Abstract
The International Circumpolar Surveillance (ICS) Program was initiated in 1999 to conduct population-based surveillance for invasive pneumococcal disease in select regions of the Arctic. An interlaboratory quality control (QC) program for pneumococcal serotyping and antibiotic susceptibility testing was incorporated into ICS by reference laboratories in northern Canada (Laboratoire de Santé Publique du Québec [LSPQ] in Sainte-Anne de Bellevue, Québec; National Centre for Streptococcus [NCS] in Edmonton, Alberta) and Alaska (Arctic Investigations Program [AIP]). The World Health Organization's Collaborating Centre for Reference and Research on Pneumococci at the Statens Serum Institute (SSI) in Copenhagen, Denmark, joined the QC program in 2004. The Iceland Reference Laboratory (IRL) in Reykjavik, Iceland, joined the QC program in 2006, but due to small sample sizes, data from IRL are not included in this report. From 1999 through 2008, 190 isolates were distributed among four laboratories (AIP, NCS, LSPQ, and SSI). The overall serotype concordance was 95.8%, and the overall serogroup concordance was 97.4%. The overall modal MIC concordance for testing by broth microdilution (BMD) and agar dilution was >96% for all the antibiotics except erythromycin (92.1%) and clindamycin (89.5%). MIC comparisons between the Etest and BMD resulted in lower concordance for erythromycin (73.9%), clindamycin (65.5%), and trimethoprim-sulfamethoxazole (80%); however, categorical concordance (susceptible, resistant) remained high at 98.6%, 89.1%, and 90.9%, respectively. Our data demonstrate a high degree of correlation of serotyping and antimicrobial susceptibility testing results between four participating laboratories.
Global concern about emerging infectious diseases served as a catalyst for initiating a collaborative program for surveillance, prevention, and control of diseases of high incidence among indigenous and nonindigenous populations in Alaska, northern Canada, and other circumpolar regions. The International Circumpolar Surveillance (ICS) Program, an infectious disease surveillance network, was established in 1999 in Canada and the United States (Alaska) and initially focused on invasive pneumococcal disease, a leading cause of pneumonia and meningitis, especially among indigenous persons (2, 10). In Alaska, prospective population-based surveillance data are collected by the Centers for Disease Control and Prevention's Arctic Investigations Program (AIP), which serves as the reference laboratory for Streptococcus pneumoniae for 23 hospitals in the state. In Canada, population-based surveillance data are collected by the Public Health Agency of Canada. The National Centre for Streptococcus (NCS) in Edmonton, Alberta, Canada, serves as the pneumococcal reference laboratory for all provinces and territories in Canada except Quebec. The Laboratoire de Santé Publique du Québec (LSPQ) in Sainte-Anne de Bellevue, Québec, Canada, serves as the pneumococcal reference laboratory for seven hospitals in Quebec and Labrador.
Questions have recently been raised regarding the comparability of data generated from international surveillance systems that often use different testing methodologies within their networks. In 1997, NCS and LSPQ established a Canadian interlaboratory quality control (QC) program for S. pneumoniae to provide an external proficiency testing mechanism for serotyping and antibiotic susceptibility testing of S. pneumoniae. In 1999, with the formation of the ICS system, this QC program was expanded to include AIP in Alaska. In 2004, the World Health Organization's Collaborating Centre for Reference and Research on Pneumococci (now known as the Neisseria and Streptococcus Reference Laboratory) at Statens Serum Institute (SSI) in Copenhagen, Denmark, joined the ICS QC program. SSI also serves as a reference laboratory for S. pneumoniae for Greenland. In September 2006, the Iceland Reference Laboratory (IRL) in Reykjavik, Iceland, which serves as the reference lab for 10 laboratories throughout Iceland, joined the QC program.
