Abstract
The interferon-γ-induced guanylate-binding protein 1 (GBP1) belongs to a special class of large GTP- binding proteins of 60–100 kDa with unique characteristics. Here we present the structure of human GBP1 in complex with the non-hydrolysable GTP analogue GppNHp. Basic features of guanine nucleotide binding, such as the P-loop orientation and the Mg2+ co-ordination, are analogous to those of Ras-related and heterotrimeric GTP-binding proteins. However, the glycosidic bond and thus the orientation of the guanine base and its interaction with the protein are very different. Furthermore, two unique regions around the base and the phosphate-binding areas, the guanine and the phosphate caps, respectively, give the nucleotide-binding site a unique appearance not found in the canonical GTP-binding proteins. The phosphate cap, which constitutes the region analogous to switch I, completely shields the phosphate-binding site from solvent such that a potential GTPase-activating protein cannot approach. This has consequences for the GTPase mechanism of hGBP1 and possibly of other large GTP-binding proteins.
Keywords: crystal structure/GTPase mechanism/GTP-binding protein/interferon
Introduction
Guanylate-binding proteins (GBPs) such as hGBP1 and hGBP2 constitute the most abundant class of proteins induced by interferon-γ, an immunomodulatory substance that induces the expression of a large number of genes to orchestrate the cellular response to the cytokine (Cheng et al., 1983, 1991). hGBP1 has recently been shown to mediate an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus (Anderson et al., 1999). hGBP1 and hGBP2 are large 67 kDa GTP-binding proteins, which are stable in the absence of nucleotides. They have a low affinity for guanine nucleotides (Praefcke et al., 1999) and show a concentration-dependent intrinsic high turnover GTPase activity with a rate of up to 80/min (Prakash et al., 2000). A peculiar feature of GBPs is their ability to hydrolyse GTP to both GDP and GMP, where the ratio of products is dependent on the assay conditions (Schwemmle and Staeheli, 1994; Neun et al., 1996).
We have recently determined the crystal structure of full-length human GBP1 at 1.8 Å resolution in the nucleotide-free state (Prakash et al., 2000). The structure depicts a modified large G (LG) domain, with a number of insertions compared with the canonical Ras structure, and a C-terminal part arranged in an extended helical domain with unique features. A long single helix at the C-terminal end of the protein runs along both the helical and the LG domain involving only a few contacts. Presumably due to the absence of guanine nucleotide, several regions of the molecule were not visible in the earlier model. Based on structural and biochemical considerations, it was proposed that hGBP1 belongs to the group of large GTP-binding proteins such as Mx and dynamin, all of which have a similar domain composition and a concentration-dependent GTPase activity, although the sequence homology is very low (Prakash et al., 2000).
Much work has been devoted to elucidating the mechanism of GTP hydrolysis of the large GTP-binding proteins, in particular for dynamin (Shpetner et al., 1992; Robinson et al., 1993; Tuma and Collins, 1994; Warnock et al., 1996), and it was proposed that the C-terminal GED (GTPase effector domain) acts as an internal GTPase-activating protein (GAP) for the G domain (Sever et al., 1999). Since hGBP1 presumably belongs to the same family and is the first protein amenable to structural analysis, it was of particular interest to look at the binding of nucleotide triphosphate to hGBP1. Here we present the crystal structure of hGBP1 in the presence of its substrate analogue, GppNHp, to extract the first clues about the characteristics of this GTPase system. It shows two regions of conformational flexibility around the nucleotide-binding pocket, which are unique to hGBP1 and presumably contribute to the concentration-dependent GTPase reaction. Possible modes of dimerization and oligomerization, which could have implications in the hydrolysis reaction, are also derived on the basis of the crystal structure.
Results and discussion
Overall structure
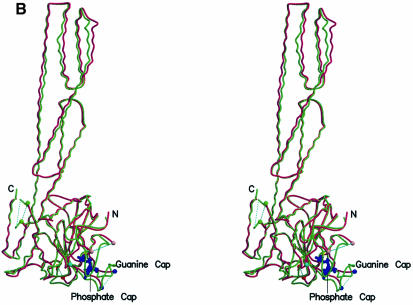
The crystal structure of full-length hGBP1 to 1.8 Å resolution has recently been determined (Prakash et al., 2000). It is a multi-domain protein and consists of an LG domain and an elongated purely α-helical domain. These two domains are connected by a short intermediate region, which is made up of a helix and a short two-stranded β-sheet. The helical domain consists of seven helices extending 90 Å away from the LG domain. The LG domain is structurally homologous to that of other GTP-binding proteins, but is distinguished by several modifications. It contains an eight-stranded β-sheet surrounded by nine helices, unlike Ras, the prototype for a minimal GTP-binding domain, which has six β-strands and five helices. Apart from two inserted helices and three β-strands, there are comparatively long insertions in the loop regions, some of which have been postulated to be involved in the interaction with the nucleotide and/or in the catalytic mechanism of GTP hydrolysis.
