Abstract
The presence of Campylobacter species and enteric RNA viruses in stools from diarrheic (n = 442) and healthy (n = 58) humans living in southwestern Alberta was examined (May to October 2005). A large number of diarrheic individuals who were culture negative for C. jejuni (n = 54) or C. coli (n = 19) were PCR positive for these taxa. Overall detection rates for C. jejuni and C. coli in diarrheic stools were 29% and 5%, respectively. In contrast, 3% and 0% of stools from healthy humans were positive for these taxa, respectively. Infection with C. jejuni was endemic over the study period. However, there was no difference in infection rates between individuals living in urban or rural locations. Stools from a large number of diarrheic (74%) and healthy (88%) individuals were positive for Campylobacter DNA. The prevalence rates of C. concisus, C. curvus, C. fetus, C. gracilis, C. helveticus, C. hominis, C. hyointestinalis, C. mucosalis, C. showae, C. sputorum, and C. upsaliensis DNA were either not significantly different or were significantly lower in stools from diarrheic than from healthy individuals. No C. lanienae or C. lari DNA was detected. Stools from 4% and 0% of diarrheic and healthy humans, respectively, were positive for rotavirus, sapovirus, or norovirus (GI/GII). Our results showed a high prevalence of diarrheic individuals living in southwestern Alberta who were infected by C. jejuni and, to a lesser extent, by C. coli. However, other Campylobacter species, norovirus, rotavirus, sapovirus, and bovine enteric calicivirus were either inconsequential pathogens during the study period or are not pathogens at all.
The former Chinook Health Region (CHR) of southwestern Alberta, Canada, is a large geographical area that possesses a high prevalence of enteritis among its human inhabitants (30). For example, the prevalence of campylobacteriosis incited by Campylobacter coli and/or Campylobacter jejuni within the CHR is substantially higher than both the provincial and national averages of ≤50 cases per 100,000 individuals. Reasons for the relatively high rates of campylobacteriosis in the CHR are currently uncertain. The CHR possesses one of the highest densities of livestock in North America (2), and an epidemiological examination indicated that one-quarter of individuals infected with C. jejuni or C. coli had been in close contact with livestock, primarily cattle (30). As in other jurisdictions (50), the majority of cases of enteritis in the CHR are not diagnosed. At the central diagnostic facility within the CHR located at the Chinook Regional Hospital (CRH) in Lethbridge, stools from humans exhibiting clinical evidence of enteritis are processed for prominent bacterial pathogens. A single method is used to isolate Campylobacter species.
While C. jejuni and to a lesser extent C. coli are thought to be the primary causes of campylobacterosis, there are 25 recognized species of Campylobacter (i.e., campylobacteria). Many Campylobacter species are fastidious and are not readily isolated using conventional media containing selective agents such as cefoperazone (39), including the medium used at the CRH; these species are commonly referred to as “cryptic” campylobacteria. The use of specialized isolation and non-culture-based methods have demonstrated that a number of cryptic taxa of Campylobacter are shed in human feces (19, 37, 41, 42, 43, 47), but the impact of these taxa on humans, including those living in the CHR, remains enigmatic. Furthermore, infection by enteric viruses is not routinely examined within the CHR, although stool samples from patients suspected to be infected by enteric viruses, primarily during outbreaks in the fall and winter, are forwarded to the Alberta Provincial Laboratory for testing (≈6% of total samples).
Considering the high rates of enteritis within the CHR, we erected the following hypotheses: a significant number of diarrheic individuals infected by C. jejuni and C. coli are not diagnosed using culture-based methods, and direct PCR detection would provide a more accurate measure of infection rates by these bacteria; conventional isolation methods are ineffective in detecting cryptic campylobacteria which infect a significant number of humans living within the CHR, thereby contributing to the high rates of enteritis in this region; enteric RNA viruses are underreported and infect a substantial number of human inhabitants of the CHR during the summer and early fall; and individuals living in rural areas within the CHR are disproportionately affected. To test these hypotheses, the following objectives were established: (i) develop and validate nested primers for C. concisus and C. upsaliensis; (ii) determine the prevalence of Campylobacter species in stools by using a direct taxon-specific PCR; (iii) contrast direct PCR detection with conventional and specialized culturing methods for campylobacteria; (iv) determine the prevalence of norovirus (NoV), sapovirus (SaV), and rotavirus (RV) in stools; (v) contrast the detection frequency of enteric campylobacteria and viruses in stool samples from diarrheic and healthy humans over a 5-month period (summer and early fall); and (vi) compare detection frequencies in diarrheic humans living in rural and urban areas within the CHR.
MATERIALS AND METHODS
Nested primers for direct detection of C. concisus and C. upsaliensis.
New nested primers for direct detection of C. concisus and C. upsaliensis DNA in stools were designed using the OLIGO primer analysis software (Molecular Biology Insights, Inc., Cascade, CO) (Table 1).
TABLE 1.
New primers to detect C. concisus and C. upsaliensis DNA in stools
| PCR target and gene | Primer | Ta (°C)a | Primer sequence (5′-3′) | Amplicon size (bases) | Reference |
|---|---|---|---|---|---|
| C. concisus 1 outside | |||||
| 23S rRNA | Ccon55U | 58 | AACGGGGCTAAAATGAGTAC | 729 | New primer |
| 23S rRNA | Ccon784L | 58 | GCTTCGCAGAGCTAACG | 729 | New primer |
| C. concisus 1 inside | |||||
| 23S rRNA | Muc1 | 60 | ATGAGTAGCGATAATTGGG | 306 | Bastyns et al. (9) |
| 23S rRNA | Con1 | 60 | CAGTATCGGCAATTCGCT | 306 | Bastyns et al. (9) |
| 23S rRNA | Con2 | 60 | GACAGTATCAAGGATTTACG | 306 | Bastyns et al. (9) |
| C. concisus 2 outside | |||||
| cpn60 | Ccon_cpn_66f | 53 | TATCGAAGTGAAACGTGGCA | 357 | New primer |
| cpn60 | Ccon_cpn_423r | 53 | GCTCAAGCACTGGCAATAAG | 357 | New primer |
| C. concisus 2 inside | |||||
| cpn60 | Ccon_cpn_72f | 53 | AGTGAAACGTGGCATGGATA | 270 | New primer |
| cpn60 | Ccon_cpn_342r | 53 | GCATCTTTTCAGGGTTTGTG | 270 | New primer |
| C. upsaliensis outside | |||||
| glyA | Cu63F | 60 | AATTGAAACTCTTGCTATCCAAAG | 482 | New primer |
| glyA | Cu545r | 60 | TACACATAATAATTCCACCTCTAG | 482 | New primer |
| C. upsaliensis inside | |||||
| glyA | Cu146F | 60 | CAAATCAAGGCGTTTATGCTG | 101 | New primer |
| glyA | Cu247R | 60 | CTTTCATACATTTTACCCGAGCT | 101 | New primer |
Ta, annealing temperature.
(i) Primer specificity and comprehensiveness.
Type strains of Campylobacter (17 species), Arcobacter butleri and Helicobacter pylori, and 38 and 49 clinical isolates of C. upsaliensis and C. concisus, respectively, were used to evaluate the comprehensiveness of the primers. To obtain biomass, all isolates were grown on Karmali agar (Oxoid Inc.) at 37°C for 48 to 72 h in a microaerobic environment (10% CO2, 30% H2, 5% O2, 55% N2). DNA was extracted using a DNeasy blood and tissue kit (Qiagen Inc.) according to the manufacturer's protocol. The PCR conditions were as follows: 1 initial denaturation cycle at 95°C for 15 min; 30 cycles of 30 s at 94°C for denaturation, 90 s at the appropriate annealing temperature (Table 1), and 60 s at 72°C for extension; and a final extension cycle of 10 min at 72°C. Reaction mixtures consisted of a total volume of 20 μl containing 1× reaction buffer, 0.2 mM deoxynucleoside triphosphates, 2 mM MgCl2, 0.05 μM each primer (Sigma-Genosys, Oakville, ON, Canada), 0.2 μg BSA (Promega, Madison, WI), and 1 U HotStar Taq polymerase (Qiagen Inc., Mississauga, ON, Canada). Each PCR was performed with a total of 2 μl of a 10−1 dilution of genomic DNA at ≈50 ng/μl. When primary and secondary primers were used together, PCR conditions remained the same except that 25 and 30 cycles were used in the primary and secondary reactions, respectively. Furthermore, the primer concentration in the secondary reaction was increased 10-fold, 1 μl of the reaction mixture from the primary amplification step was used as template, and BSA was not included. For all PCR tests, positive controls consisted of genomic DNA from reference strains and negative controls consisted of water alone. The estimated amplicon sizes are shown in Table 1.
(ii) Primer sensitivity.
To determine the sensitivity of detection, genomic DNA extracted from C. concisus (L288T) and C. upsaliensis (L270T) was quantified using a NanoDrop spectrophotometer (ND-1000; NanoDrop Technologies, Inc., Wilmington, DE), and genomic DNA was diluted in a 10-fold dilution series from 10−1 to 10−12. Primary and secondary PCRs as described above were performed.
(iii) Detection in seeded feces.
