Abstract
A high-throughput multiplex bead suspension array was developed for the rapid subgenogrouping of EV71 strains, based on single nucleotide polymorphisms observed within the VP1 region with a high sensitivity as low as 1 PFU. Of 33 viral isolates and 55 clinical samples, all EV71 strains were successfully detected and correctly subgenogrouped.
Hand, foot, and mouth disease (HFMD) mainly afflicts children and is characterized by fever and vesicles and ulcers on the mouth, palms, limbs, and buttocks. Although several enteroviruses are etiologic agents of HFMD, neurologic complications such as aseptic meningitis, brainstem encephalitis, and poliomyelitis-like paralysis have been associated with enterovirus 71 (EV71) infection. Fatalities due to EV71 infection have occurred during large outbreaks of HFMD in the Asia-Pacific region since 1997. EV71 possesses a positive single-stranded linear RNA genome, with four capsid proteins (VP1, VP2, VP3, and VP4) present on the outermost part of the nonenveloped virus (6, 11). Molecular phylogenetic analyses based on EV71 VP1 sequences have classified EV71 strains into three main genogroups, A, B, and C, which are further classified into subgenogroups B1 to B5 and C1 to C5 (2-4, 11).
Although no direct association between the severe neurologic complications of HFMD and different EV71 genetic lineages has been established, subgenogrouping of EV71 is important for epidemiologic studies of the surveillance of both endemic and epidemic HFMD and also facilitates further studies of EV71 virulence and evolution. However, subgenogrouping of EV71 has been hitherto dependent on classical reverse transcription (RT)-PCR and direct DNA sequencing, which are relatively labor-intensive for processing large numbers of samples during outbreaks. Therefore, with the ever-increasing number of HFMD outbreaks, we developed a high-throughput EV71 detection platform based on Luminex xTAG technology (5) with rapid genogrouping capability that can be applied in routine clinical diagnosis and that may be beneficial for epidemiologic surveillance of EV71 infections.
Virus isolates or viral RNA representing nine EV71 subgenogroups were initially employed as reference strains to develop the assay. However, for subgenogroups B1 and C3, viral isolates could not be obtained. Thus, corresponding plasmids incorporating the full-length VP1 genes of strains 609-AUS-74 and 009-KOR-00 were constructed using synthetic DNA cloned into the pUC57 vector (GenScript, Piscataway, NJ). Only viral RNA of strain MY104-9-SAR-97 (subgenogroup B3) was obtained, and its VP1 gene was also cloned into the pCR-XL-TOPO vector for ascertaining sensitivity. Thirty-three other viral isolates from Japan, Malaysia, and Singapore were tested for validation of the assay, with new VP1 nucleotide sequences deposited in GenBank under accession numbers HQ285091 to HQ285109. A total of 55 clinical specimens were also collected from suspected HFMD patients who presented at the National University Hospital mainly during the 2008 HFMD outbreak in Singapore (12). These samples were tested by conventional and real-time RT-PCR, by DNA sequencing (8-10, 12), and also by the multiplex subgenogrouping assay. This clinical study was approved by the Institutional Review Board of the National University of Singapore (approval no. NUS-301).
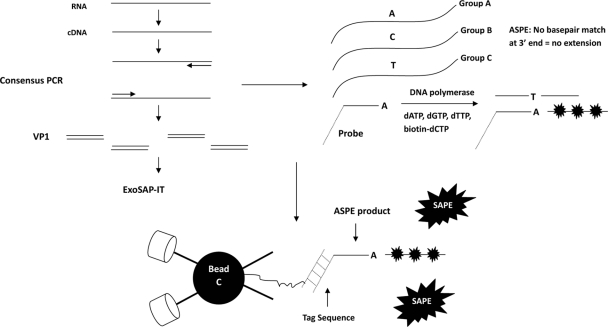