The QC program provides a means for monitoring test results and standard operating procedures across all participating laboratories. Potential problems can be identified by the QC program, and correction of these problems produces improved quality of results and, ultimately, improved prevention programs and patient care. This report describes data from the ICS interlaboratory QC program collected from 1999 through 2008.
MATERIALS AND METHODS
Distribution of isolates from 1999 to 2008.
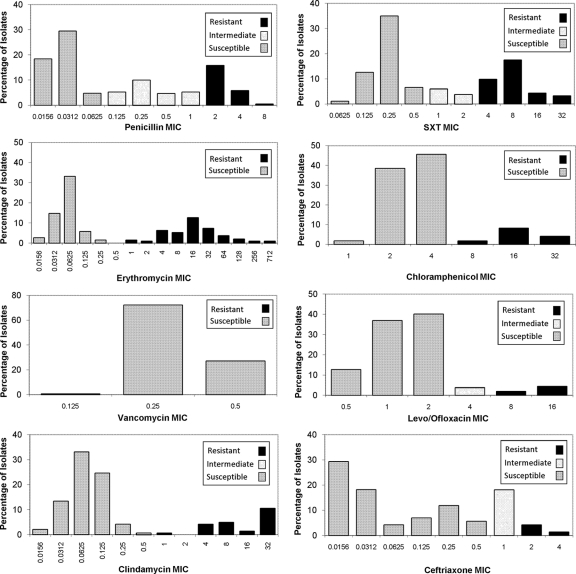
From 1999 through 2003, the participating laboratories (NCS, LSPQ, and AIP) were each responsible for the distribution of one set of seven S. pneumoniae isolates per year. Following the addition of SSI in 2004, the distribution schedule was changed to two distributions per year, with each participating laboratory being responsible for sending out one QC panel every 2 years. The distribution dates were agreed upon in advance. The pneumococcal isolates in each QC panel were selected by the distributing laboratory and represented a variety of serotypes and antibiotic susceptibility patterns (Fig. 1). Isolates were transported using charcoal transport medium, chocolate agar slants, or SSI transport medium, all of which support viability. Blood agar was not recommended for transport of S. pneumoniae, as some strains may not be viable after 2 to 3 days. The isolates were shipped according to International Air Transportation Association (IATA) regulations, and the delivery time to participating laboratories ranged from 1 to 3 days.
FIG. 1.
Number of S. pneumoniae isolates by MIC and categorical interpretation for the antimicrobial agents tested by ICS laboratories, 1999 to 2008.
Serotyping.
Serotyping was performed by the Quellung reaction using commercial antisera (SSI Diagnostica; Statens Serum Institute, Copenhagen, Denmark) (1). NCS, AIP, and SSI maintain complete sets of antisera for serotyping up to 90 serotypes; LSPQ and IRL maintain antisera for grouping and factoring the most common serotypes, including 6, 9, 18, 19, and 23. A serotype result was considered correct if it was consistent with the results identified in other laboratories using the antisera available at each of the laboratories. Serotype discrepancies between laboratories were grouped into one of three categories: those which were unexplained in occurrence, interconversion between 15B and 15C, and cross-reactions of shared factors. Unexplained discrepancies could include reporting a serotype for a nontypeable isolate or reporting a different serotype within a serogroup (factoring). The second discrepant category, interconversion between serotype 15B and 15C, has been reported in the literature (13). Cross-reactions of shared factors, the third category of discrepant results, occur when the isolate reacts with antibodies shared by more than one serogroup or serotype.
Antimicrobial susceptibility testing.
