Abstract
The HIV-1 RNA viral load is commonly used for the monitoring of disease progression and antiretroviral treatment of HIV-1-infected patients. Since the misestimating of values could lead to inappropriate therapeutical management, the comparative performances, especially the ability to span the genetic diversity of HIV-1, of available automated real-time assays need to be evaluated. We conducted a prospective study with 74 consenting patients enrolled between March 2007 and November 2008. A blood sample was obtained at the time of diagnosis of HIV seropositivity and blindly tested for HIV-1 RNA by at least 4 commercial tests: the Abbott m2000 RealTime HIV-1, bioMérieux NucliSens EasyQ HIV-1, version 1.2 (v1.2), and Cobas AmpliPrep/Cobas TaqMan (CAP/CTM) v1.0 and v2.0 assays. The means of difference were null between CAP/CTM v2.0 and Abbott for CRF02_AG subtypes but positive in favor of CAP/CTM v2.0 for genotype B and negative in favor of NucliSens for all genotypes. The standard deviation (SD) of difference ranged from 0.3 to 0.59, depending on the considered couples of assays. Reliabilities of these four tests, appreciated by the standard deviation of difference between the measurement and the estimated “true” viral load and by the coefficient of reliability, were significantly different (P < 10−4) among each other. Significant differences were also observed within each group of HIV-1 genotype. The global disparity was higher for CRF02_AG than for B subtypes. This study indicates a risk of viral load misestimating or discrepancies between techniques, depending on the HIV-1 subtype, and speaks in favor of using the same assay for the monitoring of HIV-1-infected patients.
The HIV RNA viral load in blood plasma is commonly used for the monitoring of disease progression, antiretroviral (ARV) treatment start, and survey of virological failure in HIV-infected patients. Real-time protocols have now supplanted first-generation nucleic acid amplification technologies, because they can be fully automated and faster and exhibit large dynamic ranges and high throughputs (12, 14, 23). Nowadays, several automated real-time assays are available, and their worldwide implementation is increasing, including in resource-limited countries, as recently recommended (5). Since the misestimating of the viral load could lead to inappropriate therapeutical management, the comparative performances of the assays, especially their ability to span the genetic diversity of HIV-1, need to be evaluated. This point is a major criterion to be considered for the development of assays able to amplify the different HIV-1 subtypes. Indeed, recent data have suggested viral load discrepancies between commercial quantitative assays, especially for the quantification of non-B subtype strains (2, 3, 8, 20). To achieve a high sensitivity threshold, a judicious choice of HIV-1 gene targets, primers, and probes is critical. Indeed, updated versions of the NucliSens EasyQ and Roche Cobas Ampliprep/Cobas TaqMan HIV-1 tests were recently introduced in order to improve the ability of their former versions to amplify non-B subtypes (17, 22).
In the work presented here, we conducted a prospective study comparing 4 real-time commercial tests, namely the Abbott m2000 RealTime, bioMérieux NucliSens EasyQ, version 1.2 (v1.2), and Roche Cobas AmpliPrep-Cobas TaqMan v1.0 and v2.0 assays, for HIV-1 RNA quantification in blood plasma from a cohort of patients that tested positive for HIV-1 for the first time at the University Hospitals of Lyon, Grenoble, and Saint-Etienne, France.
MATERIALS AND METHODS
Patients.
Consenting patients who tested positive for HIV-1 for the first time and were attending the Infectious Disease Departments of the University Hospital centers of Lyon, Grenoble, and Saint-Etienne were enrolled between March 2007 and November 2008. Seventy-four patients with a viral load detected by at least one technique were included in the comparison study. The study was conducted accordingly to the French Regulation Authority rules.
Sample collection.
Whole-blood samples were collected in EDTA tubes at the time of the first or second visit to the infectious disease units. Plasma was separated by centrifugation and aliquoted. All the samples were stored at −80°C until assayed in the Virobiotec collection at the Centre of Biological Resources of the University Hospital of Lyon.
HIV RNA quantification.
Each sample was blindly quantified for HIV-1 RNA by at least three of the four commercial assays, namely Abbott m2000 RealTime HIV-1 (Abbott Diagnostics) in the Saint-Etienne or Lyon laboratory, bioMérieux NucliSens HIV-1 EasyQ v1.2 in the Lyon laboratory, and the Roche Cobas AmpliPrep-Cobas TaqMan HIV-1, version 1.0 and version 2.0, (CAP/CTM v1.0 and v2.0) assays in the Grenoble laboratory.