Crystals of hGBP1 in complex with GppNHp and Mg2+ were obtained (see Materials and methods) in space group C2. The structure was determined to 1.7 Å by molecular replacement using the nucleotide-free protein as a model. The crystallographic data are summarized in Table I. The current model consists of 570 residues, one nucleotide, one Mg2+ ion and 411 water molecules, with six N- and nine C-terminal residues missing in the current structure (and residues 159–166 in I3). The overall structure of hGBP1⋅GppNHp (Figure 1) is very similar to that of the nucleotide-free form, in both the LG and the helical domains, with a medium r.m.s. deviation of 0.98 Å for 496 Cα carbon atoms, as demonstrated by the stereo view of the superposition of the Cα backbone trace of hGBP1 in the apo and GppNHp-bound forms (Figure 1B). The r.m.s. deviations (Figure 1C) are much larger for the LG than for the helical domain and are largest for the P-loop, the region around Thr75, corresponding to switch I in canonical GTP-binding proteins, around residue Val104 corresponding to switch II, and loops around residues 194 and 243. Residues 68–72 and 243–262, which form the two insert regions I1 and I5, were too flexible to be visible in the apo structure, and have now been traced with confidence. Some of the structural differences seem to be significant for nucleotide binding and/or GTP hydrolysis, as outlined below.
Table I. Crystallographic data statistics.
| Data collection and phase determination by molecular replacement method | |
| Crystal space group |
C2 (a = 161.23 Å, b = 42.65 Å, c = 91.54 Å, β = 100.2°) |
| Parameter |
Native |
| Resolution (Å) | 41.2–1.7 |
| High resolution shell (Å) | 1.8–1.7 |
| X-ray source | ID2, ESRF |
| Wavelength (Å) | 0.9903 |
| Completeness (%) | 99.2 (74) |
| Unique reflections | 67 508 |
| Redundancy | 5.2 (3.9) |
| Rsyma (%) | 8.4 (25.1) |
| I/σ | 14.9 (3.7) |
| Refinement statistics | |
| resolution | 41.2–1.7 Å |
| reflections (work set/test set) | 64 141/3366 |
| protein atoms | 4587 |
| no. of water molecules | 411 |
| average B (Å2) | 23.1 |
| Rworkb | 22.6% |
| Rfreeb | 25.5% |
| r.m.s.d. bond length | 0.005 Å |
| r.m.s.d. bond angles | 1.1° |
Values in parentheses correspond to the highest resolution shell.
Root mean square deviations (r.m.s.d.) are given as deviations from ideal values.
aRsym = Σh Σi|I(h) – Ii(h)|/ΣhΣiIi(h), where Ii(h) and I(h) are the ith and mean measurements of the intensity of reflection h.
bRwork = Σh|Fo – Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h.
Rfree is the same as Rwork, but calculated on the 5% of the data set aside from refinement.
Fig. 1. Overall structure of hGBP1⋅GppNHp and comparison with nucleotide-free protein. (A) Ribbon diagram of the structure, with GppNHp as a blue stick model and Mg2+ as a yellow sphere. (B) Stereo view of a superposition of the apo (red) and GppNHp-bound (green) structures of hGBP1, as a worm plot, highlighting important regions. The regions not visible in the apo or nucleotide-bound structures are shown by a dotted line. Coloured balls [compare with (C)] mark the beginning and end of regions invisible in the apo structure. (C) R.m.s. deviation plot for the main chain (Cα) atoms of the apo and GppNHp structures, highlighting regions that show the highest deviations and/or those that were undefined in the apo structure.