MICs were determined according to the method and antibiotics in use at each laboratory (Table 1). NCS and LSPQ used the reference broth microdilution (BMD) method with antibiotic trays prepared in-house according to Clinical and Laboratory Standards Institute (CLSI) guidelines (5). AIP used the agar dilution method until May 2000 and then changed to BMD using commercially prepared antibiotic trays (PML Microbiologicals, Wilsonville, OR). BMD and agar dilution were performed according to CLSI guidelines at AIP. SSI reported MICs obtained from the Etest (AB Biodisk, Solna, Sweden). MIC results were reported in micrograms per milliliter and, regardless of the method used, were expected to be within 1 log2 dilution of the modal MIC. The modal MIC represents the most frequently reported MIC value for a given isolate-antibiotic combination. For this report, if there was no modal MIC, then the median value was used as the reference MIC. Interpretive categorical concordance as susceptible or nonsusceptible (intermediate and resistant) was reported in addition to the modal MIC concordance for this report. Categorical interpretive errors were classified as major and very major. A major error was defined as false resistance, and a very major error was defined as false susceptibility. Etest concordance of erythromycin and clindamycin interpretations was determined according to the manufacturer's instructions. MIC results for S. pneumoniae ATCC 49619 were reported along with each QC panel and were expected to be within the range of the currently published CLSI M100 standards (4).
TABLE 1.
Antimicrobial agents tested and results reported for S. pneumoniae by each participating ICS laboratory, 1999 to 2008
| Antibiotic (yrs included in analysis) | Result was reported by: |
|||
|---|---|---|---|---|
| AIP | LSQP | NCS | SSIa | |
| Penicillin (1999-2008) | √ | √ | √ | √ |
| Cefotaxime (2002-2008) | √ | NRb | √ | √c |
| Ceftriaxone (2002-2008) | NR | √ | √ | √ |
| Chloramphenicol (1999-2008) | √ | √ | √ | NR |
| Clindamycin (2000-2008) | √ | √ | √ | √ |
| Erythromycin (1999-2008) | √ | √ | √ | √ |
| Fluoroquinolonesd (2002-2008) | √ | √ | √ | NR |
| Trimethoprim-sulfamethoxazolee (1999-2008) | √ | √ | √ | √ |
| Vancomycin (1999-2008) | √ | √ | √ | √ |
SSI joined the ICS S. pneumoniae QC program in 2004.
NR, not reported.
SSI started cefotaxime testing in 2005.
Ofloxacin and levofloxacin results were combined for this analysis.
Trimethoprim and sulfamethoxazole ratio of 1:19.
Exact MIC values for some antibiotics could not be determined due to the limited range of tested dilutions. For those results reported out as a “>” or “<” MIC, the next highest or lowest MIC was used in this analysis. These values fell within either the resistant or susceptible categories according to the CLSI document that was current at the date of testing. Etest results were rounded up to the next standard 2-fold dilution if the MIC fell in between the two dilutions, according to the manufacturers' instructions. For example, if the MIC was determined to be 0.094 μg/ml, the result would be rounded up to 0.12 μg/ml.
Comparisons were made as long as two laboratories reported results for a given antibiotic. The range of antibiotic concentrations tested varied among laboratories. Some changes occurred in the antibiotics selected for MIC testing over the study period. AIP, NCS, and LSPQ reported MIC results for penicillin, erythromycin, clindamycin, chloramphenicol, trimethoprim-sulfamethoxazole (SXT), and vancomycin consistently from the beginning (Table 1). NCS and LSPQ reported ceftriaxone results consistently from the beginning. AIP reported cefotaxime results from the beginning; NCS added cefotaxime in 2002. SSI reported both ceftriaxone and cefotaxime MICs when they joined the program in 2004. LSPQ did not report cefotaxime results during this study period.
Reporting.
A standardized report form was sent by the distributing laboratory with each QC panel. The information collected on the report form included the test method used and a table for reporting each serotype and MIC result. The report form was completed by each participating laboratory and then returned to the distributing laboratory within 6 weeks. The distributing laboratory was responsible for compiling a summary report of the results and comments when discrepancies were noted. Discussion and follow-up of the results among the laboratories were encouraged.
Ownership of isolates.
The QC isolates are considered the property of the province, state, or country from which they were distributed. The isolates could be retained for internal reference use but were not shared with other laboratories or used for research purposes by the receiving laboratories without written consent from the distributing laboratory.