Briefly, the m2000 RealTime HIV-1 assay combines an automated extraction (input volume of 0.6 ml, m2000sp apparatus), a real-time PCR amplification of the integrase gene fragment, and a noncompetitive fluorescent detection (m2000rt instrument, dynamic range of 40 to 107 copies/ml) (7). The NucliSens HIV-1 EasyQ v1.2 assay includes an automated nucleic acid extraction step (input volume of 1.0 ml, EasyMag apparatus) followed by a gag fragment nucleic acid sequence-based amplification (NASBA) associated with the detection of amplified products with molecular beacon probes (NucliSens analyzer, dynamic range of 50 to 3 × 106 copies/ml) (4). The CAP/CTM v1.0 consists of an automated RNA extraction (input volume of 1.0 ml, AmpliPrep instrument) followed by a gag fragment amplification by real-time PCR using a fluorescent TaqMan probe (Cobas TaqMan apparatus) (16). Compared to the previous version 1.0, the CAP/CTM v2.0 targets both gag and LTR regions and exhibits a dynamic range of 20 to 107 HIV RNA copies/ml (versus 40 to 107 for version 1.0) (3, 17).
HIV-1 subtype.
The HIV-1 subtype was determined by the analysis of the pol sequence at the time of the genotyping assay, by using the REGA HIV-1 Subtyping Tool (version 2.0) program.
Statistical analysis.
All assay results are reported in log copies/ml. Values quantified as ≥7 log copies/ml were changed to 7 log copies/ml. Summary statistics are presented as estimates (95% confidence intervals [CI]).
The agreement between any two assays, a1 and a2, was described using Bland-Altman analysis (1). The difference of the two assays, a2 − a1, was plotted against the average of a1 and a2: (a1 + a2)/2. The disagreement, in terms of bias and error, was calculated, respectively, as the mean m and the standard deviation (SD) s of the difference between a1 and a2; 95% limits of agreement were calculated as m ± 1.96 s.
Viral loads were then analyzed by linear regression for repeated measure determinations. In this model, each patient was supposed to have a “true” viral load value assessed by up to four measure determinations, one per assay. This model was used both to estimate the true viral load of each subject and the reliability of the four assays.
Viral load mean estimates (with CI) of the whole studied population for each assay were obtained with this linear regression for repeated measurements. This regression analysis provided a test for comparing couples of viral load means (H0 [the two means tested are equal] versus H1 [the two means are not equal]).
Reliability was used to quantify the consistency of our set of virus load measurements. The reliability was analyzed through two distinct approaches: the measurement error and the coefficient of reliability. The measurement error was expressed by standard deviation of the difference between the measurement and the estimated true viral load (SDEVL) obtained with the linear regression for repeated measures. The coefficient of reliability, calculated as σINTER2/(σINTER2 + σINTRA,a2) (where σINTER2 is the variance of virus load among subjects and σINTRA, a is the standard deviation from the estimated true viral load for assay a), ranges from 0 to 1 (1 stands for perfect reliability). The statistical significance of the differences among the coefficient of reliability of the assays was tested using a likelihood ratio test.
Similar analyses were performed for studying the influence of HIV-1 subtype on agreement or reliability. Only data from patients infected with CRF02_AG and B HIV-1 subtypes were analyzed, since sample sizes were not sufficient enough for the other subtypes.
RESULTS
Viral loads.
The viral loads obtained for each of the 74 samples by at least three of the four assays are listed in Table 1. Three samples gave at least one result under the threshold of detection (no. 45, 54, and 65). Theses three samples were analyzed separately. One sample (no. 45) was detectable by CAP/CTM v1.0 only (2.94 log copies/ml). Another sample (no. 54) was nondetectable by CAP/CTM v1.0 and positive by CAP/CTM v2.0 and m2000 RealTime (respectively, 3.81 and 3.47 log copies/ml). The third sample (no. 65) was nondetectable by CAP/CTM v1.0 only (1.99, 1.68, and 1.41 for CAP/CTM v2.0, m2000 RealTime, and NucliSens EasyQ, respectively).
TABLE 1.
