Abstract
We ran a comparative analysis of all patients for whom a positive culture of Mycobacterium tuberculosis complex was available between April 2004 and October 2005 and whose HIV serology results were known, with spoligotyping results (n = 163) split into 49 HIV-positive patients and 114 HIV-negative patients. Spoligotype international type 373 (SIT373) (T1 lineage), which was highly prevalent among the HIV+ patients, was totally absent from the HIV− population, suggesting that we had a specific clone affecting nearly 1/3 of all HIV-tuberculosis (TB)-coinfected patients. Among the LAM10-CAM sublineage strains, we had only a single strain of SIT403 among HIV− patients (0.88%), as opposed to 12.25% of the HIV+ population (χ2 = 10.77; P < 0.01), indicating a strong association between the strain and the HIV+ population. The LAM10-CAM lineage spoligotype SIT61 was prevalent among the 2 subsets (37.72% in HIV− versus 12.24% in HIV+ populations), though, with a significant difference between the 2 groups (χ2 = 10.53; P < 0.01). However, there was no significant difference for SIT53 (T1 lineage) in the 2 subsets: 6.14 versus 8.2% (χ2 = 0.22; P > 0.05). A total of 7/49, or 14.3%, other SITs among HIV+ patients were not found among the HIV− patients. When added to the most prevalent SIT among HIV+ patients (SIT373; n = 16), 23/49, or 47%, isolates among HIV-TB-coinfected patients were unique. We conclude that further studies should be carried out to investigate the evolution of these genotypes and others in the emergence of multidrug resistance and control of tuberculosis in Nigeria.
With a population of over 140 million, Nigeria is faced with a huge burden of tuberculosis (TB) and is currently rated fourth among the TB-burdened nations of the world, with a prevalence of 521 per 100,000 population in the year 2007 (27). This situation is worsened by the country's HIV/AIDS epidemic, with a HIV prevalence in 2007 of 3.6% and a TB-HIV coinfection rate of 27% (25). Although the outcome of treatment was not evaluated for a high proportion of patients in 2007, the treatment success rate was 76%. Collaborative TB-HIV activities are also being scaled up, and 32% of TB cases are now screened for HIV (27). Nonetheless, the case detection rate of new smear-positive TB cases in Nigeria, which was 18% in 2005 (23% in 2007), remained unacceptably low (http://www.stoptb.org/assets/documents/countries/GlobalReport2009/nga.pdf). Unfortunately, only limited information is available on the genotypes of the Mycobacterium tuberculosis complex (MTC) isolated from Nigeria in comparison with those obtained globally, an important factor that is useful for understanding the global spread and phylogeographical specificity of predominant circulating clones of tubercle bacilli. In addition, molecular data obtained from the MTC so far in Nigeria have not addressed the issue of TB-HIV-coinfected patients, a subpopulation involved in the spread of the disease in Africa in general and Nigeria in particular.
Molecular typing techniques have been extensively used to speciate strains of incriminating MTC involved in TB infections; among these, spoligotyping, a method based on PCR amplification of a highly polymorphic direct-repeat locus in the MTC genome, has been extensively used for simultaneous detection and typing of these organisms (16). Indeed, due to its clonal structure (24), the comparative genotype analysis of MTCs from different human populations can give unique insights into the dissemination dynamics and evolutionary genetics of this pathogen (22); consequently, spoligotyping has been used to understand the emerging problem of multidrug-resistant (MDR) TB and the virulence of certain epidemic strains of M. tuberculosis (such as the Beijing strain), as well as to better comprehend the epidemiology of TB and TB-HIV coinfection (1-6, 13, 14). A previous study on molecular analysis of MTC in Nigeria did not address the issue of TB-HIV coinfection (9). The present study, therefore, aimed to investigate the spoligotyping-based population structure of MTC clinical isolates in HIV+ and HIV− patients in Nigeria and to compare the patterns obtained with those available in the SITVIT2 proprietary database of the Pasteur Institute of Guadeloupe, which contains genotyping information on about 75,000 M. tuberculosis clinical isolates from 160 countries of origin.
MATERIALS AND METHODS
Patients and specimens.
This retrospective study included a convenience sample of frozen heat-killed cell suspensions of MTC clinical isolates (n = 163) isolated from April 2004 to October 2005 in Ibadan, Oyo State, southwestern Nigeria, and might not be fully representative of the TB population structure in Ibadan. Indeed, culture is not done systematically in various government laboratories and TB referral centers in Ibadan, Nigeria, and smear microscopy is the accepted routine methodology carried out to treat TB patients, in conjunction with clinical details and sometimes X ray. The M. tuberculosis complex isolates were recovered from 1,303 pathological samples that were cultured from suspected TB patients whose HIV serology was known. A total of 176 samples yielded positive cultures, and 163 clinical isolates from as many TB patients (one strain per patient, split into HIV+ [n = 49] and HIV− [n = 114]) were available for this study, which represented the capture of 92.6% of all available cultures. The patients were recruited from different DOTS (directly observed therapy—short course) centers in Ibadan. All of the strains were identified as MTC at the time of isolation, following culture and standard microbiological procedures at the Tuberculosis Research Laboratory of the Department of Veterinary Public Health and Preventive Medicine, University of Ibadan, Ibadan, Nigeria (9). A database maintained this information on all retrospectively stored samples.