Statistical analysis.
The data were entered into the Paradox (version 9.0) program and analyzed using Epi Info (version 6.04b) software (Centers for Disease Control and Prevention, Atlanta, GA) and SAS (version 8.0) software (SAS Institute, Cary, NC). Results from IRL are not included in this report because of the limited sample size available at the time of analysis.
RESULTS
Serotyping.
From 1999 through 2008, a total of 190 isolates representing 43 serotypes and 29 serogroups were distributed among the laboratories. The most common serogroups were 6, 9, 14, 19, and 23, which represented half (50.6%) of the distributed isolates. This was due to the fact that all laboratories had factoring antisera available for testing these serogroups and antimicrobial resistance has historically been associated with these serotypes, allowing for a wider range of MIC testing. The overall rates of concordance were 95.8% (182/190) for serotype and 97.4% (185/190) for serogroup. Three laboratories reported results for 121 isolates (64%); the rate of serotype concordance was 95.9%, and that of serogroup concordance was 96.7%. Four laboratories reported results for 69 (36%) isolates; the rate of serotype concordance was 96.7%, and that of serogroup concordance was 97.5%. One laboratory reported results for 30 isolates to the serogroup level only.
Discrepancies between the expected serotype and the reported results were noted in eight instances (Table 2). Four discrepancies were classified as unexplained, two (both serotype 15C) were due to interconversion, and two were due to cross-reactions of shared factors (29b and 38a). For one isolate, a single laboratory reported that it was not S. pneumoniae, and this isolate was excluded from this analysis. Two isolates were reported to be serotype 15C by two laboratories and serotype 15B by one laboratory. Serotypes 15B and 15C are known to interconvert in vitro and in vivo by the addition of an acetyl group to the capsular polysaccharide of serotype 15C to form an O-acetylated serotype 15B variant (7, 13). Discrepancies resulting from the incomplete serotyping of isolates where common factors are shared occurred with two isolates. These discrepancies were noted with serotype 35B and serogroup 29 and again in serogroup 38 and serotype 25A. The summary report explained that cross-reactivity between type 35B and type 29 may be expected due to the presence of factor 29b in the antigenic formula of both serotypes. Further factoring will differentiate the two serotypes. The cross-reactivity noted with serotype 25A and serotype 38 was also due to similarities in their antigenic formulae; both types carry factor 38a, and cross-reactivity with group 25 and type 38 antisera can occur (7).
TABLE 2.
Discordant S. pneumoniae serotype results reported by each participating ICS laboratory, 1999 to 2008
| Serotype result by laboratorya |
Discrepancy type | |||
|---|---|---|---|---|
| AIP | LSPQ | NCS | SSI | |
| 18B | 18C | 18B | NA | Unexplained |
| 5 | 5 | NT | NA | Unexplained |
| NT | 9V | NT | NA | Unexplained |
| 6A | 6A | 6A | 18B | Unexplained |
| 15C | 15 | 15B | 15C | Interconversion |
| 15C | 15 | 15B | 15C | Interconversion |
| 35B | 29 | 35B | NA | Shared factor |
| 38 | 38 | 25A | 38 | Shared factor |
NT, nontypeable; NA, not assessed.
Susceptibility testing.
The range of MICs reported for each antibiotic along with categorical interpretation (susceptible, intermediate, and resistant) is shown in Fig. 1. There were 190 isolates available for MIC comparisons, where three laboratories were performing BMD and agar dilution (Table 3). Comparisons to results from a fourth lab, performing MIC testing by Etest, are reported separately (Table 4). Overall modal MIC concordance for laboratories testing with BMD and agar dilution was >96% for all antibiotics except erythromycin (92.1%) and clindamycin (89.5%) (Table 3). Concordance by categorical interpretation (susceptible, resistant) was high (>94%) for all antibiotics except ceftriaxone (84.6%) when the CLSI M100-S17 (4) meningitis cutoffs were applied (Table 3).