Viral loads (in log copies/ml) of the 74 samples tested by the four commercial assays and their respective subtypea
| Sample no. | Subtype | Viral load (log copies/ml) |
|||
|---|---|---|---|---|---|
| NucliSens EasyQ | m2000 RealTime | CAP/CTM v1.0 | CAP/CTM v2.0 | ||
| 1 | CRF02_AG | 4.20 | 4.22 | 4.03 | 4.43 |
| 2 | CRF02_AG | 3.92 | 4.05 | 4.10 | 4.78 |
| 3 | B | 4.94 | 4.08 | 4.68 | 4.93 |
| 4 | B | 5.72 | 5.50 | 5.79 | 5.89 |
| 5 | CRF02_AG | 4.30 | 4.79 | 4.85 | 5.12 |
| 6 | B | 4.04 | 5.49 | 5.35 | 5.54 |
| 7 | B | 3.60 | 3.69 | 3.50 | 3.93 |
| 8 | Undet | 3.00 | 4.25 | 4.13 | 4.73 |
| 9 | B | 3.43 | 3.45 | 3.39 | 3.95 |
| 10 | CRF02_AG | 4.89 | 5.87 | 5.73 | 5.88 |
| 11 | B | 4.26 | 4.23 | 4.43 | ND |
| 12 | B | 4.08 | 4.69 | 5.02 | 5.13 |
| 13 | B | 3.59 | 3.87 | 3.80 | 4.26 |
| 14 | CRF02_AG | 3.08 | 4.20 | 3.85 | 3.89 |
| 15 | CRF01_AE | 4.04 | 4.76 | 5.13 | 5.26 |
| 16 | CRF02_AG | 5.15 | 5.29 | 4.72 | ND |
| 17 | CRF02_AG | 3.77 | 4.81 | 4.17 | ND |
| 18 | J | 4.08 | 4.85 | 4.60 | ND |
| 19 | B | 3.89 | 3.78 | 4.00 | ND |
| 20 | Undet | 3.88 | 4.35 | 4.12 | ND |
| 21 | B | 4.18 | 4.55 | 4.04 | ND |
| 22 | A | 3.41 | 5.09 | 4.84 | 5.41 |
| 23 | CRF02_AG | 3.04 | 4.71 | 4.10 | ND |
| 24 | Undet | 6.36 | 6.73 | 6.48 | ND |
| 25 | B | 3.62 | 3.99 | 4.08 | ND |
| 26 | D | 5.63 | 5.16 | 5.15 | 5.47 |
| 27 | B | 5.32 | 5.07 | 5.04 | 5.15 |
| 28 | G | 3.41 | 4.19 | 3.53 | 4.29 |
| 29 | CRF13_cpx | 6.20 | 6.12 | >7 | >7 |
| 30 | B | 4.54 | 5.27 | 5.28 | ND |
| 31 | CRF02_AG | 4.04 | 5.16 | 4.84 | ND |
| 32 | B | 5.11 | 4.84 | 4.59 | ND |
| 33 | Undet | 3.38 | 3.69 | 4.11 | 4.47 |
| 34 | CRF02_AG | 4.08 | 4.75 | 4.10 | 4.84 |
| 35 | CRF02_AG | 3.40 | 5.02 | 4.58 | 5.13 |
| 36 | G | 1.86 | 3.55 | 3.36 | ND |
| 37 | CRF02_AG | 5.08 | 5.81 | 5.05 | 5.80 |
| 38 | B | 4.26 | 4.47 | 4.85 | ND |
| 39 | J | 3.11 | 4.02 | 3.96 | 4.35 |
| 40 | CRF02_AG | 4.68 | 5.03 | 4.69 | ND |
| 41 | Undet | 3.23 | 3.78 | 3.61 | ND |
| 42 | B | 3.98 | 4.41 | 4.42 | 4.90 |
| 43 | CRF02_AG | 4.43 | 5.15 | 4.79 | 5.12 |
| 44 | B | 5.48 | 5.88 | 5.91 | 6.37 |
| 45 | ND | <1.70 | <1.60 | 2.94 | <1.30 |
| 46 | CRF02_AG | 4.08 | 4.05 | 3.72 | 4.47 |
| 47 | ND | ND | 3.62 | 3.61 | ND |
| 48 | B | ND | 3.03 | 2.95 | 3.47 |
| 49 | CRF02_AG | ND | 3.16 | 2.81 | 3.34 |
| 50 | B | ND | 4.11 | 3.60 | 4.39 |
| 51 | B | ND | 3.84 | 4.40 | 4.70 |
| 52 | CRF02_AG | ND | 5.18 | 5.02 | 5.52 |
| 53 | B | ND | 4.90 | 4.88 | 5.47 |
| 54 | CRF02_AG | ND | 3.47 | <1.60 | 3.81 |
| 55 | CRF02_AG | 3.20 | 4.16 | 3.66 | 4.04 |
| 56 | A | 3.83 | 5.18 | 4.72 | 5.02 |
| 57 | B | 4.34 | 4.88 | 5.05 | 5.18 |
| 58 | B | 3.30 | 3.72 | 3.68 | 4.07 |
| 59 | B | 4.74 | 4.81 | 4.78 | 4.83 |
| 60 | B | 3.75 | 4.02 | 4.24 | 4.20 |
| 61 | B | 3.67 | 3.42 | 3.06 | 3.72 |
| 62 | B | 5.40 | 5.53 | 5.46 | 5.42 |
| 63 | Undet | ND | 6.53 | 5.89 | 6.21 |
| 64 | CRF06_cpx | 4.04 | 4.67 | 4.57 | 4.73 |
| 65 | NA | 1.41 | 1.68 | <1.60 | 1.99 |
| 66 | CRF02_AG | 4.49 | 6.17 | 4.63 | 4.60 |
| 67 | CRF02_AG | 4.92 | 5.82 | 3.86 | 5.47 |
| 68 | C | ND | 5.21 | 4.80 | ND |
| 69 | B | 4.48 | 4.52 | 4.20 | 4.51 |
| 70 | B | 5.08 | 5.16 | 4.93 | 5.06 |
| 71 | CRF02_AG | 3.18 | 4.07 | 3.99 | 4.24 |
| 72 | B | 4.11 | 3.83 | 3.14 | 3.89 |
| 73 | B | ND | 4.49 | 4.15 | 4.70 |
| 74 | CRF02_AG | 4.15 | 5.53 | 5.15 | 5.00 |
Undet, indeterminate; NA, not amplified; ND, not done.