Nucleotide-binding interactions
The guanine nucleotide is bound to a region of the protein basically predicted from the structure of nucleotide-free hGBP1 (Prakash et al., 2000), using sequence motifs conserved between Ras and hGBP1 and biochemical considerations as guidelines (Praefcke et al., 1999). Many of the interactions between nucleotide and protein are conserved between Ras and hGBP1 (Figure 2A). From these and other structures, it is not obvious why Ras-related proteins bind with an affinity in the picomolar range (John et al., 1990) but hGBP1 (Praefcke et al., 1999) or FtsY (Moser et al., 1997) in the micromolar range. The difference may be related to the fact that Ras-related proteins are unstable in the absence of nucleotide, whereas hGBP1 and FtsY are not, and that part of the binding energy may be devoted to stabilization.
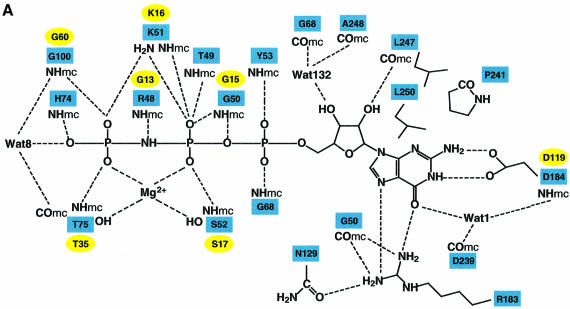
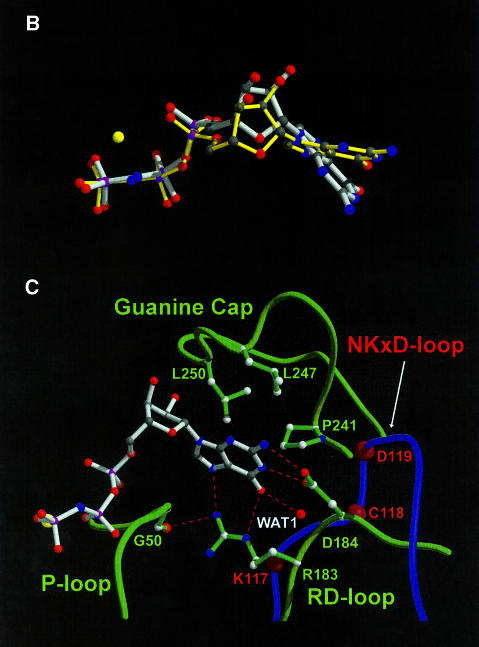
Fig. 2. Binding of nucleotide to hGBP1. (A) Schematic diagram of the interactions of GppNHp, Mg2+ and selected water molecules with the protein; dashed lines indicate contacts with distances <3.4 Å, where NHmc and COmc indicate main chain interactions. Square boxes are residues from GBP1, whereas analogous residues from Ras with similar function are in oval boxes. (B) Superposition of GppNHp bound to hGBP1 (grey) with that from the Ras⋅GppNHp structure (yellow) (Pai et al., 1990). The yellow sphere indicates the Mg2+ ion in both structures. (C) Interactions of the guanine base with selected residues and parts of the corresponding polypeptide chain of hGBP1 (in green), compared with the position of the NKCD motif in the Ras polypeptide chain (in blue), with the position of residues indicated by red balls. Red dotted lines show the interactions of the RD guanine base-binding motif.