TABLE 3.
Comparison of MIC results and concordance of modal MIC and categorical interpretations for S. pneumoniae from three ICS laboratories using broth microdilution and agar dilution, 1999 to 2008
| Antibiotic | MIC range (μg/ml) | No. of pairwise comparisons with MICs that differed from the modal (median) MIC (log2 dilutions) by: |
Total no. pairwise comparisons | Total no. of isolates | % concordance (no. of isolates with concordant results/total no. of isolates tested) by: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤−3 | −2 | −1 | Same | +1 | +2 | ≥+3 | Modal MIC (±1 log2) | Categorical interpretationa | ||||
| Penicillin | 0.015-256 | 0 | 1 | 59 | 457 | 54 | 0 | 0 | 570 | 190 | 100 (190/190) | 98.4 (187/190) |
| Vancomycin | 0.015-4 | 0 | 0 | 29 | 484 | 56 | 0 | 0 | 570 | 190 | 100 (190/190) | 100 (190/190) |
| Ceftriaxoneb | 0.015-4 | 0 | 0 | 22 | 117 | 17 | 0 | 0 | 156 | 52 | 100 (52/52) | 84.6 (46/52) |
| Ceftriaxonec | 0.015-4 | 0 | 0 | 22 | 117 | 17 | 0 | 0 | 156 | 52 | 100 (52/52) | 100 (52/52) |
| Levofloxacin | 0.5-32 | 2 | 0 | 48 | 337 | 39 | 0 | 0 | 426 | 142 | 98.6 (140/142) | 94.3 (134/142) |
| Chloramphenicol | 1-64 | 0 | 1 | 57 | 443 | 65 | 3 | 0 | 570 | 190 | 97.9 (186/190) | 96.8 (184/190) |
| SXT | 0.062-256 | 0 | 4 | 59 | 434 | 70 | 2 | 0 | 570 | 190 | 96.8 (184/190) | 96.3 (183/190) |
| Erythromycin | 0.015-256 | 3 | 6 | 116 | 343 | 94 | 8 | 0 | 570 | 190 | 92.1 (175/190) | 100 (190/190) |
| Clindamycin | 0.015-256 | 1 | 10 | 84 | 247 | 54 | 3 | 0 | 399 | 133 | 89.5 (119/133) | 99.2 (132/133) |
TABLE 4.
Concordance of MIC results for S. pneumoniae from one ICS laboratory using Etest and three ICS laboratories using broth microdilution, 1999 to 2008
| Antibiotic | MIC range (μg/ml) | Total no. of isolates | % concordance (no. of isolates) |
|||
|---|---|---|---|---|---|---|
| No CO2 adjustment for Erya and Clib |
CO2 adjustment for Erya and Clib |
|||||
| Modal MIC (±1 log2 dilution) | Interpretive category | Modal MIC | Interpretive category | |||
| Penicillin | 0.0156-4.0 | 69 | 98.6 (68) | 95.7 (66) | NAc | NA |
| Erythromycin | 0.0312-256.0 | 69 | 73.9 (51) | 98.6 (68) | 87 (60) | 100 (69) |
| SXT | 0.0625-16.0 | 55 | 80 (44) | 90.9 (50) | NA | NA |
| Clindamycin | 0.0312-256.0 | 55 | 65.6 (36) | 89.1 (49) | 76.4 (42) | 98.2 (54) |
Erythromycin (Ery) CO2 adjustment of +2 log2 dilutions according to Etest instructions.
Clindamycin (Cli) CO2 adjustment of +1 log2 dilution according to Etest instructions.
NA, not applicable.