HIV-1 subtype.
The HIV-1 subtype was determined for 71 samples. The following subtypes were obtained: B (31 samples), CRF02_AG (23 samples), A (2 samples), G (2 samples), J (2 samples), C (1 sample), D (1 sample), CRF01_AE (1 sample), CRF06_cpx (1 sample), CRF13 (1 sample), and indeterminate (6 samples).
Global comparison between the four assays. (i) Bland-Altman analysis.
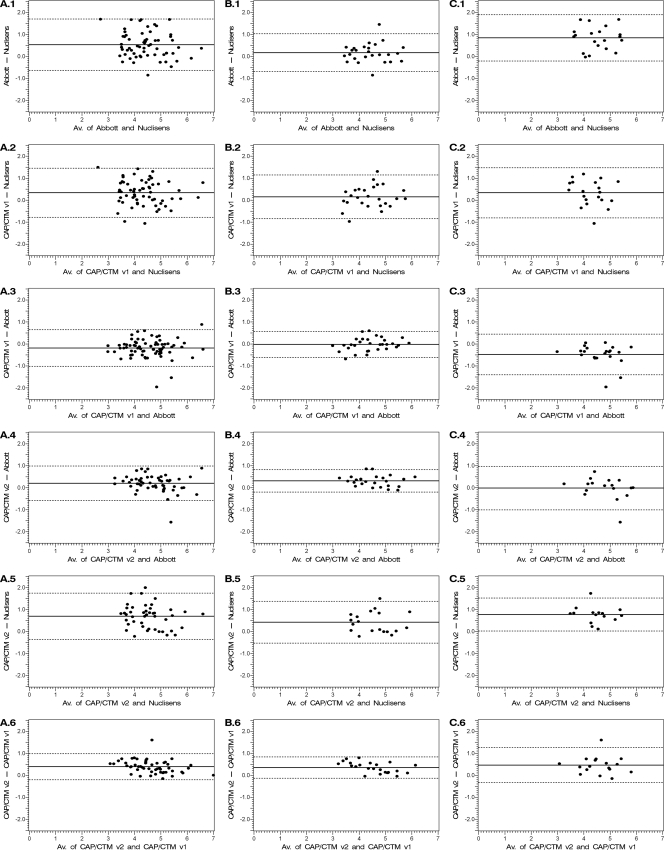
Bland-Altman plots for any possible pairs of assays are depicted in Fig. 1. For CAP/CTM v2.0 and any other assay (Fig. 1, A.4, A.5, and A.6), the bias, i.e., the mean of the difference, was always positive in favor of CAP/CTM v2.0 (0.2, 0.68, and 0.38 compared to m2000 RealTime, NucliSens EasyQ, and CAP/CTM v1.0 assays, respectively). For NucliSens EasyQ and any other assay (Fig. 1, A.1, A.2, and A.5), the bias was always negative in favor of NucliSens (−0.52 and −0.33 compared to m2000 RealTime and CAP/CTM v1.0 assays, respectively). CAP/CTM v1.0 and m2000 RealTime (Fig. 1C) exhibited the lowest bias (mean of CAP/CTM v1.0 − m2000 RealTime: −0.19) between each other. The standard deviation of the difference between any two assays ranged from 0.3 (CAP/CTM v2.0 versus CAP/CTM v1.0; limits of agreement, −0.21;0.97) to 0.59 (m2000 RealTime versus NucliSens EasyQ; limits of agreement, −0.64;1.68).
FIG. 1.
Bland-Altman analyses of the agreement between couples of HIV viral load assays (in log copies/ml): Abbott m2000 RealTime and NucliSens HIV-1 EasyQ v1.2 (row 1), CAP/CTM v1.0 and NucliSens EasyQ (row 2), CAP/CTM v1.0 and Abbott m2000 RealTime (row 3), CAP/CTM v2.0 and Abbott m2000 RealTime (row 4), CAP/CTM v2.0 and NucliSens EasyQ (row 5), and CAP/CTM v2.0 and CAP/CTM v1.0 (row 6). Bland-Altman analyses were carried out on all the patients (A), on HIV-1 B subtypes (B), and on HIV-1 CRF02_AG subtype (C). The x axis bears the average value for each sample obtained by the two techniques. The y axis bears the difference between the values obtained with the two techniques. The solid lines represent the mean of the differences between the values, and the dashed lines represent the mean of the differences ± 1.96 standard deviation (SD) (95% limits of agreement).