Spoligotyping and database comparison.
DNA was extracted by heating the isolates at 80°C for 60 min (9) and was stored at −20°C until it was subjected to spoligotyping using a previously described protocol (16). Spoligotypes in binary format were converted to an octal code for comparison with the SITVIT2 proprietary database of the Pasteur Institute of Guadeloupe, which is an updated version of the previously released SpolDB4 database (4). At the time of the present study, SITVIT2 contained genotyping information on about 75,000 M. tuberculosis clinical isolates from 160 countries of origin. In the database, a spoligotype international type (SIT) represents spoligotyping shared by 2 or more patient isolates, as opposed to “orphan,” which represents patterns reported for a single isolate. Major phylogenetic clades were assigned according to signatures provided in SpolDB4, which defined 62 genetic lineages/sublineages (4). They include specific signatures for various MTC species, such as M. bovis, M. microti, M. caprae, M. pinnipedii, and M. africanum, as well as rules defining major lineages/sublineages for M. tuberculosis sensu stricto, including the Beijing clade, the Central Asian (CAS) clade and 2 sublineages, the East African-Indian (EAI) clade and 9 sublineages, the Haarlem (H) clade and 3 sublineages, the Latin American-Mediterranean (LAM) clade and 12 sublineages, the “Manu” family and 3 sublineages, the S clade, the IS6110-low-banding X clade and 4 sublineages, and an ill-defined T clade with 5 sublineages. The worldwide distribution of spoligotypes for clustered isolates was investigated using the SITVIT2 database and recorded for countries and regions representing ≥5% of a given SIT compared to their total number in the global database. The various macrogeographical regions and subregions were defined according to United Nations (UN) specifications (http://unstats.un.org/unsd/methods/m49/m49regin.htm).
RESULTS AND DISCUSSION
This study concerned a total of 163 MTC strains isolated from both HIV+ and HIV− patients in Ibadan, Nigeria, between April 2004 and October 2005. With the population of Ibadan at 1.34 million (http://www.population.gov.ng/state/oyofinal.pdf) and an estimated incidence of new TB cases of 311/100,000 and of smear-positive TB cases of 131/100,000 (http://www.stoptb.org/assets/documents/countries/GlobalReport2009/nga.pdf), the study sample represented about 6% of all smear-positive TB cases in Ibadan. Furthermore, considering the proportion of HIV+ patients (27%) (25) and a case detection rate of 18% in 2005 (http://www.usaid.gov/our_work/global_health/id/tuberculosis/countries/africa/nigeria.pdf), the study sample represented about 13% and 15% of the HIV− and HIV+ patients newly diagnosed with TB, respectively, that were reported to the health authorities. The results and the succinct analysis obtained are summarized in Tables 1 to 3.
TABLE 1.
Orphan strains from HIV+ and HIV− patients in Ibadan, Nigeria
a Clade designations according to revised SpolDB4 rules. Unk, unknown pattern within any of the major clades described.
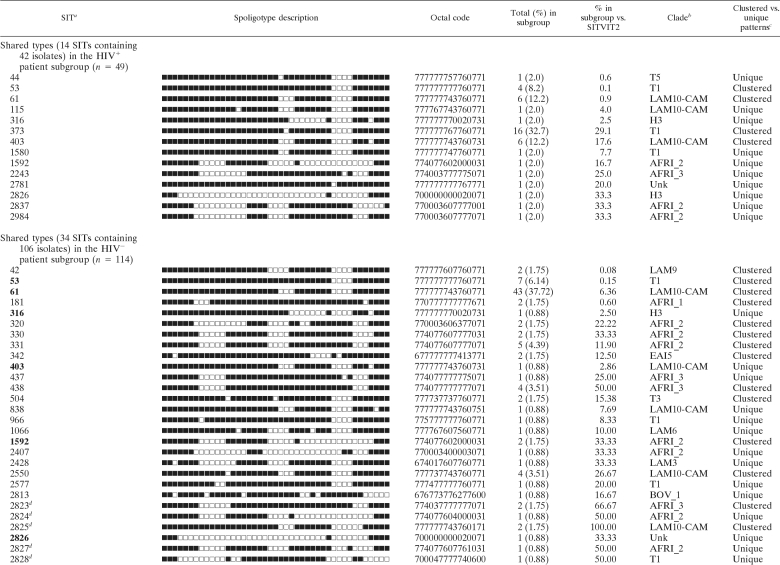
For the HIV+ patients, spoligotyping produced a total of 21 different patterns for the 49 strains studied. Seven patterns corresponded to orphan strains that were unique among all the patterns recorded in the SITVIT2 database (Table 1), as opposed to 14 patterns from 42 patients that corresponded to a SIT, i.e., an identical pattern shared by 2 or more patient isolates worldwide. For each of the SITs observed, Table 2 summarizes its binary/octal description, the total number of strains, the percentage in the HIV+ patient subgroup compared to the percentage in the SITVIT2 database, its genotypic clade designation, and information on its presence as a unique or clustered strain in the study. Among the 14 SITs found, only 4 corresponded to clustered isolates within this study (n = 32; 4 to 16 isolates per cluster) (Table 2), while the remaining isolates (n = 17, including 7 orphan strains) were unique within the HIV+ data set. Nonetheless, they accounted for a high clustering rate of 32/49, or 65.31%. Interestingly, the clustered isolates in this subgroup corresponded to only 2 phylogeographical clades, T1 (20/32, or 62.5%) and the LAM sublineage “LAM10-CAM” (12/32, or 37.5%); the latter lineage possesses a reported phylogeographical specificity for Cameroon and neighboring countries in West Africa (4, 5, 20).