Consensus primers were designed based on the VP1 region of all EV71 subgenogroups. The 1.2-kb VP1 fragment was amplified from each cDNA sample using the consensus primers, and amplicons were purified by ExoSAP-IT treatment (Affymetrix, Santa Clara, CA) to remove excess nucleotides and primers. Alignment of sequences showed that all EV71 subgenogroups (except C1) differed by only a single nucleotide at different regions within VP1 (see Fig. S1 in the supplemental material). Therefore, multiplex allele-specific primer extension (ASPE), which differentiates gene sequences by only one nucleotide difference at the 3′ end and permits amplification only of the perfectly complementary probe, was chosen instead of the direct DNA hybridization Luminex assay. Initially, the latter was analyzed, but it suffered from very low specificity. Specific ASPE probes were designed for 11 subgenogroups based on the single nucleotide disparity located at the 3′ end of each probe (see Table S1 in the supplemental material). A tag sequence was incorporated at the 5′ end of each probe to allow hybridization to the anti-tag sequence of the corresponding xTAG microspheres (Luminex, Austin, TX) as portrayed in Fig. 1. Since there was no single nucleotide difference unique for C1, we selected a nucleotide position that could distinguish C1 and C3 from the other nine subgenogroups but whose nucleotide polymorphism site was different from that of the C3 probe. The C1 probe was designed to thus identify both C1 and C3, but these subgenogroups could be differentiated by the C3 probe reading. Hence, high readings of both C1 and C3 probes indicated that the sample was from subgenogroup C3, whereas a high reading only for C1 revealed a sample belonging to subgenogroup C1. The multiplex ASPE reaction mix comprised 10 μl of PCR product, 2 μl of 10× ASPE buffer (20 mM Tris-HCl, 50 mM KCl), 0.5 μl of 50 mM MgCl2, 1 μl of 500 nM xTAG-ASPE primer mix, 1 μl each of 100 μM dATP, dGTP, and dTTP, 0.25 μl of 400 μM biotin-dCTP, 0.15 μl of Platinum GenoTYPE Tsp DNA polymerase (5 U/μl) (Invitrogen, Carlsbad, CA), and water in a total volume of 20 μl. The ASPE reactions were incubated at 96°C for 2 min, subjected to 30 cycles each at 94°C for 0.5 min, 55°C for 1 min, and 72°C for 2 min, and held at 4°C until used. ASPE products were labeled with biotin-dCTP and were hybridized with microspheres through the tag and anti-tag nucleotide sequences. Streptavidin-phycoerythrin (SAPE) was added last for the detection of biotin, and the whole microsphere-PCR complex passed through the detection chamber of the Luminex reader (Luminex, Austin, TX). The readout was the median fluorescence intensity (MFI) and is dependent on the number of viral copies present. Relative rather than absolute readings were compared; i.e., the highest sample-to-blank control ratio identified the subgenogroup of the analyte in the sample. The cutoff value (COV) specific for a particular subgenogroup was arbitrarily assigned as the ratio that was at least 10 times that of the blank sample. Each sample assay was performed at least four times, and the mean value was calculated. Virus isolates were available for subgenogroups A, B2, B4, B5, C1, C2, C4, and C5, and the assay sensitivity was based on the lowest number of PFU detected (7). Samples corresponding to 1, 5, 20, 50, and 100 PFU were subjected to RNA extraction, RT-PCR, multiplex ASPE, and hybridization. However, viral isolates were not available for three subgenogroups (B1, B3, and C3), and sensitivity was determined by the lowest number of plasmid copies detected (out of 1, 10, 100, and 1,000 copies analyzed).
FIG. 1.
Schematic representation of the multiplex suspension array system for EV71 subgenogrouping. The VP1 region was amplified by consensus primers, and the RT-PCR product was purified by ExoSAP-IT. ASPE was performed, and only perfectly complementary strands would be amplified. The ASPE products were labeled using biotin-dCTP (black stars), and hybridized to their complementary anti-tag oligonucleotide sequences on the beads or microspheres. The complex was subjected to the Luminex reader and was identified by both lasers and streptavidin-phycoerythrin (SAPE).
