, which is part of the HBV epidemiology in Sudan. The high viral load (1.3 × 106 IU/ml) observed in S81 suggested that this recombinant strain was infectious. A similar situation has been described previously in Guinea for an A3/E recombinant and in Ghana and Niger for D/E recombinant (tentatively but erroneously named D8) (11, 15). In addition, both D/E recombinants from Sudan and Niger and A3/E recombinants from Guinea shared a similar pattern of recombination breakpoints between nucleotides 1500 and 3200, confirming the presence of preferential sites of recombination across the HBV genome. While the D/E strain from Ghana (Gh16) and the D8 strains from Niger appeared genetically related (Fig. 2 and 3), the presence of an additional recombinant event within the pol gene and the lack of the 33-nucleotide-long D-specific deletion in the Pre-S1 region suggested that the D/E recombinant strains found in Sudan have a distinct origin.
In summary, this study provides clear evidence that the extension of the relatively recent HBV genotype E reaches the Red Sea in Sudan and appears to separate genotype D in the north and A1 in the east of Africa. At the junction between the boundaries of the extension of two genotypes, it is not surprising to find recombinant forms establishing new variants that should be incorporated into the epidemiology as circulating recombinant forms, as for HIV strains, rather than subgenotypes.
Acknowledgments
We thank the staff of the Laboratory of Virology, National Health Laboratory, Khartoum, Sudan, as well as the staff at the KTH and RICK blood centers, Federal Ministry of Health, Khartoum, Sudan. Sarah Abd Allah is thanked for her precious help in conducting the study.
This work was supported in part by a grant from the International Society of Blood Transfusion Foundation, and some reagents were generously provided by Roche.
Footnotes
Published ahead of print on 3 November 2010.
REFERENCES
- 1.Abou, M. A., Y. M. Eltahir, and A. S. Ali. 2009. Seroprevalence of hepatitis B virus and hepatitis C virus among blood donors in Nyala, South Dar Fur, Sudan. Virology J. 6:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain, J.-P. 2004. Occult hepatitis B virus infection: implications in transfusion. Vox Sang. 86:83-91. [DOI] [PubMed] [Google Scholar]
- 3.Allain, J.-P., D. Belkhiri, M. Vermeulen, R. Crookes, R. Cable, A. Amiri, R. Reddy, A. Bird, and D. Candotti. 2009. Characterization of occult hepatitis B virus strains in South African blood donors. Hepatology 49:1868-1876. [DOI] [PubMed] [Google Scholar]
- 4.Allain, J.-P., D. Candotti, K. Soldan, F. Sarkodie, B. Phelps, C. Giachetti, V. Shyamala, F. Yeboah, M. Anokwa, S. Owusu-Ofori, and O. Opare-Sem. 2003. The risk of hepatitis B virus infection by transfusion in Kumasi, Ghana. Blood 101:2419-2425. [DOI] [PubMed] [Google Scholar]
- 5.Behzad-Behbahani, A., A. Mafi-Nejad, S. Z. Tabei, K. B. Lankarani, A. Torab, and A. Moaddeb. 2006. Anti-HBc & HBV DNA detection in blood donors negative for hepatitis B virus surface antigen in reducing risk of transfusion associated HBV infection. Indian J. Med. Res. 123:37-42. [PubMed] [Google Scholar]
- 6.Bekondi, C., C. M. Olinger, N. Boua, A. Talarmin, C. P. Muller, A. le Faou, and V. Venard. 2007. Central African Republic is part of the West-African hepatitis B virus genotype E crescent. J. Clin. Virol. 40:31-37. [DOI] [PubMed] [Google Scholar]
- 7.Brojer, E., P. Grabarczyk, G. Liszewski, M. Mikulska, J.-P. Allain, and M. Letowska. 2006. Characterization of HBV DNA positive/HBsAg negative blood donors identified in the Polish NAT screening program. Hepatology 44:1666-1674. [DOI] [PubMed] [Google Scholar]