For C. concisus and C. upsaliensis primers, swine feces determined to be negative for Campylobacter DNA by Campylobacter genus-specific PCR (46) was used. Campylobacter concisus (L288T) and C. upsaliensis (L270T) cells were grown microaerobically on Karmali agar for 48 h at 37°C. To obtain biomass, cells were scraped from the medium and suspended in sterile Columbia broth (Oxoid Inc. Nepean, ON, Canada). The turbidity (A600) of the suspension was adjusted to 0.5 (target cell density of 9.30 log10 cells/ml). Cell densities were enumerated using a Petroff-Hausser counting chamber (Hausser Scientific, Blueball, PA), and mean cell densities for C. concisus and C. upsaliensis were 9.52 (standard error of the mean [SEM], 0.032) and 9.54 (SEM, 0.014) log10 cells/ml, respectively. However, on Columbia blood agar, densities averaged 8.05 (SEM, 0.010) and 8.10 (SEM, 0.019) log10 CFU/ml, respectively. Six 10-fold serial dilutions were prepared in Columbia broth representing a range in cell densities of ≈2 × 103 to 2 × 109 cells/ml (i.e., based on Petroff-Hausser densities). Twofold dilutions also were established at the higher dilutions to yield cell densities of ≈1 × 103 and 1 × 104 cells/ml. Feces samples were infested with each cell density in duplicate at the rate of 1 ml to 9 g (wet weight) of feces. Sterile Columbia broth was used as the control treatment. Immediately after addition of the inoculum treatments, fecal samples were thoroughly mixed with a sterile wooden splint.
DNA was extracted from two separate aliquots of each inoculated and control fecal sample by using the QIAamp DNA stool minikit (Qiagen Inc.) according to the manufacturer's protocol; an internal amplification control (IAC) was added to each sample at the rate of 10 μl per 200 mg of feces (32). Nested PCR was conducted as described above, except that 2 μl of undiluted template was used. The experiment was conducted three to four times on separate occasions.
(iv) Comparative evaluation of primers.
We examined the specificity and sensitivity of the nested primers developed in the current study relative to primers published by Samie et al. (56) and Chaban et al. (12) for direct detection of C. concisus in feces and those published by Chaban et al. (12), Eyers et al. (20), Linton et al. (46), and Wang et al. (68) for the detection of C. upsaliensis. None of these primers was nested.
Human subject information and stool collection.
Scientific and ethics approval for the study was obtained from the Regional Ethics Committee of the former CHR and from the University of Lethbridge Human Subject Research Committee. Information released with the samples included stool collection date and time and patient age, sex, and place of habitation (i.e., based on postal code). Stool samples from humans exhibiting signs of enteritis (i.e., diarrhea) from 31 May to 31 October 2005 were examined, along with stool samples from a cohort of healthy humans (processed in mid-October). The majority of stool samples from diarrheic humans were suspended in an enteric pathogen transport medium (EPT) at the time of collection and transported to the diagnostic facility at the CRH within 1 to 2 days of sample collection; the EPT used was Cary-Blair medium (11). Stool samples were processed immediately upon arrival at the CRH, and samples were placed at 4°C in ambient atmosphere until they could be transferred to the Agriculture & Agri-Food Canada (AAFC) Research Centre at Lethbridge for further processing. Samples determined to be positive for Campylobacter at the CRH by culture were not disclosed to AAFC personnel until the data collection phase of the study was completed. Stool samples from healthy humans were not placed in EPT, and samples were transferred to AAFC Lethbridge within ≈6 h of collection in the majority of instances; samples collected ≥1 h before submission were kept on ice.
Detection of campylobacteria. (i) Isolation and isolate identification.
At the CRH, a standard protocol for isolating C. jejuni and C. coli was utilized (i.e., referred to as conventional isolation here). Samples were streaked onto Campy CVA agar (Becton Dickinson, Oakville, ON, Canada), cultures were incubated at 42°C in an atmosphere consisting of 10% CO2, 10% H2, and 80% N2, and colonies exhibiting characteristic Campylobacter morphology were isolated and tested for their ability to hydrolyze hippurate by using a disc method (Dalynn Biologicals, Calgary, AB, Canada). Hippurate-positive strains were presumptively deemed to be C. jejuni, whereas hippurate-negative strains were presumptively identified as C. coli. In addition, all samples were processed for Aeromonas spp. (i.e., A. caviae, A. hydrophila, A. salmonicida, A. sobria, and A. veronii), Edwardsiella spp. (E. hoshinae and E. tarda), Escherichia coli O157:H7, Plesiomonas shigelloides, Salmonella enterica subsp. enterica (serovars Paratyphi A, Paratyphi B, Typhi, and Typhimurium), Shigella sonnei, Vibrio spp. (V. alginolyticus, V. cholerae, V. fluvialis, V. metschnikovii, V. mimicus, V. parahemolyticus, and V. vulnificus), and Yersinia spp. (Y. enterocolitica, Y. pestis, Y. pseudotuberculosis, and Y. ruckeri) using standard protocols (52). All Campylobacter isolates were frozen in 30% glycerol at −70°C at the CRH until transferred to AAFC Lethbridge for identification.
Campylobacteria were isolated from stool samples at AAFC Lethbridge by direct plating, membrane filtration, and enrichment (here referred to as specialized isolation). An incubation temperature of 37°C and a microaerobic atmosphere consisting of 5% O2, 3% H2, 10% CO2, and 82% N2 were used for all specialized isolations. For direct plating, a fecal suspension (1:10) was prepared in Columbia broth (Oxoid Inc.), and 25 μl of the suspension was spread onto Karmali agar (Oxoid Inc.) containing selective supplement SR167 (Oxoid Inc.). For each sample, 100 μl of the Columbia broth suspension also was placed on top of a hydrophobic Iso-Grid membrane (0.45 μm; Neogen Corp., Lansing, MI) situated on Mueller-Hinton agar (BD Diagnostics, Sparks, MD) containing 5% sheep blood. The suspension was allowed to filter passively for 24 h in a microaerobic environment at 37°C, after which point the membrane was removed. For enrichment, a fecal suspension (1:20) was prepared in Arcobacter enrichment broth (16). Tubes were incubated for 24 h, after which point 25 μl of the broth was spread onto Karmali agar containing selective supplement SR167 or Arcobacter agar (CM965; Oxoid Inc.) containing selective supplement SR174. In addition, the enrichment broth (100 μl) was placed centrally onto a semisolid Arcobacter selective medium (ASM) (16). All solid media were incubated for 72 h, with the exception of the membrane filtration medium, which was incubated for 96 h. Cells from colonies were examined for size, shape, and characteristic motility by using phase-contrast microscopy. Campylobacter-like isolates were then transferred onto Mueller-Hinton agar containing 5% sheep blood and incubated for 48 to 72 h. Biomass was then suspended in Columbia broth containing 30% glycerol and frozen at −80°C.
All presumptive Campylobacter isolates were tested for their ability to hydrolyze hippurate using the method described by Chapin and Lauderdale (13) with the exception that volumes were adjusted to microtiter plate volumes. Genomic DNA of isolates was extracted using an AutoGen 740 robot (Holliston, MA) according to the manufacturer's protocol. All hippurate-positive isolates were identified by PCR using a C. jejuni-specific primer set (17). Hippurate-positive but C. jejuni-negative isolates determined by PCR and all hippurate-negative isolates were tested using Campylobacter, Arcobacter, and Helicobacter genus-specific primer sets (28, 29, 46). Hippurate-negative Campylobacter isolates were tested using C. coli-specific and C. jejuni-specific primers (17, 23) followed by taxon-specific primers for C. concisus (9) and C. fetus (32).
To identify isolates of Campylobacter and Helicobacter that could not be identified by taxon-specific PCR, the near-complete 16S rRNA gene was amplified and sequenced.
(ii) Direct PCR detection.
DNA was extracted from thawed stool samples (200 ± 5 mg) to which the IAC had been added (32). All samples were subjected to nonnested PCR for the Campylobacter genus (46). Only samples in which the IAC was visualized in the absence of an amplicon for Campylobacter were deemed negatives. Samples for which the IAC and/or genus amplicon was not detected were reextracted. All Campylobacter genus-positive samples were subjected to nonnested, seminested, or nested PCR for C. coli (32), C. concisus 1 (Table 1), C. concisus 2 (Table 1), C. curvus (12), C. fetus (32), C. gracilis (12), C. helveticus (12), C. hominis (44), C. hyointestinalis (32), C. jejuni (32), C. lanienae (32), C. lari (12), C. mucosalis (12), C. showae (12), C. sputorum 1 (8), C. sputorum 2 (12), and C. upsaliensis (Table 1). PCR for each taxon was conducted a minimum of two times. The Campylobacter genus amplicon was sequenced for samples that were positive for genus but were species negative by taxon-specific PCR and/or culture. Forward primer 412F (46) and reverse primer 1111R (5′-ACGTCGTCCACACCTTCCT-3′; new primer) were used to amplify the Campylobacter genus amplicon. The 412F and 1111R primers were then used to sequence the amplicon.
(iii) Data analysis.
Comparisons of detection rates between diarrheic and healthy humans and between PCR and culture-based methods (i.e., conventional and specialized) for C. jejuni and C. coli were analyzed using the FREQ procedure of SAS and Fisher's exact test (58). If the exact test was significant, then 0-frequency values were adjusted to 0.5 and the Mantel-Haenszel and logit estimates of the odds ratios and relative risks were estimated. Temporal infection rates of humans by campylobacteria collectively, C. jejuni, and C. concisus were analyzed using the GENMOD procedure of SAS with a log-linear model, and zero-frequency values were removed from the analyses.
Detection of enteric RNA viruses.