Agreement by modal MIC was 100% for penicillin, vancomycin, and ceftriaxone (nonmeningitis cutoffs). Categorical agreement also remained high. For ceftriaxone, 52 isolates were tested by three laboratories. Categorical concordance was lower for ceftriaxone (84.6%) when the CLSI meningitis cutoffs were applied. All six isolates that lacked categorical agreement resulted from a difference of 1 log2 dilution: an MIC of 1.0 μg/ml (intermediate) compared with an MIC of 0.5 μg/ml (susceptible). Two laboratories tested an additional 115 isolates for ceftriaxone. The results for all 115 isolates were within ±1 log2 dilution of each other, and categorical concordance was 97% (111/115). Two laboratories reported cefotaxime results for 123 isolates with a 98.4% (121/123) MIC concordance and 94.3% (116/123) categorical concordance, according to the meningitis cutoffs (data not shown) (4).
For levofloxacin, chloramphenicol, and SXT, the modal MIC concordance (96 to 98%) and the categorical concordance (94 to 96%) were high (Table 3). For levofloxacin, six of eight isolates that did not agree categorically were the result of one or more labs reporting an MIC of 4.0 μg/ml (intermediate) and another lab reporting an MIC of 2.0 μg/ml (susceptible). For the two other isolates with discrepancies, one isolate had MICs of 1.0 μg/ml and 8.0 μg/ml reported, while the other had MICs of 2.0 μg/ml and 16.0 μg/ml. There were 48 additional isolates tested for levofloxacin by two laboratories. Results for all 48 isolates were within ±1 log2 dilution of each other, and categorical concordance was 90% (43/48). For SXT, six of seven categorical discrepant results were attributable to one lab reporting an MIC of 0.5 μg/ml or 0.25 μg/ml (susceptible), while another reported an MIC of 1.0 μg/ml (intermediate).
The remaining two antibiotics, erythromycin and clindamycin, had lower rates of modal MIC concordance at 92% and 89%, respectively, while their categorical concordance rate remained high at >99%. For erythromycin, 14 of 15 isolates with modal MIC discrepancies had MICs of ≥1.0 μg/ml (resistant) but not within ±1 log2 dilution of each other. For example, two laboratories reported an MIC of 32.0 μg/ml and one lab reported an MIC of 4.0 μg/ml. The majority of modal MIC discrepancies for clindamycin resulted from MICs of 0.125 μg/ml (susceptible) reported from one or more labs and 0.03 μg/ml (susceptible) reported from the others.
Very major (false susceptibility) interpretative errors occurred only for chloramphenicol (n = 6) and levofloxacin (n = 1). The absence of an intermediate category for chloramphenicol may have contributed to the six chloramphenicol results classified as very major errors.
Comparative results were available for a smaller subset of isolates where one laboratory was measuring MICs by Etest and three laboratories were using BMD. Rates of concordance by modal MIC were somewhat lower in this group, at 73.9% for erythromycin, 65.5% for clindamycin, and 80% for SXT (Table 4). Despite the low modal MIC concordance, categorical concordance remained above 89%. Adjustment of the categorical interpretations taking into account CO2 incubation, according to the manufacturer's instructions, resulted in 100% concordance for erythromycin and 92.8% for clindamycin.
DISCUSSION
Our S. pneumoniae QC data demonstrate a high degree of correlation for serotyping and antibiotic susceptibility testing between four pneumococcal reference laboratories. One hundred ninety isolates representing 43 serotypes and 29 serogroups were exchanged from 1999 through 2008, with only eight discrepant serotyping results occurring. The overall modal MIC concordance within 1 log2 dilution was 99.3% for penicillin, regardless of the method used. Overall modal MIC concordance for testing by BMD and agar dilution was >96% for all the antibiotics except erythromycin (92.1%) and clindamycin (89.5%). The greatest variance in MIC concordance occurred between the Etest and BMD methods for macrolides and SXT; however, categorical concordance remained high.