(ii) Linear regression for repeated measurements.
Roche CAP/CTM v2.0 exhibited mean values significantly higher than those of the 3 remaining assays (P < 10−4; 4.84 [4.67 to 5.02], 4.65 [4.46 to 4.84], 4.46 [4.28 to 4.64], and 4.14 [3.93 to 4.35] for CAP/CTM v2.0, m2000 RealTime, CAP/CTM v1.0, and NucliSens EasyQ assays, respectively).
Reliabilities of these four assays were significantly different (P value <10−4) among each other. The lowest SDEVL, i.e., the best precision, was obtained with CAP/CTM v2.0 (0.14 [0.09; 0.32]) followed by CAP/CTM v1.0 (0.26 [0.21;0.33]), m2000 RealTime (0.35 [0.29;0.43]), and finally NucliSens EasyQ (0.5 [0.419;0.62]). Similar results were obtained for the coefficients of reliability (0.96, 0.89, 0.82, and 0.68 for CAP/CTM v2.0, CAP/CTM v1.0, m2000 RealTime, and Nuclisens EasyQ, respectively).
Influence of the HIV-1 subtype. (i) Bland-Altman analysis.
Bland-Altman analyses were performed for CRF02_AG (Fig. 1C) and B subtypes (Fig. 1B) separately. In our study, disagreement was globally more important for CRF02_AG subtypes than for B subtypes, especially between any two of the three assays CAP/CTM v1.0, CAP/CTM v2.0, and m2000 RealTime. For the CRF02_AG subtype, the mean of the difference from CAP/CTM v2.0 − m2000 RealTime was estimated as −0.02. Standard deviation of this difference was estimated as 0.5 (95% limits of agreement, −1.01;0.96). This standard deviation might be overestimated because of the presence of an outlier (the dot at the bottom of Fig. 1, C.4). For the B subtype, the mean of the difference between CAP/CTM v2.0 and m2000 RealTime was estimated as 0.3. Standard deviation of this difference was estimated by 0.26 (95% limits of agreement, −0.21;0.8).
(ii) Linear regression for repeated measurements.
Estimates of the mean, SDEVL (with CI), and coefficient of reliability are available in Table 2. For B subtypes, the mean of viral loads measured by CAP/CTM v2.0 was significantly higher than the means measured by the three other assays (Table 2). Estimates of the means for the CAP/CTM v1.0 and m2000 RealTime assays were similar for B subtypes. NucliSens EasyQ exhibited the lowest mean viral load for B subtypes. For CRF02_AG subtypes, the CAP/CTM v2.0 and m2000 RealTime tests gave comparable means of viral loads (Table 2). The means of viral loads for these two tests appear to be significantly higher (P < 10−4) than those of the two other ones. The NucliSens EasyQ assay exhibited once again the lowest viral load mean estimate.
TABLE 2.
Estimate of the mean (with CI), standard deviation from the estimated true viral load SDEVL (with CI), and coefficient of reliability for four viral load assays (CAP/CTM v2.0, CAP/CTM v1.0, m2000 RealTime, and NucliSens EasyQ), by HIV-1 subtypes (CRF02_AG and B subtypes)
| Viral load assaya | No. tested | HIV-1 subtype | Mean (95% CI)b | SD (95% CI) | Coefficient of reliability |
|---|---|---|---|---|---|
| CAP/CTM v2.0 | 17 | CRF02_AG | 4.83 (4.54;5.13) | 0.20 (0.13;0.40) | 0.91 |
| 24 | B | 4.74 (4.50;4.98) | 0.14 (0.09;0.28) | 0.96 | |
| CAP/CTM v1.0 | 22 | CRF02_AG | 4.38 (4.08;4.69) | 0.32 (0.24;0.44) | 0.81 |
| 31 | B | 4.41 (4.17;4.65) | 0.22 (0.17;0.31) | 0.89 | |
| m2000 RealTime | 22 | CRF02_AG | 4.86 (4.56;5.17) | 0.34 (0.27;0.47) | 0.78 |
| 31 | B | 4.44 (4.19;4.68) | 0.24 (0.19;0.34) | 0.88 | |
| NucliSens EasyQ v1.2 | 20 | CRF02_AG | 4.06 (3.70;4.42) | 0.52 (0.40;0.74) | 0.61 |
| 26 | B | 4.29 (4.02;4.56) | 0.37 (0.30;0.48) | 0.76 |
Limits of detection were 1.3, 1.6, 1.6, and 1.7 log copies/ml for CAP/CTM v2.0, CAP/CTM v1.0, m2000 RealTime, and NucliSens EasyQ v1.2, respectively.