Samples were processed and RNA was extracted using the QIAamp viral RNA minikit (Qiagen Inc.) as described previously (31). Reverse transcription-PCR (RT-PCR) was used to detect human NoV genogroups I and II (GI and GII), SaV, bovine enteric calicivirus (BEC), and RV (4, 21, 25, 36, 49, 61). TaqMan quantitative PCR (qPCR) assays (for NoV GI and GII) were carried out in a 25-μl reaction volume consisting of 1 μl of RNA template and 24 μl of a master mix made with the 1-step Brilliant QRT-PCR core reagent kit (Stratagene, La Jolla, CA) and 5.0 mM MgCl2 (31). qPCR amplifications were performed in duplicate with a Stratagene Mx4000 system under the following conditions: 30 min at 45°C for reverse transcription; 10 min at 95°C for initial denaturation; 45 cycles of 15 s at 95°C and 1 min at 60°C annealing and extension. Conventional RT-PCR (for SaV, BEC, and RV) was performed with 1 μl of extracted RNA in a volume of 20 μl using the Qiagen One-Step RT-PCR kit according to the manufacturer's recommendations. Amplicons were sequenced and compared with sequences within the NCBI GenBank.
RESULTS
Nested primers for direct detection of C. concisus and C. upsaliensis. (i) Primer specificity.
The nested primer sets that we developed targeted the 23S rRNA gene of C. concisus, the cpn60 gene of C. concisus, and the glyA gene of C. upsaliensis (Table 1). The cpn60 and glyA nested primers were found to be specific for genomic DNA of the target species, whereas the primer sets targeting the C. concisus 23S rRNA gene also amplified C. showae. All 49 isolates of C. concisus evaluated provided an amplicon for the nested primers targeting the 23S rRNA and cpn60 genes. However, a weak cpn60 amplicon was observed for two strains (CHRB-3152 and CHRB-3235); neither of these two C. concisus strains belonged to genomospecies A or B (unpublished data). All 38 clinical isolates of C. upsalienesis evaluated provided an amplicon for the nested primers targeting the glyA gene. The C. concisus primer set published by Samie et al. (56) was found to be nonspecific, amplifying the gyrB gene of a variety of Campylobacter species. Furthermore, these primers were not very sensitive and only provided a weak amplicon for C. concisus that was of the wrong size. In contrast, the C. concisus primer set published by Chaban et al. (12) was specific for C. concisus. The primer sets from Eyers et al. (20) amplified C. upsaliensis but also provided an amplicon for C. hyointestinalis, C. lanienae, and C. concisus. The primers published by Linton et al. (46) and Wang et al. (68) were specific for C. upsaliensis.
(ii) Primer sensitivity.
The minimum detection limits of genomic DNA by the nested primers for C. concisus (23S rRNA gene), C. concisus (cpn60 gene), and C. upsaliensis (glyA gene) were 0.075, 0.008, and 0.016 pg, respectively.
(iii) Detection in seeded feces.
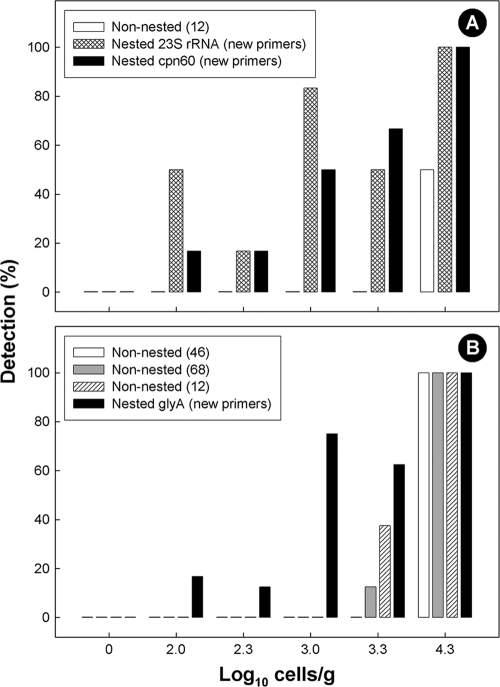
For both C. concisus and C. upsaliensis, the nested primer sets developed in the current study resulted in positive amplicons only in feces seeded with the corresponding bacterium (Fig. 1). Some minor nonspecific amplification was observed for the C. upsaliensis primers as a function of template concentration, but this did not obscure detection. For all three nested primer sets developed, the threshold of detection was 100 cells/g, but detection at cell densities lower than 2 × 104 cells/g was not absolute (Fig. 2). Nesting primers was necessary to increase sensitivity. The C. concisus outside primers targeting the 23S rRNA gene used alone exhibited a minimum detection threshold of 1 × 103 cells/g, whereas the inside primer set used alone did not produce a visible amplicon from seeded feces. For the C. concisus cpn60 primers, the minimum detection thresholds for the outside and inside primers used alone were 2 × 104 cells/g and 2 × 105 cells/g, respectively. The minimum detection limit for the C. upsaliensis inside primers used alone was 2 × 103 cells/g, and the outside primers alone did not produce an amplicon from seeded feces.
FIG. 1.
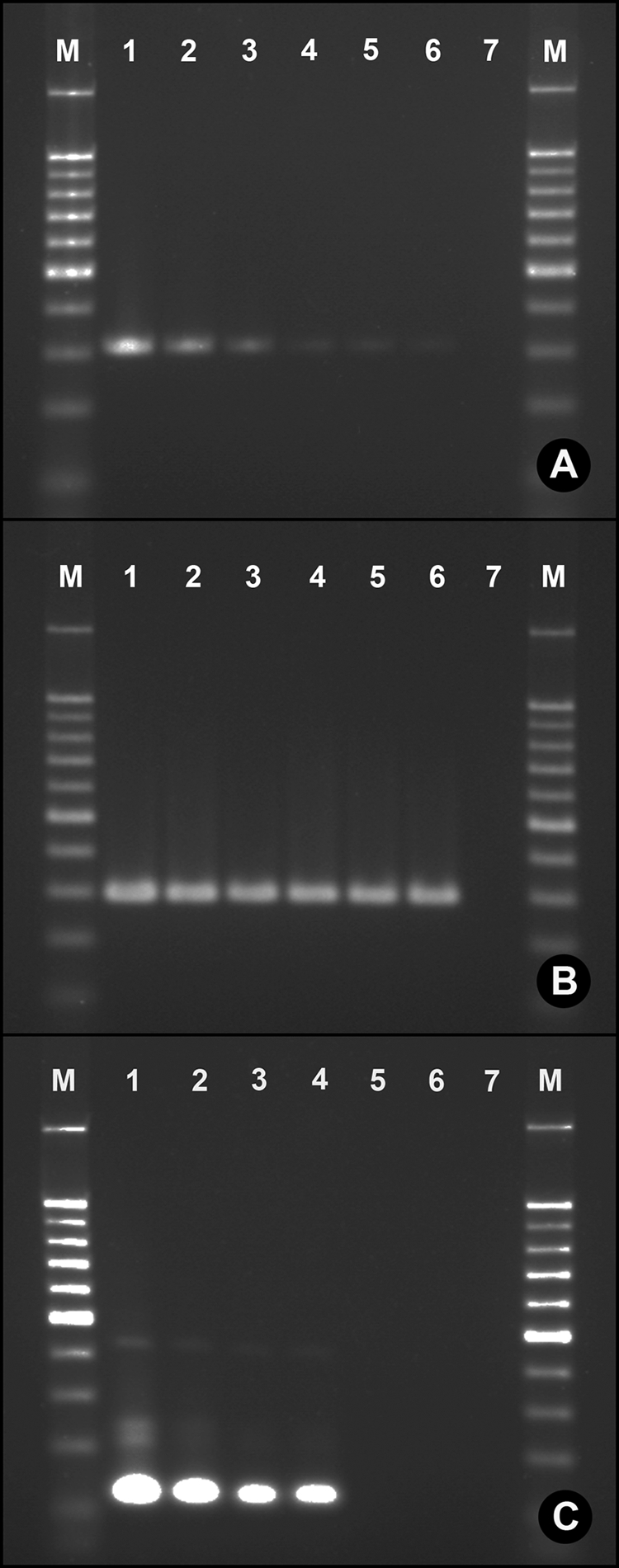
Detection of campylobacteria in seeded feces by using nested primers. (A) Campylobacter concisus 23S rRNA gene primers; (B) C. concisus cpn60 primers; (C) C. upsaliensis glyA primers. Campylobacter species were inoculated into swine feces. Lane assignments: M, 100-bp marker; 1, feces seeded at 2 × 105 cells/g; 2, feces seeded at 2 × 104 cells/g; 3, feces seeded at 2 × 103 cells/g; 4, feces seeded at 1 × 103 cells/g; 5, feces seeded at 2 × 102 cells/g; 6, feces seeded at 1 × 102 cells/g; 7, uninoculated feces. Genomic DNA of C. concisus or C. upsaliensis and water alone (instead of template) were run in all gels (data not shown). In all instances, amplicon size from seeded feces corresponded to that of the corresponding taxon, and PCR controls were always negative.
FIG. 2.
Sensitivity of published and new primers for detection of Campylobacter cells in seeded swine feces. (A) Campylobacter concisus; (B) C. upsaliensis.
The nested primer sets developed in the current study were more sensitive than previously published nonnested primers used to detect C. concisus (12) and C. upsaliensis (12, 46, 68) in feces (Fig. 2). Furthermore, we observed that the C. concisus cpn60 primers described by Chaban et al. (12) resulted in nonspecific amplification from fecal samples containing low cell densities of C. concisus and from uninoculated feces. The Samie et al. (56) primer set for C. concisus produced amplicons of the wrong size in seeded and nonseeded feces.
Human subject information.