For optimization of the system, reference strains representing the 11 subgenogroups of EV71 were tested with coxsackievirus A16 (CA16) as a negative control, and all could be detected by subgenogroup-specific probes. Nine probes yielded average ratios that were at least 100 times that of the blank control (Table 1). For probes B5 and C4, the ratios were comparatively lower but still close to 100. These relatively lower ratios may be explained by the lower GC content of B5 and C4 probes (48% and 40%, respectively) that culminates in less efficient hybridization and linear amplification (1). Compared to direct DNA sequencing of amplified products as the benchmark, the results generated from our multiplex technique were consistent and reproducible. Validation with 33 additional viral isolates demonstrated that our assay could specifically identify the subgenogroups of other EV71 strains (see Table S2 in the supplemental material). In addition, by testing 55 clinical specimens, we could reliably distinguish 11 EV71-positive samples from 44 EV71-negative samples by our assay, with the subgenogrouping data being congruent with DNA sequencing (Table 1). One drawback is that a definite conclusion cannot be made that our assay can be used for the identification of all 11 EV71 subgenogroups. Although quite a number of clinical samples and isolates were correctly identified for subgenogroups B5, C1, and C2, more samples belonging to the other subgenogroups should be tested in the future. Real-time RT-PCR can detect as few as 3 viral RNA copies and 10 plasmid copies (9, 10). Therefore, various amounts of the subgenogroups were subjected to analysis by our assay, and the sensitivity varied among the subgenogroup-specific probes (Table 2). With our multiplex assay, at least 100 PFU could be detected for all subgenogroups by plaque assay. Amounts as small as 1 PFU of C1, C2, and C5 and 5 PFU of A, B2, and B5 were detectable. Surprisingly, B4 could be detected only from 100 PFU, despite its relatively high reading in the specificity testing. The B4 probe has a GC percentage of 55%, with a melting temperature of 57.5°C, and it yielded a high reading when a larger viral load was tested for specificity. Therefore, this apparently lower sensitivity may be due to relatively poorer amplification efficiency of B4 with the consensus primers. Gel electrophoresis of amplicons supported this notion, since no bands were observed with 100 PFU of B4, whereas other subgenogroups exhibited bands with 5 or 20 PFU of template (data not shown). As expected, more virus particles were required for detection of subgenogroup C4, since the C4 probe had displayed lower binding activity. One potential limitation of determining the detection limit using the virus plaque assay is that it identifies only live virions and may thus overestimate the sensitivity. The sensitivity of the assay for the B1, B3, and C3 probes was evaluated using plasmid DNA templates, resulting in a detection limit of at least 100 copies. The use of plasmid DNA involves fewer steps and variables and may explain the smaller variations in detection limits between these three subgenogroups.
TABLE 1.
Mean sample-to-blank control ratios using EV71 subgenogroup-specific probes in the multiplex bead suspension assaya
| Reference strain or control | Sample-to-blank-control ratio for: |
Result of identification by sequencing | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EV71 subgenogroup-specific probe: |
Scrambled probe controlb | ||||||||||||
| A | B1 | B2 | B3 | B4 | B5 | C1 | C2 | C3 | C4 | C5 | |||
| Reference strains | |||||||||||||
| BrCr | 324.4 | 1.8 | 3.3 | 0.9 | 0.8 | 1.1 | 1.5 | 3.7 | 0.8 | 0.8 | 1.3 | 0.4 | A |
| 2609-AUS-74 | 7.7 | 764.9 | 199.3 | 8.2 | 10.1 | 10.7 | 8.4 | 5.5 | 4.9 | 3.1 | 1.5 | 0.5 | B1 |