For CRF02_AG subtypes, the means were significantly different when assays were compared two by two, except between m2000 RealTime and CAP/CTM v2.0. For B subtypes, the means were significantly different when assays were compared two by two, except between m2000 RealTime and CAP/CTM v1.0, between m2000 RealTime and NucliSens EasyQ v1.2, and between CAP/CTM v1.0 and NucliSens EasyQ v1.2.
Once again, with CRF02_AG or B subtypes, the reliabilities of these four assays were significantly different (P < 10−4) among each other. The best precision was obtained with CAP/CTM v2.0, followed by CAP/CTM v1.0, m2000rt RealTime, and finally NucliSens EasyQ (SDEVL estimates ranged from 0.2 to 0.52 for CRF02_AG subtype and from 0.14 to 0.37 for B subtype) (Table 2). Results concerning the coefficients of reliability showed similar trends (Table 2). Furthermore, according to our data, the reliabilities of the assays varied for patients with an HIV-1 subtype and those with another one. The estimate of SDEVL for CRF02_AG was 40% higher than for B subtype (the ratio of the SDEVL for the CRF02_AG subtype divided by the SDEVL for B subtypes was estimated at 1.41 [1.13 to 1.88]), so the reliability of the four assays was globally weaker for CRF02_AG than for B subtypes.
DISCUSSION
The quantification of HIV-1 RNA levels in blood plasma is an important tool for determining the risk of progression of infection and monitoring the response of HIV-infected patients to antiretroviral therapy (ART). HIV diversity is a critical point to be considered for the development of genomic amplification techniques, particularly for plasma viral load assays. On the one hand, from an epidemiological point of view, in the United States and Europe there is an increasing number of patients newly infected by non-B subtypes, especially by CRF02_AG strains (21). In France, the proportion of non-B subtypes increased dramatically between 1995 and 2002 and has remained stable since 2003. Indeed, almost 40% of the newly diagnosed patients are infected by a non-B subtype. The increasing diversity of HIV-1 viruses in France, even in Caucasian patients diagnosed at the time of primary infection, was recently described in the French ANRS CO06 primo cohort study (6). Moreover, in resource-limited countries, where 90% of new infections occur, there is increasing access to ART for HIV-infected people, thanks especially to action plans decided on in 2000 during the International AIDS Conference of Durban, South Africa, and to the “3 by 5′” initiative and thus an increasing need to quantify HIV viral loads. For these reasons it is of high importance to use appropriate viral load assays able to span this genetic diversity for the management of naïve and also ARV-treated patients. To verify this critical point, there is a need of comparative studies for the determination of the sensitivity of existing assays and their ability to amplify RNA, especially for non-B subtypes.
In our study, whatever the HIV-1 subtype amplified, the CAP/CTM v2.0 assay exhibited the lowest SDEVL, the best precision, and mean values significantly higher than those of the 3 other assays. This overquantification could be due to the performance of the automated extractors but more probably to the amplification step and viral load calculation since the same extractor is used for the 2.0 and 1.0 versions of the Roche test. Our findings confirm the results found by Scott et al. (17) that, for a group of predominantly HIV-1 C subtype patients, CAP/CTM v2.0 presented a positive bias of 0.33 and 0.48 log copies/ml over the m2000 RealTime and CAP/CTM v1.0 assays, respectively. In our study, an improved sensitivity of the 2.0 over the 1.0 version was also observed, in agreement with previous studies (13, 17, 18); Scott et al. also described more quantifiable results down to 20 HIV RNA copies/ml for ARV-treated patients (17, 18); since our samples originated from newly diagnosed and untreated patients, we were unable to confirm this point. In our study, the NucliSens HIV-1 EasyQ v1.2 assay was the least favorable, exhibiting significantly lower viral loads and wider discrepancies. This observation was also reported in two other studies (18, 20) using the former version 1.1. Holguin et al. (9) found that the new version of NucliSens HIV-1 EasyQ compares favorably with RNA bDNA v3.0 and Cobas AmpliPrep/Cobas TaqMan v1.0, but false-positive results obtained with the NucliSens HIV-1 EasyQ v1.2, due to human plasma, were also recently reported (22). It is nevertheless fair to mention that positive results for patients having a low viral load are stronger evidence of good sensitivity than a high mean value which might be a biased estimate. Surprisingly, B subtypes were “overquantified” by the CAP/CTM v2.0 compared to the m2000rt RealTime test. The explanation is unclear since this phenomenon was not observed for CRF02_AG strains, suggesting possible bias due to the amplification of two targets per genome for CAP/CTM v2.0, with inherent calculations. An increased number of detectable viral loads at the critical threshold of 50 copies per ml with the CAP/CTM v1.0 versus the Amplicor assay was recently reported, suggesting that caution is required in the interpretation of low viral loads obtained with this assay (11).