A total of 532 samples were obtained from humans with clinical signs of enteritis from 31 May to 31 October 2005. The majority of people submitted a single sample. However, 55 individuals submitted multiple samples. Eighteen individuals did not live within the CHR and were excluded from the study. Of the 442 individuals who lived in the CHR, 274 (62.0%) lived in an urban center and 168 (38.0%) lived in a small rural community or on a farm. A total of 271 of the diarrheic individuals submitting samples were female (61.3%) and 171 were male (38.7%). Adjusted for differential submissions, a similar proportion of females and males living in urban centers (59.8% and 65.5%, respectively) and rural areas (40.2% and 35.1%, respectively) submitted samples. The mean age of participating diarrheic individuals was 34.8 years (standard deviation [SD], 13.5). Although the majority of samples were from individuals older than 16 years of age (96%), individuals ranged in age from less than 1 year to 86 years.
An additional 58 stool samples were obtained from immunocompetent individuals living in the CHR who were free of clinical evidence of enteritis and were not taking antibiotics within 1 month before or at the time of sample submission (i.e., “healthy” human controls). Samples were collected from 16 to 24 October. Fifty (86.2%) healthy individuals lived in an urban center, and 8 (13.8%) lived in a small rural community or on a farm. Twenty-five (43.1%) of the individuals were female, and 33 (56.9%) were male. The mean age of participating healthy individuals was 47.9 years (SD, 10.3), with an age range from 28 to 76 years.
Detection of campylobacteria. (i) Isolation and isolate identification.
A total of 376 Campylobacter isolates were recovered and characterized. Campylobacter jejuni was isolated from 72 people (16.3% of samples) at the CRH (i.e., conventional isolation), and C. coli was isolated from an additional individual (0.2% of samples) (Table 2). No other campylobacteria were isolated at the CRH. The CRH policy is to retain stool samples for 7 days, and the mean time from sample collection to isolation at AAFC was 7.8 days (SD, 2.3). Despite the delay in processing of the samples at AAFC, comparable detection results were observed for C. jejuni at the CRH (i.e., using conventional isolation) and AAFC (i.e., using specialized isolation) (Table 2). However, an additional eight individuals were found to be culture positive for C. jejuni (n = 5) and C. coli (n = 3) at AAFC, but two C. jejuni culture-positive samples at the CRH were not detected at AAFC. No cryptic taxa of campylobacteria were isolated at the CRH using conventional isolation. However, isolates of C. concisus, C. curvus, C. fetus, C. showae, and Helicobacter canadensis were isolated from diarrheic patients by using specialized isolation methods, albeit at low frequencies (0.2% of samples). Other enteric bacterial pathogens were isolated relatively infrequently during the study period (Table 2). Four diarrheic individuals were infected with Aeromonas spp. (0.5%) or P. shigelloides (0.5%), and 3 and 12 individuals were infected by E. coli O157:H7 (0.7%) or S. enterica (2.7%), respectively. Samples from healthy humans were typically processed within 6 h of stool collection, but no campylobacteria were isolated using the specialized isolation methods.
TABLE 2.
Prevalence of campylobacteria and other bacteria in humans living in the CHR (May to October 2005)
| Group and species | % prevalencea (no. of positive individuals) among diarrheic and healthy groups, based on indicated test |
|||||||
|---|---|---|---|---|---|---|---|---|
| Diarrheic (n = 442)b |
Healthy (n = 58)c |
|||||||
| Conventional | Specialized | PCR | Sequenced,e | Total | PCR | Sequenced,f | Total | |
| Campylobacteria | ||||||||
| Campylobacter genus | 16.5 (73) | 18.1 (80) | 73.5 (325) | —g | 74.2 (328) | 87.9 (51)* | — | 87.9 (51) |
| Campylobacter coli | 0.2 (1) | 0.9 (4) | 5.2 (23) | 0.2 (1) | 5.4 (24) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter concisus 1 | 0.0 (0) | 0.2 (1) | 30.5 (135) | 2.5 (17) | 34.3 (152) | 56.9 (33)** | 1.7 (1) | 56.9 (33) |
| Campylobacter concisus 2 | 0.0 (0) | 0.2 (1) | 4.3 (19) | 2.5 (17) | 8.1 (36) | 19.0 (11)** | 1.7 (1) | 20.7 (12) |
| Campylobacter curvus | 0.0 (0) | 0.2 (1) | 0.9 (4) | 0.7 (3) | 1.8 (8) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter fetus | 0.0 (0) | 0.2 (1) | 0.0 (0) | 0.0 (0) | 0.2 (1) | 3.4 (2)* | 0.0 (0) | 3.4 (2) |
| Campylobacter gracilis | 0.0 (0) | 0.0 (0) | 3.4 (15) | 7.0 (31) | 9.7 (43) | 12.1 (7) | 5.2 (3) | 17.2 (10) |
| Campylobacter helveticus | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.5 (2) | 0.5 (2) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter hominis | 0.0 (0) | 0.0 (0) | 7.2 (32) | 2.7 (12) | 9.7 (43) | 17.2 (10)* | 15.5 (9) | 31.0 (18) |
| Campylobacter hyointestinalis | 0.0 (0) | 0.0 (0) | 1.8 (8) | 0.0 (0) | 1.8 (8) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter insulaenigrae | 0.0 (0) | 0.0 (0) | — | 7.2 (32) | 7.2 (32) | — | 0.0 (0) | 0.0 (0) |
| Campylobacter jejuni | 16.3 (72) | 17.0 (75) | 29.2 (128)** | 7.4 (33) | 36.9 (163) | 3.4 (2) | 0.0 (0) | 3.4 (2) |
| Campylobacter lanienae | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter lari | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter mucosalis | 0.0 (0) | 0.0 (0) | 0.2 (1) | 0.0 (0) | 0.2 (1) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter showae | 0.0 (0) | 0.2 (1) | 5.4 (24) | 0.9 (4) | 6.6 (29) | 6.9 (4) | 3.4 (2) | 8.6 (5) |
| Campylobacter sputorum 1 | 0.0 (0) | 0.0 (0) | 3.2 (14) | 0.2 (1) | 3.4 (15) | 12.1 (7)** | 0.0 (0) | 12.7 (7) |
| Campylobacter sputorum 2 | 0.0 (0) | 0.0 (0) | 0.0 (0) | 0.2 (1) | 0.2 (1) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter upsalienesis | 0.0 (0) | 0.0 (0) | 0.7 (3) | 0.0 (0) | 0.7 (3) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Campylobacter ureolyticus | 0.0 (0) | 0.0 (0) | — | 2.9 (13) | 2.9 (13) | — | 1.7 (1) | 1.7 (1) |
| Other bacteria | ||||||||
| Aeromonas spp. | 0.5 (2) | — | — | — | 0.5 (2) | — | — | — |
| Escherichia coli O157:H7 | 0.7 (3) | — | — | — | 0.7 (3) | — | — | — |
| Helicobacter canadensis | 0.0 (0) | 0.2 (1) | — | — | 0.2 (1) | — | — | — |
| Plesiomonas shigelloides | 0.5 (2) | — | — | — | 0.5 (2) | — | — | — |
| Salmonella enterica | 2.7 (12) | — | — | — | 2.7 (12) | — | — | — |
*, detection rate was significantly higher (P ≤ 0.05 but >0.01) relative to the corresponding diarrheic or healthy human control treatment; **, detection rate was significantly higher (P ≤ 0.01) relative to the corresponding diarrheic or healthy human control treatment.
Bacteria in stools from diarrheic individuals were detected by direct PCR or specialized and/or conventional isolation (see text for additional information).
No campylobacteria were isolated from the stools of healthy individuals.
Based on sequencing of Campylobacter genus amplicons (i.e., those that were PCR negative for species).
Sequence-based identification was definite for 0/1 C. coli, 15/17 C. concisus, 1/3 C. curvus, 29/31 C. gracilis, 0/2 C. helveticus, 12/12 C. hominis, 0/32 C. insulaenigrae, 0/33 C. jejuni, 0/4 C. showae, 0/1 C. sputorum, and 13/13 C. ureolyticus isolates.
Sequence-based identification was definite for 1/1 C. concisus, 2/3 C. gracilis, 0/2 C. showae, and 1/1 C. ureolyticus isolates.
—, not determined.
(ii) Direct PCR detection.
In excess of 22,000 PCRs were conducted on 590 stool samples. A large number of stool samples from diarrheic (73.5%; n = 325) and healthy (87.9%; n = 51) individuals were positive for Campylobacter DNA (Table 2). The prevalence of Campylobacter DNA in stools was slightly higher for healthy humans than for diarrheic humans (P = 0.015).
Using direct PCR, C. jejuni and C. coli DNA was detected in 29.2% (n = 128) and 5.2% (n = 23) of stool samples from diarrheic individuals, respectively (Table 2). An increase (P < 0.002) in the number of individuals infected by C. jejuni and C. coli was detected using PCR relative to culture detection. A total of 54 and 19 individuals that were culture negative for C. jejuni and C. coli were PCR positive for these two taxa, respectively. PCR provided a negative result for only three individuals deemed to be infected with C. jejuni by culture (2.3% of individuals positive for the bacterium) and for no individuals that were culture positive for C. coli. Significantly more diarrheic than healthy humans were positive for C. jejuni (P < 0.001), and individuals in the diarrheic relative to the control group were at an eight-times-higher risk of infection with C. jejuni. Infection with C. jejuni during the course of the study (i.e., by PCR) was 11 times more common than infection by S. enterica and 44 times more common than infection by E. coli O157:H7. Two healthy individuals (3.4%) were positive for C. jejuni DNA. One of these individuals had been confirmed infected by C. jejuni ≈6 months previous to the sample submission; the infection occurred during travel outside Canada. The second positive control individual had never been diagnosed with campylobacteriosis. Although none of the samples provided by healthy volunteers was positive for C. coli DNA, there was no difference (P = 0.094) in infection rates between the two groups. Five diarrheic individuals (1.1%) were determined by PCR to be infected with both C. jejuni and C. coli. Of the 19 individuals that submitted multiple stool samples (i.e., in which at least one of the samples was positive for C. jejuni by culture or PCR), C. jejuni was not detected in all submitted samples for a majority of these individuals (n = 14).