| 7423/MS/87 | 1.7 | 18.9 | 833.1 | 1.5 | 1.5 | 2.6 | 2.4 | 1.4 | 1.6 | 1.0 | 1.1 | 0.4 | B2 |
| MY104-9-SAR-97 | 1.6 | 13.5 | 74.3 | 561.5 | 1.9 | 4.0 | 2.1 | 1.6 | 1.2 | 1.1 | 1.1 | 0.9 | B3 |
| 5865/SIN/000009 | 2.3 | 13.5 | 49.4 | 2.1 | 390.9 | 5.6 | 3.7 | 1.7 | 1.6 | 1.5 | 1.1 | 0.5 | B4 |
| 2933-Yamagata-03 | 2.3 | 4.6 | 54.9 | 1.5 | 1.8 | 97.7 | 2.0 | 1.5 | 1.1 | 1.0 | 1.2 | 0.4 | B5 |
| S10862-SAR-98 | 2.3 | 2.6 | 1.6 | 1.2 | 0.9 | 1.5 | 476.1 | 10.8 | 1.0 | 1.0 | 2.4 | 0.0 | C1 |
| Y97-1188 | 2.5 | 4.8 | 2.3 | 1.1 | 2.3 | 3.1 | 19.9 | 265.4 | 1.4 | 0.9 | 5.0 | 0.5 | C2 |
| 009-KOR-00 | 12.2 | 12.9 | 5.3 | 2.3 | 3.8 | 8.1 | 998.4 | 2.2 | 229.6 | 2.6 | 4.3 | 0.8 | C3 |
| 75-Yamagata-03 | 2.6 | 5.3 | 2.3 | 35.5 | 2.9 | 4.2 | 5.7 | 8.7 | 1.4 | 91.7 | 1.9 | 0.8 | C4 |
| 3437/SIN/06 | 2.9 | 70.6 | 4.0 | 1.8 | 2.9 | 2.7 | 41.5 | 7.7 | 3.1 | 1.6 | 176.4 | 0.5 | C5 |
| CA16 control | 2.5 | 0.6 | 0.0 | 0.3 | 1.7 | 0.0 | 0.0 | 1.2 | 0.6 | 0.1 | 1.3 | 0.6 | CA16 |
| Clinical specimens from 2008 outbreak | |||||||||||||
| NUH0029/SIN/08 | 1.6 | 8.3 | 10.1 | 1.5 | 96.1 | 1.0 | 0.0 | 1.1 | 7.6 | 2.9 | 0.7 | 1.0 | B4 |
| NUH0012/SIN/08 | 4.0 | 1.0 | 2.3 | 3.0 | 1.0 | 86.7 | 1.7 | 0.5 | 0.8 | 0.3 | 0.9 | 0.8 | B5 |
| NUH0037/SIN/08 | 4.5 | 1.3 | 2.1 | 2.3 | 0.1 | 60.3 | 1.3 | 0.7 | 0.6 | 0.3 | 0.9 | 0.7 | B5 |
| NUH0043/SIN/08 | 3.5 | 2.3 | 4.8 | 5.5 | 1.1 | 103.6 | 2.7 | 0.6 | 2.2 | 1.2 | 0.9 | 0.7 | B5 |
| NUH0047/SIN/08 | 2.3 | 0.0 | 1.9 | 5.0 | 2.3 | 27.5 | 4.9 | 1.3 | 0.5 | 0.2 | 1.5 | 2.2 | B5 |
| NUH0049/SIN/08 | 3.8 | 1.4 | 1.7 | 3.0 | 0.6 | 47.0 | 1.1 | 0.3 | 0.8 | 1.0 | 0.8 | 1.4 | B5 |
| NUH0083/SIN/08 | 3.8 | 0.5 | 2.5 | 5.7 | 1.0 | 68.6 | 1.4 | 0.8 | 0.8 | 1.2 | 0.9 | 0.9 | B5 |
| NUH0085/SIN/08 | 1.8 | 1.1 | 1.5 | 2.7 | 1.2 | 27.2 | 1.4 | 0.9 | 0.7 | 1.1 | 0.8 | 0.8 | B5 |
| NUH0086/SIN/08 | 0.0 | 1.2 | 1.2 | 8.7 | 0.9 | 25.4 | 1.4 | 0.9 | 0.5 | 1.1 | 0.9 | 1.8 | B5 |
| NUH0013/SIN/08 | 3.5 | 2.5 | 0.8 | 4.0 | 1.7 | 0.2 | 3.7 | 151.3 | 1.2 | 5.1 | 0.5 | 0.7 | C2 |
| NUH0075/SIN/08 | 3.3 | 2.1 | 0.8 | 5.3 | 1.1 | 3.9 | 4.1 | 160.3 | 1.3 | 6.5 | 1.2 | 1.2 | C2 |
The blank control is the background signal obtained with the respective probe in the absence of consensus PCR product. Numbers in boldface accurately identify the subgenogroups which correlated with the sequencing data of the 11 EV71 reference strains and 11 EV71-positive clinical specimens from the 2008 HFMD outbreak in Singapore.
The scrambled probe control was designed with a random or scrambled sequence.
TABLE 2.
Mean sample-to-blank control ratios using EV71 subgenogroup-specific probes for determination of assay sensitivitya
| Probe | Reference strain | Sample-to-blank-control ratio for virus PFU |
Ratio for no. of plasmid copies |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 20 | 50 | 100 | 1 | 10 | 100 | 1,000 | ||
| A | BrCr | 0.8 | 56.1 | 91.6 | 174.6 | |||||
| B2 | 7423/MS/87 | 3.6 | 94.1 | 89.1 | 207.6 | |||||
| B4 | 5865/SIN/000009 | 0.7 | 1.1 | 1.3 | 2.2 | 12.4 | ||||
| B5 | 2933-Yamagata-03 | 2.8 | 29.4 | 70.8 | 76.3 | |||||
| C1 | 90-3761 | 18.8 | 143.3 | 449.3 | 511.9 | |||||
| C2 | Y97-1188 | 18.6 | 103.3 | 126.5 | 141.1 | |||||
| C4 | 75-Yamagata-03 | 1.4 | 1.7 | 4.3 | 38.5 | |||||
| C5 | 3437/SIN/06 | 13.9 | 94.2 | 196.5 | 212.4 | |||||
| B1 | 2609-AUS-74 | 0.5 | 2.1 | 82.0 | 223.2 | |||||
| B3 | MY104-9-SAR-97 | 0.6 | 0.9 | 63.7 | 180.2 | |||||
| C3 | 009-KOR-00 | 0.9 | 1.6 | 11.7 | 142.4 | |||||
The blank control is the background signal obtained with the respective probe in the absence of consensus PCR product. Shown are mean sample-to-blank control ratios using EV71 subgenogroup-specific probes for the determination of assay sensitivity using reference strains. Numbers in boldface depict the detection limit, expressed as either the number of virus PFU or the number of plasmid copies.