TABLE 2.
Spoligotyping shared types from HIV+ and HIV− patient subgroups in Ibadan, Nigeria
a Unique strains matching a preexisting pattern in the SITVIT2 database are classified as SITs, whereas in cases where there was no match, they are designated “orphan” (Table 1). SITs in the HIV− subgroup that were simultaneously present in the HIV+ subgroup are in boldface.
b Clade designations according to SITVIT2 using revised SpolDB4 rules. Unk, unknown patterns within any of the major clades described.
c Clustered strains correspond to a similar spoligotype pattern shared by 2 or more strains within this study as opposed to unique strains harboring a spoligotype pattern that does not match another strain from this study.
dNewly created shared type (none among 14 SITs from HIV+ patients as opposed to 11 new SITs observed among 34 SITs from HIV− patients). In the latter subgroup, these new SITs were created due to 2 or more strains belonging to an identical new pattern within this study (SIT2825, n = 2; SIT2829, n = 2; SIT2831, n = 3), a unique strain from this study matching another orphan in the database (SIT2824 matched an orphan from metropolitan France; SIT2830, SIT2832, and SIT2834 each matched an orphan from the United States), or 2 or more strains from this study that matched an existing orphan from Nigeria (SIT2823, n = 2; SIT2827, n = 1; SIT2828, n = 1; SIT2835, n = 3).
Spoligotyping of the HIV− patient subgroup (n = 114) resulted in 42 different patterns; 8 patterns corresponded to orphan strains that were unique compared to those in the SITVIT2 database (Table 1), as opposed to 34 patterns from 106 patients that corresponded to shared types (Table 2). Among the 34 SITs found, 17 corresponded to clustered isolates within this study (n = 89; 2 to 43 isolates per cluster) (Table 2), while the remaining isolates (n = 25, including 8 orphan strains), or 21.93%, were unique within the data set. This corresponded to a very high clustering rate of 89/114, or 78.07%. Unlike the HIV+ subgroup, the clustered isolates in the HIV− patient subgroup corresponded to several phylogeographical clades, including LAM9, LAM10-CAM, T1, T3, AFRI_1, AFRI_2, AFRI_3, EAI, and EAI5; the LAM sublineage LAM10-CAM, with phylogeographical specificity for Cameroon and neighboring countries in West Africa (4, 5, 20), accounted for the majority of isolates (52/89, or 58.43%).
When the overall repartition of strains according to major M. tuberculosis genotypic families was defined (i.e., by adding all the shared types for each individual family, as well as the orphan strains), the major circulating M. tuberculosis clades among the HIV+ patients ranked in the following order: T1, 22/49, or 44.9%; LAM10-CAM, 13/49, or 26.5%; AFRI, 4/49, or 8.2%; Manu, 3/49, or 6.1%; and H3, 2/49, or 4.1% (Tables 1 and 2). For the HIV− patient subgroup, the ranking was in this order: LAM10-CAM, 59/114, or 51.8%; AFRI, 27/114, or 23.7%; T1, 10/114, or 8.8%; H3, 4/114, or 3.5%; EAI, 3/114, or 2.6%; EAI5, 2/114, or 1.8%; BOVIS, 2/114, or 1.8%; T3, 2/114, or 1.8%; LAM9, 2/114, or 1.8%; LAM6, 1/114, or 0.9%; and LAM3, 1/114, or 0.9% (Tables 1 and 2). Note that 1 strain per subgroup studied could not be classified in any specific lineage, since the spoligotyping pattern did not correspond to any of the major clades described in the SITVIT2 database. It should be mentioned that the “T” genotype does not represent a clade in a strict evolutionary sense, since it was defined by default to include strains that may not be classified in one of the established genotypic lineages with well-established phylogeographical specificity (4).