Direct sequencing of Campylobacter genus amplicons (i.e., that were negative for taxon-specific PCR) indicated that an additional 33 individuals may have been infected with C. jejuni, resulting in an overall infection rate of 36.9% (n = 163) (Table 2). However, it was not possible to definitely distinguish these amplicons from C. insulaenigrae.
DNA of C. concisus, C. curvus, C. fetus, C. gracilis, C. helveticus, C. hominis, C. hyointestinalis, C. insulaenigrae, C. mucosalis, C. showae, C. sputorum, C. upsaliensis, and C. ureolyticus was detected at various frequencies in stools from diarrheic and healthy individuals (Table 2). For both C. concisus primer sets, a higher number (P < 0.001) of healthy than diarrheic humans were positive for DNA of this bacterium. The prevalence of C. curvus, C. fetus, C. gracilis, C. helveticus, C. hominis, C. hyointestinalis, C. mucosalis, C. showae, C. sputorum, and C. upsaliensis was either not significantly different (P > 0.05) or it was significantly lower (P ≤ 0.05) for diarrheic compared to healthy individuals. No C. lanienae or C. lari DNA was detected in stools of either group. A relatively high prevalence of samples from diarrheic individuals (25.6%; n = 113) were Campylobacter genus positive but species negative by taxon-specific PCR. DNA sequencing of these genus amplicons presumptively identified them as C. concisus (n = 17), C. curvus (n = 3), C. gracilis (n = 31), C. helveticus (n = 2), C. hominis (n = 12), C. insulaenigrae (n = 32), C. showae (n = 4), C. sputorum (n = 1), and C. ureolyticus (n = 13) (Table 2).
(iii) Spatial and temporal prevalence of campylobacteriosis.
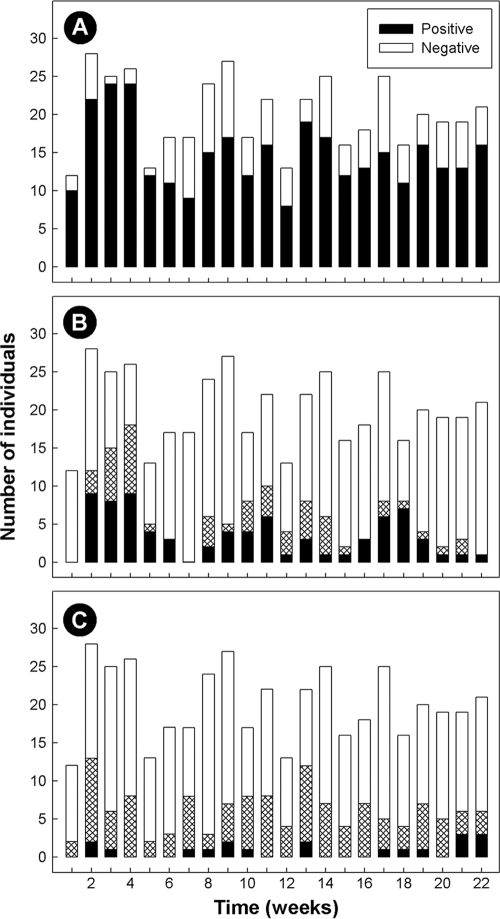
The majority of individuals infected with C. jejuni lived in the three urban centers within the CHR (63.4%; n = 83), primarily Lethbridge (51.9%; n = 68). Although the majority of samples submitted to the CRH during the study period came from urbanites, C. jejuni infection rates adjusted by sample submissions were nearly identical for individuals living in urban (30.4%) versus rural (28.6%) locations. Infection with C. jejuni was endemic over the study period, but infection rates varied over time (P = 0.005). A larger number of infections with C. jejuni occurred during weeks 3 and 4 (Fig. 3). In contrast to C. jejuni, detection rates for campylobacteria collectively (P = 0.99) or C. concisus (P = 0.56) did not vary over time. The prevalence of C. concisus/C. showae in stools was similar for people living in urban (32.0%) versus rural (29.1%) locations.
FIG. 3.
Temporal Campylobacter detection rates in humans living in the former Chinook Health Region from 31 May to 31 October 2005 (n = 442). (A) Campylobacter genus; (B) C. jejuni; (C) C. concisus and/or C. showae. In panel B, black bars indicate culture-positive detection of C. jejuni, hatched bars indicate PCR-positive but culture-negative results for C. jejuni, and open bars indicate negative individuals. In panel C, black bars indicate positive PCR detection of C. concisus based on the nested cpn60 primers, hatched bars indicate positive PCR detection of C. concisus/C. showae using the 23S rRNA primers, and open bars indicate negative individuals.
Detection of enteric RNA viruses.
None of the healthy humans was positive for enteric RNA viruses (Table 3). Stools from 0.2 to 2.3% of diarrheic humans were confirmed positive for RV, SaV/NoV, and GI and GII NoV during the study period. All of the GI or GII NoV TaqMan RT-qPCR positive samples were confirmed positive. In contrast, conventional RT-PCR rendered a substantive number of false-negative results (i.e., 7 to 23 individuals). The single confirmed RV infection occurred in early June, and the confirmed NoV infections occurred from 20 July to 17 October. No diarrheic humans were positive for bovine enteric calicivirus (BEC). None of the individuals that were culture positive for bacterial pathogens was positive for viral RNA. However, two samples that were positive for NoV GII were also PCR positive for C. jejuni.
TABLE 3.
Prevalence of enteric RNA viruses in humans living in the CHR (May to October 2005)
| Virus | % prevalence (no. of individuals) with indicated result among diarrheic and healthy groups |
|||
|---|---|---|---|---|
| Diarrheic (n = 442) |
Healthy (n = 58) |
|||
| Positive | Confirmeda | Positive | Confirmeda | |
| Sapovirus/norovirusb | 4.5 (20)c | 1.6 (7)e | 0.0 (0) | —i |
| Norovirus GId | 0.2 (1) | 0.2 (1)f | 0.0 (0) | — |
| Norovirus GIId | 2.3 (10) | 2.3 (10)g | 0.0 (0) | — |
| Rotavirusb | 1.8 (8) | 0.2 (1)h | 0.0 (0) | — |
| Bovine enteric calicivirus (CBECU)b | 4.5 (20) | 0.0 (0) | 0.0 (0) | — |
| Bovine enteric calicivirus (NBU)b | 5.2 (23) | 0.0 (0) | 0.0 (0) | — |
DNA sequence was confirmed.
Viruses were detected using conventional RT-PCR with the CBECU or NBU primers.
Virus was detected using TaqMan RT-qPCR.
Sequence was confirmed as NoV GII (four individuals) or NoV GII.4 (three individuals).
Sequence was confirmed as NoV GI.
Sequence was confirmed as NoV GII (six individuals) or NoV GII.4 (four individuals).
Sequence was confirmed as RV group A.
—, not applicable.
DISCUSSION
Primer development and validation.
Direct detection of nucleic acids of specific taxa in feces by PCR is not subject to the inherent limitations of culture-based diagnostic methods, and for this reason the use of PCR detection methods is a very attractive alternative to culture-based methods to detect campylobacteria. While PCR possesses a number of conspicuous advantages to culture-based diagnosis, the Achilles heel of PCR-based diagnostics is the occurrence of both false-positive and false-negative results. Regarding the latter, the inclusion of an IAC is imperative to ensure PCR inhibitors have been adequately removed (7), and we used a qualitative IAC in concert with primers targeting the 16S rRNA gene to detect Campylobacter genus DNA (32, 46). The specificity and sensitivity of PCR are essential components of diagnostic PCR. We chose to use nested PCR to address both specificity and sensitivity. Nested PCR has been used with various degrees of success to increase the sensitivity of detection, particularly for genes with one or a small number of copies (32, 34, 57, 66). All PCR possesses a detection threshold, and we observed that the nested primers that we developed for C. concisus and C. upsaliensis were very sensitive (i.e., a detection threshold of 102 CFU/g, or ≈20 cells in a 200-mg subsample of stool) and were substantially more sensitive (≈10 to 100 times) than the nonnested PCR primers described by others (12, 46, 68). The initial primer set that we developed for C. concisus targeted the 23S rRNA gene, and although these primers were very sensitive, they did not distinguish C. concisus from C. showae. The subsequent nested primer set that we developed targeted the cpn60 gene, which is present as a single copy in most organisms (18), as opposed to the multicopy 23S rRNA gene. Consistent with increased sensitivity based on gene copy, the cpn60 primer sets were found to be somewhat less sensitive than the nested primers that targeted the multicopy 23S rRNA gene. It is thus likely that detection rates of both C. concisus and C. showae were underestimated by targeting the cpn60 gene, and we observed that C. concisus and C. showae were detected at rates of 4.3% and 5.4%, respectively (≈10%, collectively), which was approximately three times less than carriage rates indicated by the nested primers targeting the 23S rRNA gene. High sensitivity is particularly important, given that DNA must be extracted from relatively small subsamples from stools (e.g., due to the presence of PCR inhibitors), that Campylobacter cells are not uniformly distributed throughout the stool and subsamples may thus contain a low density of cells, and that cells may be shed periodically. Consistent with the two latter points, we observed inconsistent detection of C. jejuni in stool samples submitted by the same individual on separate occasions. Collectively, our findings disagree with those of Chaban et al. (12), who concluded that low primer sensitivity would be compensated for by the large numbers of Campylobacter cells that are shed from infected animals. While the high sensitivity of PCR is beneficial, it is also important to emphasize that elevated sensitivity can be a concern. For example, the introduction of spurious template DNA from the primary into the secondary reaction mixture will result in false positives. Thus, the inclusion of negative template controls is mandatory, and extreme care must always be exercised in handling the template from the primary reaction. Primer comprehensiveness is also an important issue to be considered for diagnostic PCR to guard against false negatives. This is particularly important, given that we currently possess an incomplete understanding of the phylogenetics of most Campylobacter species. For example, C. concisus is a genetically complex taxon (1), and we found that the nested primers targeting the C. concisus cpn60 gene only provided weak amplicons for strains not belonging to genomospecies A or B. Although it is unlikely that nested PCR will ever be used in diagnostic facilities as a routine detection tool (for logistical reasons), our study demonstrates that nested PCR is an effective research tool due to its sensitivity and specificity.