Whereas an overlay of the bound nucleotide from Ras and hGBP1 depicts similar positions for the atoms of the ribose and the phosphates, a significant difference is seen for the guanine base, which occupies a different position with respect to the ribose. While all other GTP-binding proteins such as EF-Tu, Gα subunits and Ras-like proteins show a similar N-glycosidic bond angle of around –110°, corresponding to a pronounced anti-conformation of the base, hGBP1 shows a significant deviation from this conformation with an angle of –75.9° (Figure 2B; Table II). As a result of the different orientation, the guanine base interacts differently with the protein. The N/TKxD motif of the canonical GTP-binding proteins contacts the guanine base via hydrogen bonds of the conserved asparagine/threonine and aspartate residues, and by hydrophobic interactions with the hydrophobic part of lysine. This motif was originally believed to be absent in GBPs (Cheng et al., 1991) and was later proposed to be the 181TLRD184 motif (Praefcke et al., 1999). This was based on the D184N mutation, which changed the specificity of nucleotide binding from guanine to xanthine nucleotides, similar to what has been found for Gα (Yu et al., 1997), EF-Tu (Weijland et al., 1994), Ras (Feig et al., 1986; Zhong et al., 1995; Schmidt et al., 1996) and Rab5 (Hoffenberg et al., 1995; Simon et al., 1996). Asp184 is indeed involved in a double hydrogen bond to the endocyclic and exocyclic nitrogen of the base, as in other systems, but, due to the different glycosidic angle, Asp184 does not overlay with Asp from the N/TKxD motif, using Asp119 from the Ras⋅GppNHp complex as an example (Figure 2C). Instead, it is rather close to the position of the Cys118 from the NKCD motif in Ras. A highly conserved hydrogen bond between the guanine O6 atom and a main chain NH from an alanine residue, Ala146 in the SAK/L motif in Ras, is often seen in Ras-like GTP-binding proteins and in Gα proteins. In hGBP1, there is no such interaction (Figure 2C). Instead, the guanine O6 is stabilized by two hydrogen bonds, contacting the side chain nitrogen Nε of Arg183 and a buried water molecule (Wat1), which in turn is positioned by interaction with backbone atoms of Asp184 and Asp239, and with OD1 from Asp184. Arg183, which in the Ras–hGBP1 overlay is close to Lys117 from the N/TKxD motif, has the additional role of binding the N7 of the guanine base, and is also involved in a hydrogen bond to the main chain O from Gly50, thus connecting the base-binding area with the P-loop, similar to the role of N116 in Ras, and it also forms a hydrogen bond with Asn129 in β4. The hydrophobic interaction of a lysine residue with one side of the base is absent in hGBP1. No functional role can be assigned to T180 and L181, which point away from the binding site. Thus, the N/TKxD motif of the canonical GTP-binding proteins, including dynamin and Mx proteins, is replaced by an RD guanine base-binding motif conserved in GBPs.
Table II. Distribution of the torsion angles along the glycosidic bond observed in GTP-binding proteins.
| Molecule | Nucleotide | χ(O4′-C1′-N9-C4) | Reference |
|---|---|---|---|
| hGBP1 | GppNHp | –75.86 | this work |
| Ras | GppNHp | –112.39 | Pai et al. (1990) |
| EF-Tu | GppNHp | –110.85 | Berchtold et al. (1993) |
| Giα | GppNHp | –108.95 | Coleman et al. (1994) |
| Rac1 | GppNHp | –115.36 | Hirshberg et al. (1997) |
| Rho | GTPγS | –110.49 | Ihara et al. (1998) |
| SRP | GDP | –122.33 | Freymann et al. (1999) |
| Transducin | GTPγS | –109.39 | Noel et al. (1993) |
While the guanine base-binding site in Ras is open to the solvent, in hGBP1 it is mostly covered by the I5 region, which was disordered in the nucleotide-free structure. The I5 insert is arranged in two short helices, α5A and α5B, which form a cap (the guanine cap) parallel to the flat side of the guanine base. The guanine cap contains 19 residues, some of which are conserved, and forms a hydrophobic pocket for the guanine base, which in Ras is provided by Phe28 in switch I (Figure 2C). An analogous scenario is observed in the SRP GTPases where residues from the ‘closing loop’ give rise to similar hydrophobic contacts (Freymann et al., 1999). If the guanine was bound in an extended (anti) conformation as in other GTP-binding proteins, it would clash with the position of the guanine cap, suggesting that the cap is also involved in directing the different conformation of bound GTP.