The worldwide distribution of the clustered spoligotypes in our study (Table 3) showed that, with the exception of the ubiquitous spoligotype T1, the HIV+ patients in Nigeria mainly harbored the M. tuberculosis spoligotypes SIT61 and SIT403 (LAM10-CAM sublineage) prevailing in Middle and Western Africa. A countrywide distribution showed that they were essentially shared by Nigeria's immediate neighbors, Cameroon (Table 3) and the Benin Republic (1). The latter study showed that the LAM10-CAM prototype, SIT61, was the predominant spoligotype in Cotonou, Benin Republic; however, unlike Benin, where the Beijing genotype represented more than 10% of all strains (1), no Beijing-type strains were isolated from the HIV+ or HIV− TB patients from our data set. As summarized in Table 2, in our study, the LAM10-CAM sublineage was essentially made up of its prototype, SIT61 (6/13 LAM10-CAM strains in the HIV+ subgroup and 43/59 strains in the HIV− subgroup), and related strains clearly derived from it, e.g., SIT115, which evolved from the prototype SIT61 by loss of spacer 15 (a single strain in the HIV+ subset), and SIT403, which evolved by loss of spacer 40 (6/13 LAM10-CAM strains in the HIV+ subgroup and 1/59 in the HIV− subgroup).
TABLE 3.
Description of clustered M. tuberculosis complex isolates (2 or more isolates in this study) from HIV+ and HIV- patients in Ibadan, Nigeria, and their worldwide distribution in the SITVIT2 database
| SIT (clade)a | Total (%) in subgroup | % in subgroup vs. SITVIT2 | Distribution (%) in regions with: |
|
|---|---|---|---|---|
| ≥5% of a given SITb | ≥5% of a given SITc | |||
| Clustered isolates in the HIV+ patient subgroup (n = 49) | ||||
| 53 (T1) | 4 (8.2) | 0.1 | AMER-N, 19.95; AMER-S, 14.67; EURO-W, 13.0; EURO-S, 10.16; ASIA-W, 8.81; AFRI-S, 6.04 | USA, 17.56; ZAF, 5.89; ITA, 5.20 |
| 61 (LAM10-CAM) | 6 (12.2) | 0.9 | AFRI-M, 27.59; AFRI-W, 25.39; AMER-N, 12.54; EURO-W, 10.97; ASIA-W, 6.90 | CMR, 26.49; NGA, 22.57; USA, 12.54; SAU, 6.90; FRA, 6.27 |
| 373 (T1) | 16 (32.7) | 29.1 | AFRI-W, 29.09; EURO-W, 16.36; AFRI-S, 14.55; AMER-S, 10.91; EURO-S, 9.09; EURO-E, 9.09; AMER-N, 7.27 | NGA, 29.09; ZAF, 14.55; CZE, 9.09; USA, 7.27; BEL, 7.27; PER, 5.45; AUT, 5.45; BRA, 5.45 |
| 403 (LAM10-CAM) | 6 (12.2) | 17.6 | AFRI-M, 44.12; AFRI-W, 26.47; AMER-N, 8.82; EURO-W, 8.82; ASIA-S, 5.88 | CMR, 44.12; NGA, 26.47; USA, 8.82; IND, 5.88 |
| Clustered isolates in the HIV− patient subgroup (n = 114) | ||||
| 42 (LAM9) | 2 (1.75) | 0.08 | AMER-S, 29.83; AMER-N, 16.28; EURO-S, 12.78; EURO-W, 7.02; AFRI-N, 5.07 | USA, 15.24; BRA, 10.33; COL, 7.87; ITA, 6.72 |
| 53 (T1) | 7 (6.14) | 0.15 | AMER-N, 19.95; AMER-S, 14.67; EURO-W, 13.0; EURO-S, 10.16; ASIA-W, 8.81; AFRI-S, 6.04 | USA, 17.56; ZAF, 5.89; ITA, 5.20 |
| 61 (LAM10-CAM) | 43 (37.72) | 6.36 | AFRI-M, 27.59; AFRI-W, 25.39; AMER-N, 12.54; EURO-W, 10.97; ASIA-W, 6.90 | CMR, 26.49; NGA, 22.57; USA, 12.54; SAU, 6.90; FRA, 6.27 |
| 181 (AFRI_1) | 2 (1.75) | 0.60 | AFRI-W, 80.06; AMER-N, 6.34; EURO-W, 6.04 | GNB, 51.06; GMB, 25.38; USA, 6.34 |