- 8.Candotti, D., K. Danso, and J.-P. Allain. 2007. Maternofetal transmission of hepatitis B virus genotype E in Ghana, west Africa. J. Gen. Virol. 88:2686-2695. [DOI] [PubMed] [Google Scholar]
- 9.Candotti, D., P. Grabarczyk, P. Ghiazza, R. Roig, N. Casamitjana, P. Iudicone, M. Schmidt, A. Bird, R. Crookes, E. Brojer, M. Miceli, A. Amiri, C. Li, and J.-P. Allain. 2008. Characterization of occult hepatitis B virus from blood donors carrying genotype A2 or genotype D strains. J. Hepatol. 49:537-547. [DOI] [PubMed] [Google Scholar]
- 10.Candotti, D., O. Opare-Sem, H. Rezvan, F. Sarkodie, and J.-P. Allain. 2006. Molecular and serological characterization of hepatitis B virus in deferred Ghanaian blood donors with and without elevated alanine aminotransferase. J. Viral. Hepat. 13:715-724. [DOI] [PubMed] [Google Scholar]
- 11.Chekaraou, M. A., S. Brichler, W. Mansour, F. Le Gal, A. Garba, P. Deny, and E. Gordien. 2010. A new hepatitis B virus subgenotype D (D8) strain, resulting from recombination between genotypes D and E, is circulating in Niger along with HBV/E strains. J. Gen. Virol. doi: 10.1099/vir.0.018127-0. [DOI] [PubMed]
- 12.El Ekiaby, M., N. Lelie, and J.-P. Allain. 2010. Nucleic acid testing (NAT) in high prevalence-low resource settings. Biologicals 38:59-64. [DOI] [PubMed] [Google Scholar]
- 13.Elsheikh, R. M., A. A. Daak, M. A. Elsheikh, M. S. Karsany, and I. Adam. 2007. Hepatitis B virus and hepatitis C virus in pregnant Sudanese women. Virol. J. 4:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Zayadi, A. R., E. H. Ibrahim, H. M. Badran, A. Saeid, N. A. Moneib, M. A. Shemis, R. M. Abdel-Sattar, A. M. Ahmady, and A. El-Nakeed. 2008. Anti-HBc screening in Egyptian blood donors reduces the risk of hepatitis B virus transmission. Transfusion Med. 18:55-61. [DOI] [PubMed] [Google Scholar]
- 15.Garmiri, P., A. Loua, N. Haba, D. Candotti, and J.-P. Allain. 2009. Deletions and recombinations in the core region of hepatitis B virus genotype E strains from asymptomatic blood donors in Guinea, west Africa. J. Gen. Virol. 90:2442-2451. [DOI] [PubMed] [Google Scholar]
- 16.Garmiri, P., H. Rezvan, H. Abolghasemi, and J.-P. Allain. Full genome characterization of hepatitis B virus strains from blood donors in Iran. J. Med. Virol., in press. [DOI] [PubMed]
- 17.Grabarczyk, P., P. Garmiri, G. Liszewski, D. Doucet, E. Sulkowska, E. Brojer, J.-P. Allain, and Polish Blood Transfusion Centres Viral Study Group. 2009. Molecular and serological characterization of hepatitis B virus genotype A and D infected blood donors in Poland. J. Viral. Hepat. doi: 10.1111/j.1365-2893.2009.01192.x. [DOI] [PubMed]
- 18.Hübschen, J. M., I. E. Andernach, and C. P. Muller. 2008. Hepatitis B virus genotype E variability in Africa. J. Clin. Virol. 43:376-380. [DOI] [PubMed] [Google Scholar]
- 19.Jayaraman, S., Z. Chalabi, P. Perel, C. Guerriero, and I. Roberts. 2010. The risk of transfusion-transmitted infections in sub-Saharan Africa. Transfusion 50:433-442. [DOI] [PubMed] [Google Scholar]
- 20.Kramvis, A., and M. C. Kew. 2007. Epidemiology of hepatitis B virus in Africa, its genotypes and clinical associations of genotypes. Hepatol. Res. 37:S9-S19. [DOI] [PubMed] [Google Scholar]
- 21.Kramvis, A., K. Restorp, H. Norder, J. F. Botha, L. O. Magnius, and M. C. Kew. 2005. Full genome analysis of hepatitis B virus genotype E strains from South-Western Africa and Madagascar reveals low genetic variability. J. Med. Virol. 77:47-52. [DOI] [PubMed] [Google Scholar]
- 22.Kurbanov, F., Y. Tanaka, and M. Mizokami. 2010. Geographical and genetic diversity of the human hepatitis B virus. Hepatol. Res. 40:14-30. [DOI] [PubMed] [Google Scholar]