The conformations of the ribose and phosphate moieties in the overlay are strikingly similar to those of Ras. The ribose is less exposed to the solvent, such that only the 3′ position would be available for modification with reporter groups, as the mant group has been used successfully for following nucleotide binding to hGBP1 (Praefcke et al., 1999). The O′3 atom of the sugar forms a hydrogen bond with the main chain O of Leu247. The P-loop in hGBP1 is wrapped around the phosphates in the canonical fashion observed in GTP- and many ATP-binding proteins (Vetter and Wittinghofer, 1999). A number of its main chain amide groups form hydrogen bonds with the phosphate oxygens. Lys51 from the P-loop contacts one pair of β,γ-phosphate oxygens, while the Mg2+ ion co-ordinates the other. The Mg2+ ion is co-ordinated octahedrally by the O atoms of Pβ and Pγ, the side chain hydroxyls of Thr75 from the switch I region and Ser52 from the P-loop, and two water molecules, exactly as is found in canonical GTP-binding proteins. In comparison with the apo structure, the protein shows several rearrangements to accommodate Mg2+⋅GppNHp. As predicted from the apo structure, a peptide flip involving residues Tyr47 and Arg48 in the P-loop moves the carbonyl oxygen of Arg48 away from the nucleotide and brings the backbone nitrogen into a position suitable to interact with the nitrogen of GppNHp (which would be an oxygen in GTP), similarly to Gly13 in Ras at the corresponding position in the P-loop. These changes in the P-loop also reorient Lys51, which in the nucleotide-free state interacts with Thr98 in switch II. Two γ-phosphate oxygens interact additionally with the main chain oxygens of Thr75 and Gly100, which correspond to the totally invariant threonine (Thr35 in Ras) and glycine from the DxxG motif (Gly60 in Ras) in the canonical GTP-binding proteins, an indication that the conformational change between the GTP- and GDP-bound state is likely to be triggered in a similar way as in canonical GTP-binding proteins. The most dramatic difference from Ras-like proteins is the presence of a region covering the phosphates (the phosphate cap), corresponding to the switch I region, which presumably has significant consequences for the mechanism of GTP hydrolysis, as will be outlined below.
Nucleotide-dependent oligomerization
One of the intriguing aspects of some of the proteins belonging to the large GTPase family is their ability to oligomerize depending on the nucleotide-bound state, and the subsequent concentration-dependent GTP hydrolysis (Warnock et al., 1996). We have shown that hGBP1 also shows a concentration-dependent increase in the GTPase activity of at least 20-fold and that hGBP1 is a monomer in the absence of nucleotide and in complex with GMP/GDP, but a dimer in the presence of GppNHp and GTP. Furthermore, it aggregates to approximately twice the molecular weight of the dimer in the presence of GDP and AlFx (Prakash et al., 2000). Since the GDP–AlFx-bound state of the protein mimics the transition state of the reaction, we can infer that GTP hydrolysis necessitates a transient higher order oligomerization.
The present GppNHp-bound structure was determined in space group C2, with one molecule in the asymmetric unit. This could indicate either that the dimer in solution corresponds to two molecules related by the crystallographic symmetry, or that hGBP1 does not form a dimer under the crystallization conditions [high salt, polyethylene glycol (PEG)] and that the dimer observed in the crystal represents a crystallographic artefact. However, two of the interactions between symmetry-related molecules show a large buried surface area (Table III) and might thus be taken as tentative models for the dimer (which may or may not be valid). Dimer A, with 2891 Å2 of buried surface area (Figure 3A), shows head-to-tail contacts, whereas dimer B, with 2140 Å2, is formed by a head-to-head contact (Figure 3B).
Table III. Buried surface areas for closest crystallographic neighbours calculated by using CNS with a probe radius of 1.4 Å.
| Dimers |
Buried surface area (Å2) |
| Dimer A | 2891 |
| Dimer B | 2140 |
| Dimer C | 1479 |
| Dimer D | 810 |
| Dimer E | 522 |
| Dimer F | 412 |
| Dimer G | 283 |
Fig. 3. Crystal contacts of hGBP1⋅GppNHp. (A) The head-to-tail dimer A, which buries 2890 Å2 of surface area, where a lip from the LG domain is close to the helical domain in helices α10 and the long helix α12. The nucleotide is shown in ball and stick representation with the yellow sphere representing the Mg2+ ion. (B) The head-to-head dimer B with 2140 Å2 of buried surface, using a similar colour code.
Since the interaction of the head of monomer 1 with the helical domain of monomer 2 in dimer A (Figure 3A) is not reciprocal, it would result in the formation of a polymeric assembly of hGBP1 in the crystal. Since all interactions between the subunits of the polymer are similar, it would be difficult to understand why those contacts should lead to dimer formation in solution. In dimer B, the regions close to the nucleotide-binding sites of the monomers interact with each other in a symmetrical fashion, and the two monomers are related by a 2-fold symmetry axis (Figure 3B). Compared with dimer A, this reciprocal interaction across the dimer interface is more likely to alter the properties of the active site or, vice versa, the dimerization could be induced by changes in the nucleotide-binding site. Dimer B also more closely resembles the model of the SRP–SR heterodimer where the head-to-head interaction is believed to be responsible for reciprocal stimulation of the GTPase (Montoya et al., 2000). However, the electrostatic potential does not favour the formation of this dimer since it is strongly negative at the site of the interaction and would result in a repulsive force. In contrast, an interaction as observed in dimer A would be facilitated by the charge distributions. Furthermore, preliminary equilibrium ultracentrifugation studies suggest a compact structure more appropriate for dimer A. In summary, more detailed biochemical and structural studies need to be performed in order to understand the oligomerization reaction.