| 320 (AFRI_2) | 2 (1.75) | 22.22 | AFRI-W, 44.44; AMER-N, 22.22; AFRI-M, 22.22; EURO-S, 11.11 | NGA, 44.44; USA, 22.22; CMR, 22.22; ESP, 11.11 |
| 330 (AFRI_2) | 2 (1.75) | 33.33 | AFRI-W, 33.33; AFRI-M, 33.33; EURO-W, 16.67; AFRI-N, 16.67 | NGA, 33.33; CAF, 16.67; CMR, 16.67; TUN, 16.67; BEL, 16.67 |
| 331 (AFRI_2) | 5 (4.39) | 11.90 | AFRI-W, 64.29; AMER-N, 23.81; AFRI-M, 7.14 | NGA, 54.76; USA, 23.81; CIV, 7.14; CMR, 7.14 |
| 342 (EAI5) | 2 (1.75) | 12.50 | AFRI-W, 43.75; EURO-W, 25.00; EURO-N, 18.75; AMER-N, 12.50 | NGA, 31.25; GBR, 12.50; GMB, 12.50; USA, 12.50; BEL, 12.50; NLD, 6.25; DNK, 6.25; DEU, 6.25 |
| 438 (AFRI_3) | 4 (3.51) | 50.00 | AFRI-W, 75.00; AMER-N, 25.00 | NGA, 62.50; USA, 25.00; SLE, 12.50 |
| 504 (T3) | 2 (1.75) | 15.38 | AMER-N, 53.85; AFRI-W, 15.38; EURO-W, 7.69; EURO-S, 7.69; ASIA-S, 7.69; AFRI-E, 7.69 | USA, 53.85; NGA, 15.38; TZA, 7.69; FXX, 7.69; IND, 7.69; ITA, 7.69 |
| 1592 (AFRI_2) | 2 (1.75) | 33.33 | AFRI-W, 66.67; EURO-W, 33.33 | NGA, 66.67; DEU, 16.67; FRA, 16.67 |
| 2550 (LAM10-CAM) | 4 (3.51) | 26.67 | CARI, 46.67; AFRI-W, 26.67; ASIA-W, 6.67; AMER-S, 6.67; AMER-N, 6.67; AFRI-E, 6.67 | TTO, 46.67; NGA, 26.67; USA, 6.67; BRA, 6.67; TUR, 6.67; MDG, 6.67 |
| 2823d (AFRI_3) | 2 (1.75) | 66.67 | AFRI-W, 100.00 | NGA, 100.00 |
| 2825d (LAM10-CAM) | 2 (1.75) | 100.00 | AFRI-W, 100.00 | NGA, 100.00 |
| 2829d (AFRI_2) | 2 (1.75) | 100.00 | AFRI-W, 100.00 | NGA, 100.00 |
| 2831d (LAM10-CAM) | 3 (2.63) | 100.00 | AFRI-W, 100.00 | NGA, 100.00 |
| 2835d (EAI) | 3 (2.63) | 75.00 | AFRI-W, 100.00 | NGA, 100.00 |
SITs simultaneously present in HIV-positive and HIV-negative subgroups are in boldface.
Worldwide distribution is reported for regions with ≥5% of a given SIT compared to the total number in the SITVIT2 database. The definitions of regions and subregions are according to those of the United Nations (http://unstats.un.org/unsd/methods/m49/m49regin.htm): regions, AFRI (Africa), AMER (Americas), ASIA (Asia), EURO (Europe), and OCE (Oceania), subdivided into E (eastern), M (middle), C (central), N (northern), S (southern), SE (southeastern), and W (western). Furthermore, CARIB (Caribbean) belongs to the Americas, while Oceania is subdivided into 4 subregions, AUST (Australasia), MEL (Melanesia), MIC (Micronesia), and POLY (Polynesia). Note that in our classification scheme, Russia has been assigned a new subregion by itself (Northern Asia) instead of including it with the rest of Eastern Europe. This reflects its geographical location, as well as the similarity of specific TB genotypes circulating in Russia (a majority of Beijing genotypes) to those prevalent in Central, Eastern, and Southeast Asia.
The 3-letter country codes are defined in wikipedia (http://en.wikipedia.org/wiki/ISO_3166-1_alpha-3); countrywide distribution is shown only for SITs with ≥5% compared to their total number in the SITVIT2 database.
Newly created shared type (for details, see note d to Table 2).