Previous reports describing international proficiency testing exercises for antimicrobial susceptibility testing, particularly for the detection of penicillin-nonsusceptible pneumococci, have used the oxacillin disk diffusion screening method (3, 12). The ICS QC program did not report results for oxacillin disk diffusion screening; however, the rates of modal MIC and categorical concordance were 100% and 99%, respectively, for penicillin, regardless of test method. Similar high levels of concordance were noted in phase II of a Latin American study, where the rates of MIC and categorical concordance for penicillin were 93.3% and 93.6%, respectively, (9). In another large international quality assurance exercise distributed by the United Kingdom National External Quality Assurance Scheme to the European Antimicrobial Resistance Surveillance System (EARSS) laboratories, concordance for penicillin-nonsusceptible isolates using the MIC method was 94%, whereas it was 79% for non-MIC-based methods (3). Concordance in the EARSS quality assurance exercise was based on two categories: susceptible and nonsusceptible.
Differences in methodologies while using the BMD and Etest resulted in lower rates of modal MIC concordance for erythromycin (73.9%), clindamycin (65.5%), and SXT (80%). Similar results for erythromycin (68.7%) were reported in the Latin American study (9). The wide variation in MICs was most likely due to the CO2 environment required for testing of S. pneumoniae with the Etest. A decrease in the pH of the agar resulting from the CO2 environment decreases the activity of macrolides, producing an elevated MIC (6).
Detection of emerging antibiotic resistance is an important function of reference laboratories. Participation in international external quality assurance (EQA) programs may provide insight into resistance rate variability between countries by examining the microbiological procedures being performed in each lab (11). The isolates selected for the ICS QC panels represented a wide range of MICs to ensure that each laboratory is able to detect resistance and accurately determine MICs, regardless of the method used. Comparison of data is more difficult when different methods are used; however, our MIC data demonstrated very good categorical concordance on the basis of CLSI breakpoints. In the ICS QC program, MIC data (as well as the test method used) are collected to ensure that analysis of the data will be possible over time as changes in the antibiotic breakpoints occur.
Reports describing programs that evaluate pneumococcal serotyping are limited (8, 9). Konradsen et al. (8) evaluated the quality of serotyping of pneumococci among 11 reference laboratories in Europe and found a serotype concordance of 95%. An international EQA program developed to monitor and support ongoing laboratory performance of serotyping and antibiotic susceptibility testing for S. pneumoniae in Latin America found overall rates of serotype concordance of 88% and 93.8% for phase I (1994 to 1998) and phase II (1999 to 2005) of the program, respectively (9). These results are comparable to the overall serotyping concordance of 95.8% that we identified in the ICS QC program. A total of 38 different serotypes, including three nonencapsulated strains, were selected for the European study (8), in contrast to the 43 serotypes represented in the ICS comparisons. The types of discrepancies noted in the European study were similar to those reported here and included the assignment of the wrong serotype within a serogroup, the misidentification of nontypeable or rough strains of S. pneumoniae, not screening for all possible serotypes when cross-reactions with shared factors occur, and some unexplained discrepancies. Few factoring errors were noted in the Latin American EQA program. The two shared-factor serotype discrepancies reported in this study emphasize the importance of screening isolates in all relevant antisera.
Serotyping of invasive isolates provides valuable information concerning the dynamics of pneumococcal disease in relation to vaccine uptake (8). The assignment of the correct serotype within a serogroup remains vital, as not all serotypes are represented in the current vaccines (8). Serotype distribution will continue to be an important outcome measure as new generations of capsule-based vaccines are introduced.
The results reported here demonstrate that the ICS S. pneumoniae QC program is a successful collaboration of reference labs which has proven to be sustainable over a span of 9 years. Sharing the workload associated with the selection, distribution, and analysis of the survey panels among the participants is a significant factor in the sustainability of this program. High concordance of both the serotyping and susceptibility results indicates that the reference laboratories are producing high-quality results which are comparable internationally and can detect seroprevalence and antibiotic resistance changes over time.