Mechanism of GTP hydrolysis
GTP hydrolysis by canonical GTP-binding proteins has a number of structural and mechanistic features in common. The intrinsic hydrolysis reaction has been proposed to proceed by in-line attack of a nucleophilic water molecule (Feuerstein et al., 1989), which has been identified in all available triphosphate structures (Wittinghofer, 2000). For Ras, it has been assumed additionally that the γ-phosphate acts as a general (or specific) base that activates the nucleophilic water, and indeed the structure shows hydrogen bond distance between a γ-phosphate oxygen and the nucleophilic water molecule (Schweins et al., 1995). An essential residue for catalysis in Gα proteins and Ras-related proteins (except Rap) is a glutamine residue, which has been postulated to stabilize the transition state by orienting the relative positions of the nucleophile and the γ-phosphate (Prive et al., 1992), as supported by the structure of the Ras–RasGAP complex (Scheffzek et al., 1997). Further activation of the reaction rate is achieved by the presence of an arginine, which is intrinsic in the case of Gα proteins and is supplied in trans by GAPs, as shown for RasGAPs (Ahmadian et al., 1997; Scheffzek et al., 1997) and RhoGAPs (Rittinger et al., 1997a; Hoffman et al., 1998; Nassar et al., 1998; Graham et al., 1999). GAPs and RGS proteins, the GAPs for Gα proteins, in addition stabilize the catalytic machinery, such as the critical glutamine (Tesmer et al., 1997). However, proteins of the family of large GTP-binding proteins lack a conserved catalytic residue following the DxxG motif, which strongly suggests a different mechanism for GTP hydrolysis. The fact that members of this family of GTP-hydrolysing proteins show a much higher intrinsic hydrolysis rate, which in the case of hGBP1 and dynamin has been shown to be concentration dependent, is another indication that the mechanism of catalysis is bound to be different.
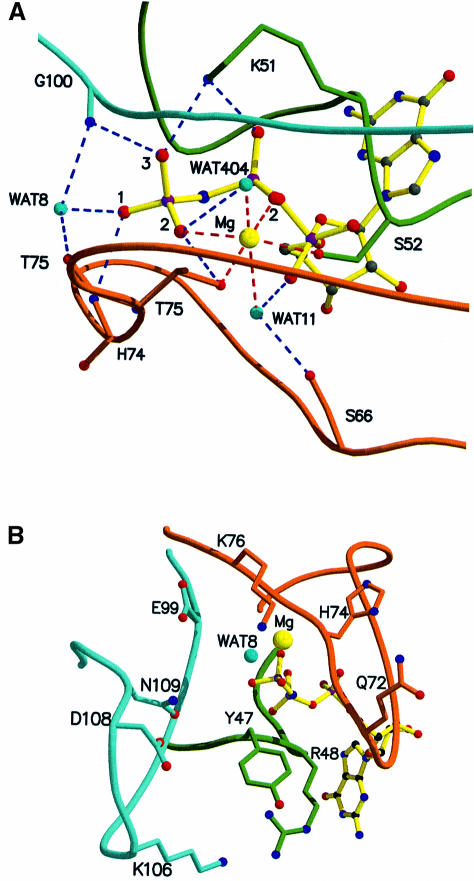
In the present structure, a nucleophilic water molecule (Wat8) is positioned 3.5 Å away from the γ-phosphate (Figure 4A). The γ-phosphate oxygens are engaged in hydrogen bonds with the main chain NH of Thr75 and Gly100, two invariant residues, in the same way as found in Ras-related and Gα proteins (Wittinghofer, 2000). The major difference in the active site as compared with the small Ras-related proteins is the presence of the switch I loop (phosphate cap) including the conserved Thr75, which forms a β-hairpin-like cap, stabilized by internal main chain and side chain interactions, shielding the phosphates of GppNHp and Wat8 from the solvent (Figure 4B). Another consequence of the phosphate cap is that it provides, via the main chain NH of His74, another strong hydrogen bond to the γ-phosphate. In Ras and Rho, the phosphate-binding site is open to the solvent (Figure 4C) and becomes shielded from water by the corresponding RasGAP and RhoGAP (Rittinger et al., 1997a; Scheffzek et al., 1997; Nassar et al., 1998). In contrast, in hGBP1, the active site is shielded from the bulk solvent by the phosphate cap such that only the nucleophilic water has access to the γ-phosphate (Figure 4D). GAPs for Ras and Rho use a protruding arginine residue, the arginine finger, to interact with the only phosphate oxygen not co-ordinated to the protein, the same one which in hGBP1 is contacted by another main chain NH. For Rho, it has been shown that the contact with GAP occurs only in the transition state mimic presented by the Rho⋅GDP⋅AlFx⋅GAP complex, and not in the ground state of the reaction (Rittinger et al., 1997a,b), and mutational and NMR evidence suggest that the same is true for the Ras–RasGAP system (Bollag et al., 1991; Gideon et al., 1992; Geyer et al., 1996). In hGBP1, the contact of the intrinsic main chain NH of His74 with the third phosphate oxygen might thus be responsible for increased probability of nucleophilic attack by water and an increased stability of the transition state.