In the HIV− subset, there were many other strains of the LAM10-CAM sublineage derived from SIT61 that evolved by loss of one or more spacers (SIT838, -2550, -2825, -2830, -2831, -2832, and -2834). Among the LAM10-CAM members, we had only a single strain of SIT403 from the HIV− patients (1/114, or 0.88%), as opposed to 6/49, or 12.25%, in the HIV+ population, suggesting a strong correlation between the strain and the HIV+ subset. On the other hand, the LAM10-CAM lineage spoligotype SIT61 was prevalent in the 2 subsets (43/114, or 37.72%, in the HIV− population versus 6/49, or 12.24%, in the HIV+ population). This was also true for SIT53 (T1 lineage) (6.14% versus 8.2%). Similarly, the T1 sublineage was represented by its prototype, SIT53 (n = 4), but the major prevailing spoligotype among the HIV+ patients, SIT373, apparently derived from the prototype by loss of spacer 24 (n = 16), as well as a minor pattern, SIT1580 (cumulative loss of spacers 23 and 24; n = 1). As can be seen, SIT373 (T1 lineage), which was highly prevalent among HIV+ patients, was totally absent from the HIV-negative population, suggesting that we had a specific clone affecting nearly 1/3 of all HIV-TB-coinfected patients in our setting.
Another interesting feature was the smaller presence of M. africanum in the HIV+ population (4/49, or 8.2%); nonetheless, it is the first reported case of this kind among TB-HIV-coinfected patients in Nigeria. Three of the strains were similar (i.e., they lacked spacers 7 to 16 and had most of the spacers from 25 to 36 intact). The HIV− subgroup was characterized by a larger presence of this genotype (27/114, or 23.7%). The preponderance of M. africanum in the HIV− subset could be seen from the presence of 3 main sublineages (AFRI-1, AFRI-2, and AFRI-3), with the occurrence of two “orphan” strains. The presence of this clade is not surprising based on earlier findings about TB patients in Nigeria (9) and other West African countries (11, 20, 21). Similar reports about this sublineage have also been made in some East African countries, though with phenotypic traits linked to M. tuberculosis (4, 6, 18, 19).
Quite interestingly, two strains of M. bovis were found in the HIV− population, with none identified in the HIV+ subset, though this reflects the zoonotic transmission of the disease in humans, judging from the high prevalence of bovine tuberculosis in Nigeria (10). It is noteworthy, however, that there is a high potential for human infection with this genotype, judging from the lack of a control and eradication program for bovine tuberculosis in Nigeria, where people still consume unpasteurized milk and milk products, along with other risk factors (8, 9).
In our study, the Haarlem clade (specifically, the H3 sublineage) also featured in both subsets, making up a very small percentage of the entire data set (i.e., 1/49, or 2%, and 4/114, or 3.5%, respectively, in the HIV+ and HIV− groups); however, with a majority (3/5, or 60%) of the strains being orphans and restricted to Nigeria, not much can be concluded about its significance. Again, strains of the ancestral EAI clade (4), which are very characteristic in terms of their spoligotype patterns and the presence of genomic regions of difference (with both TbD1 and RD9 not deleted), were isolated from the HIV− subset. However, the smaller percentage of these strains in the HIV− subset (5/114, or 4.4%) may explain its absence in the HIV+ population in our data set. EAI represents the most ancestral clade of the M. tuberculosis complex (except for the M. canettii ancestor [12]). Recently, an EAI5 strain was isolated from a slaughtered female goat in an abattoir within the same study area (7). Whether this animal infection resulted from cross-infection due to close human and animal cohabitation is not known, since this was the only report of its kind in our setting.
Another remarkable finding was the presence of 3 ancestral Manu lineage strains limited to orphan patterns in the HIV+ population (2 were classified as Manu2 and 1 as Manu3). Incidentally, no Manu strain was found in the HIV− subgroup. The Manu lineage was initially described as a new family from India in 2004 (23), and later, similar strains in small numbers were reported in a study from Madagascar (13). Soon afterwards, it was tentatively subdivided into the Manu1 (deletion of spacer 34), Manu2 (deletion of spacers 33 and 34), and Manu3 (deletion of spacers 34 to 36) sublineages, and it was suggested that it could represent an ancestral clone of principal genetic group (PGG) 1 strains (4). Manu lineage strains were reported from Saudi Arabia (2) and Tunisia (17) and more recently in a study from Egypt, where it represented as much as 27% of all isolates (15). However, according to a recent report, a fraction of Manu2 patterns could also arise due to mixed infection by a strain belonging to evolutionarily recent H, LAM, X, and T lineages with a Beijing strain, a risk that may be excluded by looking for genomic deletion of region of difference RD105, which is characteristic of the Beijing lineage (26). However, the complete absence of Beijing strains in our setting argues against this possibility. Nonetheless, the presence of the ancestral Manu lineage strains, supposed to be a missing link from the split between ancestral and modern tubercle bacilli during M. tuberculosis evolution (15), should be further investigated within larger data sets to discover the contribution of Africa to the origin, evolution, and spread of tuberculosis.