The ICS QC program has expanded to include other organisms. In 2005, the ICS S. pneumoniae QC program was used as a model to establish the ICS QC program for serotyping of Haemophilus influenzae and serogrouping of Neisseria meningitidis. These QC programs ensure that the quality of microbiological testing of these three important pathogens, all of which cause vaccine-preventable disease, remains high. External QC programs will continue to be a key component of quality surveillance systems.
Acknowledgments
We gratefully acknowledge the technical staff of all the reference laboratories who provide the necessary expertise and support to participate in the ICS S. pneumoniae QC program.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 3 November 2010.
REFERENCES
- 1.Austrian, R. 1976. The Quellung reaction, a neglected microbiological technique. Mt. Sinai J. Med. 43:699-709. [PubMed] [Google Scholar]
- 2.Bruce, M. G., S. L. Deeks, T. Zulz, D. Bruden, C. Navarro, M. Lovgren, L. Jette, K. Kristinsson, G. Sigmundsdottir, K. B. Jensen, O. Lovoll, J. P. Nuorti, E. Herva, A. Nystedt, A. Sjostedt, A. Koch, T. W. Hennessy, and A. J. Parkinson. 2008. International Circumpolar Surveillance System for invasive pneumococcal disease, 1999-2005. Emerg. Infect. Dis. 14:25-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaitram, J. M., L. A. Jevitt, S. Lary, and F. C. Tenover. 2003. The World Health Organization's External Quality Assurance System Proficiency Testing Program has improved the accuracy of antimicrobial susceptibility testing and reporting among participating laboratories using NCCLS methods. J. Clin. Microbiol. 41:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing. Seventeenth informational supplement M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.CLSI/NCCLS. 2003. Methods for dilution antimicrobial susceptibility testing, 6th ed. Approved standard M7-A6. CLSI/NCCLS, Wayne, PA.
- 6.Doern, G. V., A. B. Brueggemann, M. A. Pfaller, and R. N. Jones. 1999. Assessment of laboratory performance with Streptococcus pneumoniae antimicrobial susceptibility testing in the United States: a report from the College of American Pathologists Microbiology Proficiency Survey Program. Arch. Pathol. Lab. Med. 123:285-289. [DOI] [PubMed] [Google Scholar]
- 7.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konradsen, H. B. 2005. Validation of serotyping of Streptococcus pneumoniae in Europe. Vaccine 23:1368-1373. [DOI] [PubMed] [Google Scholar]
- 9.Lovgren, M., J. A. Talbot, M. C. Brandileone, S. T. Casagrande, C. I. Agudelo, E. Castaneda, M. Regueira, A. Corso, I. Heitmann, A. Maldonado, G. Echaniz-Aviles, A. Soto-Nogueron, M. Hortal, T. Camou, J. M. Gabastou, and J. L. Di Fabio. 2007. Evolution of an international external quality assurance model to support laboratory investigation of Streptococcus pneumoniae, developed for the SIREVA project in Latin America, from 1993 to 2005. J. Clin. Microbiol. 45:3184-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkinson, A. J., M. G. Bruce, and T. Zulz. 2008. International Circumpolar Surveillance, an Arctic network for the surveillance of infectious diseases. Emerg. Infect. Dis. 14:18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snell, J. J., and D. F. Brown. 2001. External quality assessment of antimicrobial susceptibility testing in Europe. J. Antimicrob. Chemother. 47:801-810. [DOI] [PubMed] [Google Scholar]
- 12.Tenover, F. C., M. J. Mohammed, J. Stelling, T. O'Brien, and R. Williams. 2001. Ability of laboratories to detect emerging antimicrobial resistance: proficiency testing and quality control results from the World Health Organization's external quality assurance system for antimicrobial susceptibility testing. J. Clin. Microbiol. 39:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Selm, S., L. M. van Cann, M. A. Kolkman, B. A. van der Zeijst, and J. P. van Putten. 2003. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect. Immun. 71:6192-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]