Infections with C. jejuni and C. coli in the Chinook Health Region.
Campylobacter jejuni is recognized as the most common cause of bacterial enteritis in Canada (55). The majority of cases of campylobacteriosis (≥95%) in the CHR are attributed to C. jejuni, with the remainder attributed to C. coli (based on conventional isolation). We hypothesized that a significant number of diarrheic individuals infected by C. jejuni and C. coli are not diagnosed using culture-based methods, and direct PCR detection would provide a more accurate measure of infection rates by these bacteria. In the current study, we contrasted conventional and specialized isolation methods with PCR to detect C. jejuni and C. coli in stools. Our findings indicated that there was no advantage to utilizing specialized isolation methods to detect C. jejuni and C. coli relative to the culture-based method currently used in the CHR. However, PCR-based detection revealed that a substantive number of infections by C. jejuni (n = 54; 12.2%) and C. coli (n = 19; 4.3%) were missed by culture, with overall infection rates of 29.6% and 5.2%, respectively. Others have also demonstrated that PCR detects a significant number of infections with C. jejuni and C. coli that are missed by culture (3, 59), albeit at much lower rates than we observed in the CHR. In contrast, Kulkarni et al. (37) and Lawson et al. (43) observed no significant difference between PCR-based and culture-based methods for detection of C. jejuni and C. coli. Different PCR methods were used in the above studies, confounding direct comparisons. However, it is possible that our use of nested PCR, which is typically much more sensitive than nonnested PCR methods, contributed to the very high number of cases of infection that were missed by culture-based detection. Some evidence indicated that the nested PCR that we used to detect C. jejuni and C. coli was not absolute. Sequence-based identification of Campylobacter genus amplicons (i.e., for samples in which no species were identified by PCR or culture) indicated that at least an additional 34 individuals may have been infected with C. jejuni or C. coli. Our findings using PCR indicated that a significant number of infections with C. jejuni and C. coli are missed by culture, and they affirm the importance of C. jejuni and to a lesser extent C. coli as enteric pathogens of humans in the CHR. Although the current study was conducted during a time of the year when campylobacteriosis typically peaks in temperate climates (51, 54), the exceptionally high rates of C. jejuni and C. coli infection of humans living within the CHR during this period (≈40% of diarrheic individuals submitting stools for diagnosis) are cause for concern.
Direct detection of C. jejuni and C. coli in stools by PCR revealed unique information on coinfections and carriage by healthy humans. Using PCR, we detected five culture-negative individuals (1.1%) who were infected by both C. jejuni and C. coli. Although Lawson et al. (43) did not observe a significant difference between PCR-based and culture-based detection, they did conclude that PCR provided unique information on mixed infections, consistent with our findings. We also observed that two healthy control individuals (3.4%) were positive for C. jejuni by PCR but not by culture; one individual had been confirmed infected with C. jejuni approximately 6 months previous to the sample submission date. Although the majority of cases of C. jejuni are considered self-limiting, evidence suggests that a relatively small percentage of asymptomatic individuals living in the Western world are positive for the bacterium. For example, Amar et al. (3) observed that ≈1% of 2,205 healthy individuals examined in England were positive for C. jejuni. In developing countries, infection in asymptomatic individuals can be much higher (15). Furthermore, recrudescent infection by C. jejuni has been documented in immunocompetent individuals (6). These observations raise questions regarding the importance of asymptomatic and/or recrudescently infected humans in the epidemiology of campylobacteriosis, as well as to the complexity of host responses to C. jejuni and C. coli taxa, and in particular host responses to specific genotypes.
Epidemiology of campylobacteriosis in the Chinook Health Region.
The epidemiology of campylobacteriosis is poorly understood at present. As indicated previously, the CHR consistently possesses a relatively high rate of campylobacteriosis among its human inhabitants. For example, culture-based infection rates for C. jejuni and C. coli in the CHR from 1988 to 2005 ranged from 60 to 100 cases per 100,000 individuals and averaged 215 cases per 100,000 individuals from 2004 to 2006 (26). These rates are comparable to culture-based infection rates observed in Australia (64), but they are substantially lower than the rate of campylobacteriosis observed in New Zealand (5). In addition to the high rate of infection by C. jejuni that we observed in the current study (≥29%), infections by this bacterium were found to be endemic during the research period. Reasons for the high rates and endemicity of campylobacteriosis within the CHR are currently unknown. Food-borne infection alone does not explain this occurrence, as food habits among people living in the northern portion of Alberta (e.g., in the Capital Health Region, ≈500 km north of the CHR), where campylobacteriosis rates are much lower (26), are similar to people living in the southern portion of the province. The CHR covers a large area (≈26,000 km2) and contains a population of ≈150,000 humans living in a blend of rural (54%) and urban (46%) areas (14). A unique characteristic of the CHR is the large population of livestock, in particular, beef cattle. There are ≈1.28 million cattle in the CHR at any given time, with ≈700,000 cattle in confined feeding operations (2). We estimate that this number of cattle produces ≈40 million kg (fresh weight) of manure per day, and this amount constitutes ≈96% of the manure produced in the CHR relative to swine, poultry, and humans (data not presented). Beef cattle readily shed C. jejuni cells in their feces (33), and an epidemiological study indicated that direct contact with cattle was a significant risk factor for campylobacteriosis in the CHR (30). This led us to hypothesize that individuals living in rural areas would be infected with C. jejuni at a higher rate than those living in urban centers. Although the majority of infections occurred in people living in urban centers, contrary to our hypothesis, we observed that campylobacteriosis rates (adjusted by differential sample size) were the same among individuals living in rural (28.6%) or urban (30.4%) locations. A limitation of our study design was our inability to follow up with diarrheic individuals and, thus, we had no way of ascertaining the degree of animal contact. Nonetheless, our findings raise questions with regard to the epidemiology of campylobacteriosis within the CHR and other health regions. In particular, what role do beef cattle play in disease and what are the mechanisms by which C. jejuni is transmitted to humans?
Prevalence of cryptic campylobacteria in stools.
While C. jejuni and to a lesser extent C. coli are recognized as the primary causes of campylobacteriosis, the clinical relevance of other Campylobacter species, including cryptic campylobacteria, is currently uncertain. We hypothesized that conventional isolation methods are ineffective in detecting cryptic campylobacteria, which infect a significant number of humans living within the CHR, thereby contributing to the high rates of enteritis in this region. The application of specialized isolation strategies has demonstrated that a variety of Campylobacter species are common in stools from diarrheic patients (19, 41, 42, 47). For example, the use of a membrane filtration method to isolate campylobacteria from stools of pediatric patients with enteritis demonstrated that C. concisus and C. upsaliensis were isolated at similar frequencies to C. jejuni (39). However, the pathogenicity of cryptic campylobacteria is poorly understood, confounded by both the lack of efficient isolation media and the lack of controlled clinical studies. Although the merits of membrane filtration for isolating fastidious campylobacteria are recognized, it must be realized that this method is not a panacea, as the cells must be highly motile and the method is still subject to many of the inherent limitations of culture-based detection. This was demonstrated by the comparatively low frequency at which we isolated campylobacteria using membrane filtration in the current study. It is possible that the delay in processing diarrheic samples, the low hydrogen isolation atmosphere (3%), the type of membrane that we used, and the slow growth of cryptic campylobacteria all contributed to our poor success in isolating these fastidious bacteria (A. J. Lastovica, personal communication). Other factors may also have contributed to our poor culturing success. For example, the vast majority of stools were maintained in Cary-Blair transport medium at low temperatures before processing. Our findings emphasize that no one medium or method will provide an accurate measure of cryptic campylobacteria in stools.