In general, we noticed some cross-reactivity between certain subgenogroups, e.g., with reference strains and viral isolates. These samples carry relatively higher virus titers or plasmid copy numbers, which may explain the cross-reactivity, whereas virtually no cross-reactivity was observed in clinical specimens that had relatively lower viral loads. Moreover, a mixed infection is not commonly encountered in clinical settings due to viral interference that favors one dominant strain. In addition, despite some cross-reactivity observed for certain reference strains and viral isolates, the specific ratios were relatively much higher than nonspecific signals. However, in the titration experiments, the numbers of viral copies were at relatively low levels, and thus ratios that were 10-fold greater than the blanks were considered positive.
In conclusion, the multiplex suspension assay represents a specific, sensitive, reliable, and high-throughput method for EV71 detection and subgenogrouping. Its application was shown to be sensitive enough for clinical specimens and would greatly facilitate subgenogrouping to enhance epidemiologic analyses of EV71 infections.
Nucleotide sequence accession numbers.
The new VP1 nucleotide sequences were deposited in GenBank under accession numbers HQ285091 to HQ285109.
Supplementary Material
Acknowledgments
We are grateful to the patients and their parents, our clinical colleagues in the National University Hospital for obtaining specimens, S. H. Koo for technical advice, S. H. Lau for technical assistance, and FMC Asia Pacific, Hong Kong, for providing Avicel RC-591 for the viral plaque assays.
This study was supported by a grant from the Academic Research Fund, Ministry of Education, Singapore (R178-000-146-112).
Footnotes
Published ahead of print on 17 November 2010.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Baskaran, N., et al. 1996. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 6:633-638. [DOI] [PubMed] [Google Scholar]
- 2.Brown, B. A., M. S. Oberste, J. P. Alexander, M. L. Kennett, and M. A. Pallansch. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969-9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardosa, M. J., et al. 2003. Molecular epidemiology of human enterovirus 71 strains and recent outbreaks in the Asia-Pacific region: comparative analysis of the VP1 and VP4 genes. Emerg. Infect. Dis. 9:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan, K. P., et al. 2003. Epidemic hand, foot and mouth disease caused by human enterovirus 71, Singapore. Emerg. Infect. Dis. 9:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginocchio, C. C., and K. St. George. 2009. Likelihood that an unsubtypeable influenza A virus result obtained with the Luminex xTAG respiratory virus panel is indicative of infection with novel A/H1N1 (swine-like) influenza virus. J. Clin. Microbiol. 47:2347-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMinn, P., et al. 2001. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J. Virol. 75:7732-7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sim, A. C., A. Luhur, T. M. Tan, V. T. Chow, and C. L. Poh. 2005. RNA interference against enterovirus 71 infection. Virology 341:72-79. [DOI] [PubMed] [Google Scholar]
- 8.Singh, S., V. T. Chow, M. C. Phoon, K. P. Chan, and C. L. Poh. 2002. Direct detection of enterovirus 71 (EV71) in clinical specimens from a hand, foot, and mouth disease outbreak in Singapore by reverse transcription-PCR with universal enterovirus and EV71-specific primers. J. Clin. Microbiol. 40:2823-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan, E. L., et al. 2008. Rapid detection of enterovirus 71 by real-time TaqMan RT-PCR. J. Clin. Virol. 42:203-206. [DOI] [PubMed] [Google Scholar]
- 10.Tan, E. L., V. T. Chow, S. H. Quak, W. C. Yeo, and C. L. Poh. 2008. Development of multiplex real-time hybridization probe reverse transcriptase polymerase chain reaction for specific detection and differentiation of Enterovirus 71 and Coxsackievirus A16. Diagn. Microbiol. Infect. Dis. 61:294-301. [DOI] [PubMed] [Google Scholar]
- 11.Tee, K. K., et al. 2010. Evolutionary genetics of human enterovirus 71: origin, population dynamics, natural selection, and seasonal periodicity of the VP1 gene. J. Virol. 84:3339-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu, Y., et al. 2010. The largest outbreak of hand, foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int. J. Infect. Dis. 14:e1076-e1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.