Although only 23 strains were tested, CRF02_AG subtype samples yielded significantly higher results when viral loads were measured with the CAP/CTM HIV-1 v2.0 and m2000 RealTime assays than with the CAP/CTM HIV-1 v1.0 and NucliSens EasyQ ones. Similar results were previously obtained for the subtype C and CRF02_AG strains (13, 17). The m2000 RealTime assay also compared favorably with NucliSens EasyQ and CAP/CTM v1.0 for the G and CRF02_AG strains in another study (18). It is interesting to notice that the means of the viral loads assessed by the m2000rt RealTime and CAP/CTM v2.0 assays were comparable for the CRF02_AG subtype, whereas the mean of viral loads estimated by m2000rt RealTime was significantly lower than the mean estimated with CAP/CTM v2.0 for B subtypes. This could be due, at least in part, to genetic differences between integrase and gag/LTR regions. The poor recognition by CAP/CTM v1.0 of non-B subtypes was previously described by others (8, 10), and De Bel et al. recently also reported an underquantification with some B subtype strains (3). Indeed, the ability of the CAP/CTM assay to recognize non-B subtypes was dramatically improved between v1.0 and v2.0 (17; this study). Recently, the ability of another real-time assay, namely, the generic HIV viral load assay, compared to the Amplicor HIV-1 Monitor v1.5 and NucliSens EasyQ v1.2 to better quantify non-B strains was established by Rouet et al. (15). Until now, no comparison of this generic assay with the CAP/CTM v2.0 and the m2000 RealTime tests had been done.
Statistically significant wider discrepancies were observed for CRF02_AG than for B strains when comparing results obtained with the four assays tested in the present study, suggesting a weaker ability of these assays to quantify these non-B strains. Holguin et al. (9) also showed differences or discrepancies among non-B HIV-1 subtypes when comparing the performances of the following three assays: Versant HIV-1 RNA bDNA v3.0, CAP/CTM v2.0, and NucliSens EasyQ v1.2. This observation emphasizes the need to improve the design of primers and probes and could be linked, at least in part, to differences of genetic variability among strains belonging to the same subtype.
Our study is focused on samples taken from a French cohort, including B- and CRF02_AG-infected patients. Globally, NucliSens EasyQ v1.2 seems to differ from CAP/CTM v1.0, CAP/CTM v2.0, and m2000 RealTime. However, our results also underline the limitation of automated real-time protocols for non-B HIV-1 subtypes, despite a recent improvement of the sensitivity of these assays with these strains, with the possibility of wide discrepancies and the misestimating of the viral load. As a whole, these observations speak in favor of using the same assay for monitoring treatment of HIV-1-infected patients, for resistance studies, and for clinical trials, eventually, after testing for the most appropriate assay for non-B subtypes.
Acknowledgments
We thank Patricia Barbot from Virobiotec and the Center for Biological Resources, Hospices Civils de Lyon, for her support in the management of the samples.
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Altman, D. G., and J. M. Bland. 1983. Measurement in medicine: the analysis of method comparison studies. Statistician 32:307-317. [Google Scholar]
- 2.Damond, F., B. Roquebert, A. Benard, G. Collin, M. Miceli, P. Yeni, F. Brun-Vezinet, and D. Descamps. 2007. Human immunodeficiency virus type 1 (HIV-1) plasma load discrepancies between the Roche COBAS AMPLICOR HIV-1 MONITOR version 1.5 and the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 assays. J. Clin. Microbiol. 45:3436-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Bel, A., D. Marissens, L. Debaisieux, C. Liesnard, S. Van den Winjngaert, S. Lauwers, and D. Piérard. 2010. Correction of underquantification of human immunodeficiency virus type 1 load with the second version of the Roche Cobas AmpliPrep/Cobas TaqMan Assay. J. Clin. Microbiol. 48:1337-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Mendoza, C., M. Koppelman, B. Montes, V. Ferre, V. Soriano, H. Cuypers, M. Segondy, and T. Oosterlaken. 2005. Multicenter evaluation of the NucliSens EasyQ HIV-1 v1.1 assay for the quantitative detection of HIV-1 RNA in plasma. J. Virol. Methods 127:54-57. [DOI] [PubMed] [Google Scholar]