Very limited information is available on the pathogenicity of cryptic taxa of campylobacteria. In the current study, we used direct PCR to contrast the prevalence of C. concisus, C. curvus, C. fetus, C. gracilis, C. helveticus, C. hominis, C. hyointestinalis, C. lanienae, C. lari, C. mucosalis, C. showae, C. sputorum, and C. upsaliensis in stools from diarrheic and healthy adults living in the CHR. We observed that a majority of individuals examined were positive for Campylobacter DNA (≥73.5%) and that a higher prevalence of healthy versus diarrheic humans was positive for Campylobacter DNA in their stools. These observations are inconsistent with a pathogenic role for campylobacteria collectively. Campylobacter concisus has been suggested to incite enteritis in children (40), and recent evidence has suggested that C. concisus may be involved in the etiology of Crohn's disease in Australian children (48, 69). We observed no differences in carriage of C. concisus between diarrheic and healthy humans, and consistent with our findings, Engberg et al. (19) and Van Etterijck et al. (65) concluded that C. concisus is a commensal in the human intestine. However, given that C. concisus is a genetically heterogenous taxon (1), our findings do not preclude the possibility that pathogenic genotypes exist within this heterogenous species. In this regard, we recently observed that C. concisus genomospecies differed in their pathogenic potential (unpublished data). Further studies are needed to examine the molecular genetics and pathogenicity (e.g., in vivo assessments) of C. concisus. Campylobacter upsaliensis has been commonly isolated from children in Sweden (45) and South Africa (39), and this bacterium is considered to be an emerging cause of enteritis, albeit currently of unknown significance (24, 35, 38, 42). While no C. upsaliensis was detected in the stools of healthy individuals in the current study, only three diarrheic individuals (0.7%) were positive for the bacterium. Campylobacter lanienae and C. lari were not detected in either group, and the prevalence of C. curvus, C. fetus, C. gracilis, C. helveticus, C. hominis, C. hyointestinalis, C. mucosalis, C. showae, and C. sputorum in stools was either not significantly different between the diarrheic and healthy groups or detection rates were significantly higher in the healthy group. Thus, our data indicate that C. concisus, C. curvus, C. fetus, C. gracilis, C. helveticus, C. hominis, C. hyointestinalis, C. lanienae, C. lari, C. mucosalis, C. showae, C. sputorum, and C. upsaliensis were not enteric pathogens of significance from a community perspective among adults living in the CHR during the study period.
Infections by enteric viruses in the Chinook Health Region.
We hypothesized that enteric RNA viruses are underreported and infect a substantial number of human inhabitants of the CHR during the summer and early fall. Over the study period, we detected relatively low rates of infection by enteric viruses. However, 19 undiagnosed individuals were determined to be infected by RNA enteric viruses. The majority of NoV and RV infections occur in fall and winter months (i.e., October through March) (62, 63). The relatively low prevalence of NoV GI and GII and of RV infections (2.7%) that we detected in diarrheic stool samples during the study period are consistent with low rates of infection observed in the summer and early fall in temperate climates. However, summer peaks of NoV infection (June to September) have been observed in children in Brazil, Peru, Spain, and Tunisia (10, 53, 60, 67). NoV and RV are highly infectious (with ≈10 to 1,000 virus particles) and are readily transmitted via person-to-person contact (22, 27). The current study showed that infected individuals exist within the community during the summer and early fall, and given the highly infectious nature of these viruses it is uncertain why NoV and RV outbreaks do not occur during the spring and summer within the CHR and elsewhere. As indicated previously, the CHR contains a very high beef cattle density (2), yet we did not detect any BEC in diarrheic or healthy individuals in the current study. Whether this indicates that zoonotic transmission of BEC strains to humans does not occur, or occurs at very low frequencies, is uncertain and warrants investigation.
Conclusions.
We examined the presence of Campylobacter species and enteric RNA viruses in stools of diarrheic and healthy humans living in southwestern Alberta over a 5-month period (May to October 2005). Specialized isolation and direct nested PCR were employed to detect campylobacteria, and in excess of 22,000 PCRs were conducted. We observed a very high rate of infection with C. jejuni (29% of diarrheic individuals) and to a lesser extent by C. coli (5%), and we found that a high number of infections by these bacteria (≥68 individuals) were not detected using a conventional culture-based method. Infection by C. jejuni was endemic during the study period, and further research is needed to elucidate the epidemiology of campylobacteriosis in the CHR, including the identification of important reservoirs of infectious cells and the mechanisms of their transmission to humans. In contrast to C. jejuni, evidence indicated that C. concisus, C. curvus, C. fetus, C. gracilis, C. helveticus, C. hominis, C. hyointestinalis, C. lanienae, C. lari, C. mucosalis, C. showae, C. sputorum, C. upsaliensis, rotavirus, sapovirus, bovine enteric calicivirus, and norovirus (GI and GII) were not significant pathogens of adults within the CHR during the study period. However, further research is needed to examine infection rates over a protracted period and to empirically examine the pathogenicity of cryptic Campylobacter species.
Acknowledgments
We thank the following people for their assistance: Kathaleen House (AAFC, Lethbridge) for evaluating the new primers, extracting DNA from stool samples, conducting the diagnostic PCR and sequencing of amplicons, and for assisting with the isolation and identification of campylobacteria; Jenny Gusse (AAFC, Lethbridge) for designing and evaluating the primers for C. concisus and C. upsaliensis; Danielle Leblanc (AAFC, St. Hyacinthe) for RNA extraction and conducting conventional RT-PCR for RNA viruses; Elyse Poitras (AAFC, St. Hyacinthe) for conducting qPCR for NoV GI and GII; Pierre Ward (AAFC, St. Hyacinthe) for cloning, sequencing, and confirmation of viral amplicons; Judy Baxter, Deborah Sweeny, and the staff of the CRH Department of Laboratory Medicine for their enthusiastic assistance with the project; Lisa Vandergouwe and Rod MacKay (CRH) for approving and supporting the project; Toby Entz (AAFC, Lethbridge) for statistical advice; Albert Lastovica (University of the Western Cape, Cape Town, South Africa) for kindly providing clinical isolates of C. concisus and C. upsaliensis; Brent Selinger (University of Lethbridge) for facilitating access to the University of Lethbridge Human Subject Research Committee; and all of the human volunteers for providing stool samples and for their interest in the study.
Financial support for this project was provided by grants from AAFC (Growing Forward) to G.D.I. and A.H., the Advanced Foods and Materials Network to G.D.I., and the Canada-Alberta Beef Industry Development Fund to G.D.I.
Footnotes
Published ahead of print on 24 November 2010.
REFERENCES
- 1.Aabenhus, R., S. L. On, B. L. Siemer, H. Permin, and L. P. Andersen. 2005. Delineation of Campylobacter concisus genomospecies by amplified fragment length polymorphism analysis and correlation of results with clinical data. J. Clin. Microbiol. 43:5091-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberta Agriculture, Food and Rural Development. 2001. 2001 census of agriculture for Alberta, I.D., M.D., and county data by region. Agdex 852-1. Government of Alberta, Agriculture and Rural Development, Edmonton, Alberta, Canada.
- 3.Amar, C. F., et al. 2007. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993-1996). Eur. J. Clin. Microbiol. Infect. Dis. 26:311-323. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, A. D., et al. 2003. A waterborne outbreak of Norwalk-like virus among snowmobilers—Wyoming 2001. J. Infect. Dis. 187:303-306. [DOI] [PubMed] [Google Scholar]
- 5.Baker, M. G., E. Sneyd, and N. A. Wilson. 2007. Is the major increase in notified campylobacteriosis in New Zealand real? Epidemiol. Infect. 135:163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baqar, S., et al. 2010. Recrudescent Campylobacter jejuni infection in an immunocompetent adult following experimental infection with a well-characterized organism. Clin. Vaccine Immunol. 17:80-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkham, T., and J. Hoorfar. 2004. Internal amplification control for PCR should not be mandatory in the clinical medical environment. J. Clin. Microbiol. 42:3379-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastyns, K., S. Chapelle, P. Vandamme, H. Goossens, and R. de Wachter. 1994. Species-specific detection of campylobacters important in veterinary medicine by PCR amplification of 23S rDNA areas. Syst. Appl. Microbiol. 17:563-568. [Google Scholar]
- 9.Bastyns, K., S. Chapelle, P. Vandamme, H. Goossens, and R. De Wachter. 1995. Specific detection of Campylobacter concisus by PCR amplification of 23S rDNA areas. Mol. Cell. Probes 9:247-250. [DOI] [PubMed] [Google Scholar]
- 10.Boga, J. A., et al. 2004. Etiology of sporadic cases of pediatric acute gastroenteritis in Asturias, Spain, and genotyping and characterization of norovirus strains involved. J. Clin. Microbiol. 42:2668-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cary, S. G., and E. B. Blair. 1964. New transport medium for shipment of clinical specimens. I. Fecal specimens. J. Bacteriol. 88:96-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaban, B., K. M. Musil, C. G. Himsworth, and J. E. Hill. 2009. Development of cpn60-based real-time quantitative PCR assays for the detection of 14 Campylobacter species and application to screening of canine fecal samples. Appl. Environ. Microbiol. 75:3055-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapin, K. C., and T. L. Lauderdale. 2003. Reagents, stains and media: bacteriology, p. 354-383. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th 3ed. American Society for Microbiology, Washington, DC.
- 14.Chinook Health Region. 2003. Annual report 2002-2003. Chinook Health Region, Lethbridge, AB, Canada. http://www.chr.ab.ca/about_chr/chr%20annual%20report%2002-03.pdf.