In conclusion, our study shows that TB among HIV+ and HIV− patients in Ibadan, southwestern Nigeria, is essentially caused by predominant genotypes characteristic of the western and middle African regions and, more specifically, the LAM10-CAM sublineage, which is also the predominant spoligotype in the neighboring countries of Cameroon and Benin. In the same vein, the M. africanum clade was found to play a prominent role in both subsets, a scenario that had been reported earlier in neighboring West African countries, providing further incentive to carry out detailed investigations of the clade in Nigeria. The presence of the EAI clade essentially in the HIV− population provided more insight into the diversity of the M. tuberculosis complex strains in Nigeria; coupled with this is the identification of the Haarlem genotype in both subsets, a situation that indicates the interaction of Nigerians with people from other countries, possibly due to trade links and the presence of Indo-Asians in Nigeria. Nonetheless, the complete absence of the Beijing genotype in our study population as opposed to a recent report from Benin (1) is noteworthy. Our study provides a good platform to work through the diversity of M. tuberculosis complex strains among TB patients in Nigeria, providing a basis for further epidemiological investigations into the evolution and control of TB in the country.
Acknowledgments
We thank M. Lawal Oyewole (TB Control Officer, Oyo State, Nigeria) and other collaborators who provided technical support in Nigeria in carrying out this work. We also thank Martin Vordermeier and Glyn Hewinson for facilitating S.C.'s visit to the Veterinary Laboratories Agency (VLA), Weybridge, United Kingdom, to carry out part of the molecular analysis. We are also grateful to Thierry Zozio (Institut Pasteur de la Guadeloupe) for his help in the interpretation of the spoligotyping data.
Work at VLA was funded by the Department for Environment, Food, and Rural Affairs. V.H. was awarded a Ph.D. fellowship by the European Social Funds through the Regional Council of Guadeloupe. The work done at the Pasteur Institute of Guadeloupe was financed by the Regional Council of Guadeloupe (CR/08-1612: Biodiversité et Risque Infectieux dans les Modèles Insulaires).
Footnotes
Published ahead of print on 3 November 2010.
REFERENCES
- 1.Affolabi, D., G. Anyo, F. Faïhun, N. Sanoussi, I. C. Shamputa, L. Rigouts, L. Kestens, S. Anagonou, and F. Portaels. 2009. First molecular epidemiological study of tuberculosis in Benin. Int. J. Tuberc. Lung Dis. 13:317-322. [PubMed] [Google Scholar]
- 2.Al-Hajoj, S. A., T. Zozio, F. Al-Rabiah, V. Mohammad, M. Al-Nasser, C. Sola, and N. Rastogi. 2007. First insight into the population structure of Mycobacterium tuberculosis in Saudi Arabia. J. Clin. Microbiol. 45:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 4.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, C. L. Aristimuño, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Guttierez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, M. L. Ho, C. Martin, I. Mokrousov, O. Narvskaïa, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rüsch-Gerdes, A. Sajduda, S. Samper, I. G. Shemyakin, U. B. Singh, A. Somoskovi, R. A. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. M. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brudey, K., I. Filliol, S. Ferdinand, V. Guernier, P. Duval, B. Maubert, C. Sola, and N. Rastogi. 2006. Long-term population-based genotyping study of Mycobacterium tuberculosis complex isolates in the French departments of the Americas. J. Clin. Microbiol. 44:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brudey, K., C. Gutierrez, V. Vincent, L. M. Parsons, M. Salfinger, N. Rastogi, and C. Sola. 2004. Mycobacterium africanum genotyping using novel spacer oligonucleotide in the direct repeat locus. J. Clin. Micobiol. 42:5053-5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadmus, S. I., H. K. Adesokan, A. O. Jenkins, and D. van Soolingen. 2009. Mycobacterium bovis and M. tuberculosis in goats, Nigeria. Emerg. Infect. Dis. doi: 10.3201/eid1512.090319. [DOI] [PMC free article] [PubMed]
- 8.Cadmus, S. I. B., H. K. Adesokan, A. F. Adepoju, and E. B. Otesile. 2008. Zoonotic risks and transmission of Mycobacteria species from cows' milk and slaughtered cattle to man in Ibadan: role of butchers. Niger. Vet. J. 291:30-39. [Google Scholar]
- 9.Cadmus, S., S. Palmer, M. Okker, J. Dale, K. Gover, N. Smith, K. Jahans, R. G. Hewinson, and S. V. Gordon. 2006. Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J. Clin. Microbiol. 44:29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cadmus, S. I. B., A. A. Atsanda, S. O. Oni, and E. E. U. Akang. 2004. Bovine tuberculosis in one cattle herd in Ibadan in Nigeria. Vet. Med. Czech. 49:406-412. [Google Scholar]