- 5.Fiscus, S. A., B. Cheng, S. M. Crowe, L. Demeter, C. Jennings, V. Miller, R. Respess, and W. Stevens. 2006. HIV-1 viral load assays for resource-limited settings. PLoS Med. 3:e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frange, P., J. Galimand, N. Vidal, C. Goujard, C. Deveau, F. Souala, M. Peeters, L. Meyer, C. Rouzioux, and M. L. Chaix. 2008. New and old complex recombinant HIV-1 strains among patients with primary infection in 1996-2006 in France: the French ANRS CO06 primo cohort study. Retrovirology 5:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Diaz, A., G. S. Clewly, C. L. Booth, W. Labett, N. McAllister, and A. M. Geretti. 2006. Comparative evaluation of the performance of the Abbott real-time human immunodeficiency virus type 1 (HIV-1) assay for measurement of HIV-1 plasma viral load following automated specimen preparation. J. Clin. Microbiol. 44:1788-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gueudin, M., J. C. Plantier, V. Lemee, M. P. Schmitt, L. Chartier, T. Bourlet, A. Ruffaut, F. Damond, M. Vray, and F. Simon. 2007. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 44:500-505. [DOI] [PubMed] [Google Scholar]
- 9.Holguin, A., M. Lopez, M. Molinero, and V. Soriano. 2008. Performance of three commercial viral load assays, Versant human immunodeficiency virus type 1 (HIV-1) RNA bDNA, v3.0, Cobas AmpliPrep/Cobas TaqMan HIV-1, and NucliSens HIV-1 EasyQ v1.2, testing HIV-1 non-B subtypes and recombinant variants. J. Clin. Microbiol. 46:2918-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labbett, W., A. Garcia-Diaz, Z. Fox, G. S. Clewlay, T. Fernandez, M. Johnson, and A. M. Geretti. 2009. Comparative evaluation of the ExaVirTM load version 3 reverse transcriptase assay for the measurement of HIV-1 plasma viral load. J. Clin. Microbiol. 47:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lima, V., R. Harrigton, and J. S. G. Montaner. 2009. Increased reporting of detectable plasma HIV-1 RNA levels at the critical threshold of 50 copies per milliliter with the Taqman assay in comparison to the Amplicor assay. J. Acquir. Immune Defic. Syndr. 51:3-6. [DOI] [PubMed] [Google Scholar]
- 12.MacDougall, D. S. 1996. Quantitative measurement of HIV RNA: techniques and clinical applications. J. Int. Assoc. Physicians AIDS Care 2:9-14. [PubMed] [Google Scholar]
- 13.Pas, S., J. W. A. Rossen, D. Schoener, D. Thamke, A. Pettersson, R. Babiel, and M. Schutten. 2010. Performance evaluation of the new Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 test version 2.0 for quantification of human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 48:1195-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peter, J. B., and R. A. Blum. 2002. Monitoring HIV viral loads in the United States: recent trends and methodologies. J. Acquir. Immune Defic. Syndr. 30:261-262. [DOI] [PubMed] [Google Scholar]
- 15.Rouet, F., V. Foulongne, J. Viljoen, K. Steegen, P. Becquart, D. Valea, S. Danaviah, M. Segondy, C. Verhofstede, and P. Van de Perre, for the WHO/ANRS 1289 Kesho Bora Study Group. 2010. Comparison of the generic HIV Viral Load®, Amplicor™ HIV-1 Monitor v1.5 and Nuclisens HIV-1 EasyQ® v1.2 techniques for plasma HIV-1 RNA quantitation of non-B subtypes: the Kesho Bora preparatory study. J. Virol. Methods 163:253-257. [DOI] [PubMed] [Google Scholar]
- 16.Schumacher, W., E. Frick, M. Kauselmann, V. Maier-Hoyle, R. Van der Vliet, and R. Babiel. 2007. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J. Clin. Virol. 38:304-312. [DOI] [PubMed] [Google Scholar]
- 17.Scott, L., S. Carmona, and W. Stevens. 2009. The performance of the new Roche COBAS Ampli/COBAS TaqMan HIV-1 version 2.0 assay. J. Clin. Microbiol. 47:3400-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott, L. E., L. D. Noble, J. Moloi, L. Erasmus, W. D. F. Venter, and W. Stevens. 2009b. Evaluation of the Abbott m2000 RealTime human immunodeficiency virus type 1 (HIV-1) assay for HIV load monitoring in South Africa compared to the Roche Cobas AmpliPrep-Cobas Amplicor, Roche Cobas AmpliPrep-Cobas TaqMan HIV-1, and BioMerieux NucliSENS EasyQ HIV-1 assays. J. Clin. Microbiol. 47:2209-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Swanson, P., C. de Mendoza, Y. Joshi, A. Golden, R. L. Hodinka, V. Soriano, S. G. Devare, and J. Hackett, Jr. 2005. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J. Clin. Microbiol. 43:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UNAIDS. 2008. Report on the global AIDS epidemic. UNAIDS, Geneva, Switzerland. http://www.unaids.org/en/KnowledgeCentre/HIVData/GlobalReport/2008/.
- 22.van Zyl, G. U., S. N. J. Korsman, L. Maree, and W. Preiser. 2010. NucliSens EasyQ® HIV-1 V1.2 system: detection of human plasma-derived background signal. J. Virol. Methods 165:318-319. [DOI] [PubMed] [Google Scholar]
- 23.Wittek, M., M. Sturmer, H. W. Doerr, and A. Berger. 2007. Molecular assays for monitoring HIV infection and antiretroviral therapy. Expert Rev. Mol. Diagn. 7:237-246. [DOI] [PubMed] [Google Scholar]