- 23.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. S. S. Gadkari, G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makroo, R. N., N. Choudhury, L. Jagannathan, M. Parihar-Malhotra, V. Raina, R. K. Chaudhary, N. Marwaha, N. K. Bhatia, and A. K. Ganguly. 2008. Multicenter evaluation of individual donor nucleic acid testing (NAT) for simultaneous detection of human immunodeficiency virus-1 & hepatitis B & C viruses in Indian blood donors. Indian J. Med. Res. 127:140-147. [PubMed] [Google Scholar]
- 25.McCarthy, M. C., A. el-Tigani, I. O. Khalid, and K. C. Hyams. 1994. Hepatitis B and C in Juba, southern Sudan: results of a serosurvey. Trans. R. Soc. Trop. Med. Hyg. 88:534-536. [DOI] [PubMed] [Google Scholar]
- 26.Meldal, B. H., N. M. Moula, I. H. Barnes, K. Boukef, and J.-P. Allain. 2009. A novel hepatitis B virus subgenotype, D7, in Tunisian blood donors. J. Gen. Virol. 90:1622-1628. [DOI] [PubMed] [Google Scholar]
- 27.Meldal, B. H., A. H. Bon, D. Prati, Y. Ayob, and J.-P. Allain. 2010. Diversity of hepatitis B virus infecting Malaysian candidate blood donors is driven by viral and host factors. J. Viral. Hepat. doi: 10.1111/j.1365-2893.2010.01282.x. [DOI] [PubMed]
- 28.Mudawi, H. M. Y., H. M. Smith, S. A. Rahoud, I. A. Fletcher, O. K. Saeed, and S. S. Fedail. 2007. Prevalence of hepatitis B virus infection in the Gezira State of Central Sudan. Saudi J. Gastroenterol. 13:81-83. [DOI] [PubMed] [Google Scholar]
- 29.Mulders, M. N., V. Venard, M. Njayou, A. P. Edorh, A. O. Bola Oyefolu, M. O. Kehinde, J. J. Muyembe Tamfum, Y. K. Nebie, I. Maiga, W. Ammerlaan, F. Fack, S. A. Omilabu, A. Le Faou, and C. P. Muller. 2004. Low genetic diversity despite hyperendemicity of hepatitis B virus genotype E throughout West Africa. J. Infect. Dis. 190:400-408. [DOI] [PubMed] [Google Scholar]
- 30.Olinger, C. M., V. Venard, M. Njayou, A. O. Oyefolu, I. Maiga, A. J. Kemp, S. A. Omilabu, A. le Faou, and C. P. Muller. 2006. Phylogenetic analysis of the precore/core gene of hepatitis B virus genotypes E and A in West Africa: new subtypes, mixed infections, and recombinations. J. Gen. Virol. 87:1163-1173. [DOI] [PubMed] [Google Scholar]
- 31.Omer, E. E., P. Van't Veer, A. M. Kadaru, E. Kampman, I. M. el Khidir, S. S. Fedail, and F. J. Kok. 2001. The role of hepatitis B and hepatitis C viral infections in the incidence of hepatocellular carcinoma in Sudan. Trans. R. Soc. Trop. Med. Hyg. 95:487-491. [DOI] [PubMed] [Google Scholar]
- 32.Owusu-Ofori, S., J. Temple, F. Sarkodie, M. Anokwa, D. Candotti, and J.-P. Allain. 2005. Predonation screening of blood donors with rapid tests: implementation and efficacy of a novel approach to blood safety in resource-poor settings. Transfusion 45:133-140. [DOI] [PubMed] [Google Scholar]
- 33.Phikulsod, S., S. Oota, T. Tirawatnapong, T. Sakuldamrongpanich, W. Chalermchan, S. Louisirirotchanakul, S. Tanprasert, V. Chongkolwatana, P. Kitpoka, P. Phanuphak, C. Wasi, C. Nuchprayoon, and Working Group for NAT Study in Thai Blood Donations. 2009. One-year experience of nucleic acid technology testing for human immunodeficiency virus type 1, hepatitis C virus, and hepatitis B virus in Thai blood donations. Transfusion 49:1126-1135. [DOI] [PubMed] [Google Scholar]
- 34.Swofford, D. L. 2003. Phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 35.Yu, H., Q. Yuan, S. X. Ge, H. Y. Wang, Y. L. Zhang, Q. R. Chen, J. Zhang, P. J. Chen, and N. S. Xia. 2010. Molecular and phylogenetic analyses suggest an additional hepatitis B virus genotype “I.” PLoS One 5:e9297. [DOI] [PMC free article] [PubMed]
- 36.Zahn, A., C. Li, K. Danso, D. Candotti, S. Owusu-Ofori, J. Temple, and J.-P. Allain. 2008. Molecular characterization of occult hepatitis B virus in genotype E-infected subjects. J. Gen. Virol. 89:409-418. [DOI] [PubMed] [Google Scholar]