- 15.Coker, A. O., et al. 2002. Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Boer, E., J. J. Tilburg, D. L. Woodward, H. Lior, and W. M. Johnson. 1996. A selective medium for the isolation of Arcobacter from meats. Lett. Appl. Microbiol. 23:64-66. [DOI] [PubMed] [Google Scholar]
- 17.Denis, M., et al. 1999. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29:406-410. [DOI] [PubMed] [Google Scholar]
- 18.Dumonceaux, T. J., et al. 2006. Enumeration of specific bacterial populations in complex intestinal communities using quantitative PCR based on the chaperonin-60 target. J. Microbiol. Methods 64:46-62. [DOI] [PubMed] [Google Scholar]
- 19.Engberg, J., S. L. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eyers, M., S. Chapelle, G. Van Camp, H. Goossens, and R. De Wachter. 1993. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J. Clin. Microbiol. 31:3340-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farkas, T., et al. 2004. Genetic diversity among sapoviruses. Arch. Virol. 149:1309-1323. [DOI] [PubMed] [Google Scholar]
- 22.Glass, R. I., U. D. Parashar, and M. K. Estes. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez, I., K. A. Grant, P. T. Richardson, S. F. Park, and M. D. Collins. 1997. Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J. Clin. Microbiol. 35:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goossens, H., et al. 1990. Is “Campylobacter upsaliensis” an unrecognised cause of human diarrhoea? Lancet 335:584-586. [DOI] [PubMed] [Google Scholar]
- 25.Gouvea, V., et al. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Government of Alberta Health and Wellness. 2007. Health trends in Alberta. Alberta Health and Wellness, Edmonton, AB, Canada. http://www.health.alberta.ca/newsroom/health-trends.html.
- 27.Graham, D. Y., G. R. Dufour, and M. K. Estes. 1987. Minimal infective dose of rotavirus. Arch. Virol. 92:261-271. [DOI] [PubMed] [Google Scholar]
- 28.Harmon, K. M., and I. V. Wesley. 1996. Identification of Arcobacter isolates by PCR. Lett. Appl. Microbiol. 23:241-244. [DOI] [PubMed] [Google Scholar]
- 29.Harper, C. M. G., et al. 2000. Isolation and characterization of a Helicobacter sp. from the gastric mucosa of dolphins, Lagenorhynchus acutus and Delphinus delphis. Appl. Environ. Microbiol. 66:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasselback, P. 2002. Feedlot alley and enteric illness: are they related or is southern Alberta just a wonderful place for humans, cattle and bugs to live? Canadian Laboratory Medicine Congress, Calgary, AB, Canada.
- 31.Houde, A., et al. 2006. Comparative evaluation of RT-PCR, nucleic acid sequence based amplification (NASBA) and real-time RT-PCR for detection of noroviruses in faecal material. J. Virol. Methods 135:163-172. [DOI] [PubMed] [Google Scholar]
- 32.Inglis, G. D., and L. D. Kalischuk. 2003. Use of PCR for direct detection of Campylobacter species in bovine feces. Appl. Environ. Microbiol. 69:3435-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inglis, G. D., et al. 2006. Antimicrobial resistance of Campylobacter species from beef cattle in Alberta feedlots. Appl. Environ. Microbiol. 72:4088-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang, C., et al. 1998. Identification of H. pylori in saliva by a nested PCR assay derived from a newly cloned DNA probe. Dig. Dis. Sci. 43:1211-1218. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez, S. G., R. G. Heine, P. B. Ward, and R. M. Robins-Browne. 1999. Campylobacter upsaliensis gastroenteritis in childhood. Pediatr. Infect. Dis. J. 18:988-992. [DOI] [PubMed] [Google Scholar]
- 36.Kageyama, T., et al. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulkarni, S. P., et al. 2002. Detection of Camplylobacter species: a comparison of culture and polymerase chain reaction based methods. J. Clin. Pathol. 55:749-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labarca, J. A., et al. 2002. Campylobacter upsaliensis: another pathogen for consideration in the United States. Clin. Infect. Dis. 34:E59-E60. [DOI] [PubMed] [Google Scholar]
- 39.Lastovica, A. J. 2006. Emerging Campylobacter spp.: the tip of the iceberg. Clin. Microbiol. Newslett. 28:49-55. [Google Scholar]
- 40.Lastovica, A. J. 2009. Clinical relevance of Campylobacter concisus isolated from pediatric patients. J. Clin. Microbiol. 47:2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lastovica, A. J., and E. Le Roux. 2000. Efficient isolation of campylobacteria from stools. J. Clin. Microbiol. 38:2798-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lastovica, A. J., and E. Le Roux. 2001. Efficient isolation of Campylobacter upsaliensis from stools. J. Clin. Microbiol. 39:4222-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawson, A. J., J. M. Logan, G. L. O'Neill, M. Desai, and J. Stanley. 1999. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3860-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawson, A. J., M. S. Shafi, K. Pathak, and J. Stanley. 1998. Detection of campylobacter in gastroenteritis: comparison of direct PCR assay of faecal samples with selective culture. Epidemiol. Infect. 121:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindblom, G. B., E. Sjögren, J. Hansson-Westerberg, and B. Kaijser. 1995. Campylobacter upsaliensis, C. sputorum sputorum and C. concisus as common causes of diarrhoea in Swedish children. Scand. J. Infect. Dis. 27:187-188. [DOI] [PubMed] [Google Scholar]
- 46.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 47.Maher, M., et al. 2003. Evaluation of culture methods and a DNA probe-based PCR assay for detection of Campylobacter species in clinical specimens of feces. J. Clin. Microbiol. 41:2980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Man, S. M., et al. 2010. Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn's disease. Inflamm. Bowel Dis. 16:1008-1016. [DOI] [PubMed] [Google Scholar]
- 49.Martella, V., et al. 2003. Molecular analysis of the VP7, VP4, VP6, NSP4, and NSP5/6 genes of a buffalo rotavirus strain: identification of the rare P[3] rhesus rotavirus-like VP4 gene allele. J. Clin. Microbiol. 41:5665-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mead, P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meldrum, R. J., J. K. Griffiths, R. M. M. Smith, and M. R. Evans. 2005. The seasonality of human campylobacter infection and Campylobacter isolates from fresh, retail chicken in Wales. Epidemiol. Infect. 133:49-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray, P. R., E. J. Baron, J. Jorgensen, M. Pfaller, and M. L. Landry (ed.). 2007. Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 53.Parashawar, U. D., et al. 2004. Human caliciviruses as a cause of severe gastroenteritis in Peruvian children. J. Infect. Dis. 190:1088-1092. [DOI] [PubMed] [Google Scholar]
- 54.Patrick, M. E., et al. 2004. Effects of climate on incidence of Campylobacter spp. in humans and prevalence in broiler flocks in Denmark. Appl. Environ. Microbiol. 70:7474-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Public Health Agency of Canada. 2010. Notifiable diseases on-line. Public Health Agency of Canada, Ottawa, Canada. http://dsol-smed.phac-aspc.gc.ca/dsol-smed/ndis/index-eng.php.
- 56.Samie, A., C. L. Obi, L. J. Barrett, S. M. Powell, and R. L. Guerrant. 2007. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa: studies using molecular diagnostic methods. J. Infect. 54:558-566. [DOI] [PubMed] [Google Scholar]
- 57.Saruta, K., et al. 1997. Simultaneous detection of Streptococcus pneumoniae and Haemophilus influenzae by nested PCR amplification from cerebrospinal fluid samples. FEMS Immunol. Med. Microbiol. 19:151-157. [DOI] [PubMed] [Google Scholar]
- 58.SAS Institute. 2005. SAS OnlineDoc 9.1.3. SAS Institute, Cary, NC.
- 59.Schuurman, T., et al. 2007. Feasibility of a molecular screening method for detection of Salmonella enterica and Campylobacter jejuni in a routine community-based clinical microbiology laboratory. J. Clin. Microbiol. 45:3692-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sdiri-Loulizi, K., et al. 2008. Acute infantile gastroenteritis associated with human enteric viruses in Tunisia. J. Clin. Microbiol. 46:1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smiley, J. R., A. E. Hoet, M. Traven, H. Tsunemitsu, and L. J. Saif. 2003. Reverse transcription-PCR assay for detection of bovine enteric caliciviruses (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J. Clin. Microbiol. 41:3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svraka, S., et al. 2007. Etiological role of viruses in outbreaks of acute gastroenteritis in The Netherlands from 1994 through 2005. J. Clin. Microbiol. 45:1389-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tran, A., et al. 2010. Prevalence of rotavirus, adenovirus, norovirus, and astrovirus infections and coinfections among hospitalized children in northern France. J. Clin. Microbiol. 48:1943-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vally, H., G. Hall, E. Scallan, M. D. Kirk, and F. J. Angulo. 2009. Higher rate of culture-confirmed Campylobacter infections in Australia than in the USA: is this due to differences in healthcare-seeking behavior or stool culture frequency? Epidemiol. Infect. 137:1751-1758. [DOI] [PubMed] [Google Scholar]
- 65.Van Etterijck, R., et al. 1996. Isolation of Campylobacter concisus from feces of children with and without diarrhea. J. Clin. Microbiol. 34:2304-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verdin, E., C. Saillard, A. Labbe, J. M. Bove, and M. Kobisch. 2000. A nested PCR assay for detection of Mycoplasma hyopneumoniae in tracheobronchiolar washings from pigs. Vet. Microbiol. 76:31-40. [DOI] [PubMed] [Google Scholar]
- 67.Victoria, M., F. A. Carvalho-Costa, M. B. Heinemann, J. P. Leite, and M. Miagostovich. 2007. Prevalence and molecular epidemiology of noroviruses in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil, 2004. Pediatr. Infect. Dis. J. 26:602-606. [DOI] [PubMed] [Google Scholar]
- 68.Wang, G., et al. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40:4744-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, L., et al. 2009. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn's disease. J. Clin. Microbiol. 47:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]