- 11.de Jong, B. C., P. C. Hill, R. H. Brookes, J. K. Otu, K. L. Peterson, P. M. Small, and R. A. Adegbola. 2005. Mycobacterium africanum: a new opportunistic pathogen in HIV. AIDS 19:1714-1715. [DOI] [PubMed] [Google Scholar]
- 12.Fabre, M., J. L. Koeck, P. Le Fleche, F. Simon, V. Herve, G. Vergnaud, and C. Pourcel. 2004. High genetic diversity revealed by variable-number tandem repeat genotyping and analysis of hsp65 gene polymorphism in a large collection of “Mycobacterium canettii” strains indicates that the M. tuberculosis complex is a recently emerged clone of “M. Canettii”. J. Clin. Microbiol. 42:3248-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferdinand, S., C. Sola, S. Chanteau, H. Ramarokoto, T. Rasolonavalona, V. Rasolofo-Razanamparany, and N. Rastogi. 2005. A study of spoligotyping-defined Mycobacterium tuberculosis clades in relation to the origin of peopling and the demographic history in Madagascar. Infect. Genet. Evol. 5:340-348. [DOI] [PubMed] [Google Scholar]
- 14.Groenen, P. M. A., A. E. Bunschoten, D. van Soolingen, and J. D. A. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 15.Helal, Z. H., M. S. Ashour, S. A. Eissa, G. Abd-Elatef, T. Zozio, S. Babapoor, N. Rastogi, and M. I. Khan. 2009. Unexpectedly high proportion of ancestral Manu genotype Mycobacterium tuberculosis strains cultured from tuberculosis patients in Egypt. J. Clin. Microbiol. 47:2794-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Namouchi, A., A. Karboul, B. Mhenni, N. Khabouchi, R. Haltiti, H. R. Ben, B. Louzir, A. Chabbou, and H. Mardassi. 2008. Genetic profiling of Mycobacterium tuberculosis in Tunisia: predominance and evidence for the establishment of a few genotypes. J. Med. Microbiol. 57:864-872. [DOI] [PubMed] [Google Scholar]
- 18.Niemann, S., T. Kubica, F. C. Bange, O. Adjei, E. N. Browne, M. A. Chinbuah, R. Diel, J. Gyapong, R. D. Horstmann, M. L. Joloba, C. G. Meyer, R. D. Mugerwa, A. Okwera, I. Osei, E. Owusu-Darbo, S. K. Schwander, and S. Rusch-Gerdes. 2004. The species Mycobacterium africanum in the light of new molecular markers. J. Clin. Microbiol. 42:3958-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niemann, S., S. Rusch-Gerdes, M. L. Joloba, C. C. Whelan, D. Guwatudde, J. J. Ellner, K. Eisenach, N. Fumokong, J. I. Johnson, T. Aisu, R. D. Mugerwa, A. Okwera, and S. K. Schwander. 2002. Mycobacterium africanum subtype II is associated with two distinct genotype and is a major cause of human tuberculosis in Kampala, Uganda. J. Clin. Microbiol. 40:3398-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, J. Thonnon, V. Vincent, and M. C. Gutierrez. 2004. Molecular characteristics of strains of the Cameroon family, the major group of Mycobacterium tuberculosis in a country with a high prevalence of tuberculosis. J. Clin. Microbiol. 42:5029-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, P. Cunin, J. Thonnon, C. Sola, N. Rastogi, V. Vincent, and M. C. Gutierrez. 2003. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 41:2547-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rastogi, N., and C. Sola. 2007. Molecular evolution of the Mycobacterium tuberculosis complex, p. 53-91. In J. C. Palomino, S. Leao, and V. Ritacco (ed.), Tuberculosis 2007: from basic science to patient care. Amedeo Online Textbooks. http://www.tuberculosistextbook.com/index.htm.
- 23.Singh, U. B., N. Suresh, N. V. Bhanu, J. Arora, H. Pant, S. Sinha, R. C. Aggarwal, S. Singh, J. N. Pande, C. Sola, N. Rastogi, and P. Seth. 2004. Predominant tuberculosis spoligotypes, Delhi, India. Emerg. Infect. Dis. 10:1138-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittman, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc. Natl. Acad. Sci. U. S. A. 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.UNAIDS. 2009. AIDS epidemic update, December 2009. WHO Library Cataloguing-in-Publication, Geneva, Switzerland. http://data.unaids.org/pub/Report/2009/jc1700_epi_update_2009_en.pdf.
- 26.Viegas, S. O., A. Machado, R. Groenheit, S. Ghebremichael, A. Pennhag, P. S. Gudo, Z. Cuna, P. Miotto, V. Hill, T. Marrufo, D. M. Cirillo, N. Rastogi, G. Källenius, and T. Koivula. 2010. Molecular diversity of Mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in Mozambique. BMC Microbiol. 10:195. http://www.biomedcentral.com/1471-2180/10/195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO. 2009. Global tuberculosis control: epidemiology, strategy, financing: WHO, Geneva, Switzerland., http://whqlibdoc.who.int/publications/2009/9789241598866_eng.pdf.