Fig. 4. The phosphate-binding region and implications for GTP hydrolysis. (A) Interactions of the phosphate oxygens and Mg2+ with the P-loop (green), the switch I/phosphate cap (brown) and the switch II region (maroon). Wat8 is in a homologous position to the nucleophilic water found in other structures of GTP-binding proteins. In contrast to those, there are three main chain NH interactions of the protein with the γ-phosphate. (B) Potential catalytic residues around the active site that could modify the rate of the GTPase reaction in an oligomerization-dependent manner, without directly participating in catalysis. (C and D) van der Waals surface representation of the region of the active site of Ras (C) and hGBP1 (D) in the GppNHp-bound state, the surface being coloured according to the electrostatic potential, as calculated with GRASP (Nicholls et al., 1991). In hGBP1, only the base is open to the solvent.
We have shown that the rate of hydrolysis of hGBP1 is increased by increasing concentrations of the protein and that, in the presence of GDP and AlFx, a higher oligomeric state as compared with the GppNHp state is formed. Although, for technical reasons, we cannot determine the GTPase hydrolysis rate at infinitely low concentrations, the rate is increased at least 20-fold. We can thus conclude that the present structural architecture of the active site shows features relevant to the intrinsic reaction but possibly not to the stimulated reaction machinery. We can infer from the closed nature of the active site, however, that the oligomerization-induced GTPase reaction is unlikely to contribute directly GAP-type residues such as an arginine finger into the active site to stabilize developing negative charge on the phosphate, unless major conformational changes were induced by oligomerization.
Residues situated close to the active site of the hGBP1⋅GppNHp complex might, however, constitute part of the catalytic machinery in the oligomerized state. Such residues are Glu72, His74 and Lys76 on the phosphate cap, Glu99 preceding the invariant Gly100 in the DxxG motif, and Lys106, Asp108 and Asn109 in the insertion I2 with the highly conserved 103DxEKGD108 motif, which forms a tight loop conspicuously close to the γ-phosphate (Figure 4B). The B-factors indicate a high mobility for this region, which suggests that such residues could move into a catalytically competent position upon oligomerization. Arg48 in the P-loop, which is conserved in GBPs and not found in other GTP-binding proteins, could potentially also play a catalytic role. An arginine at a homologous position (GxxRxGKS) in type I thymidylate kinase has been shown to be required for fast phosphoryl transfer (Lavie et al., 1998).
For dynamin and other members of the family of large GTP-binding proteins, a nucleotide-dependent oligomerization has been demonstrated (Hinshaw et al., 1995). In addition, for dynamin, a concentration-dependent GTPase reaction has been observed (Tuma and Collins, 1995; Warnock et al., 1997; Barylko et al., 1998) that is dependent on the presence of a C-terminal domain, which has been called the GED, just as the C-terminal 60 residues of Mx are required for its GTPase reaction (Schwemmle et al., 1995). From the studies using the isolated GED added in trans to the G domain of dynamin, it was concluded that it acts as an assembly-dependent internal GAP, which supplies a catalytic arginine residue (Sever et al., 1999). We have proposed that Mx and dynamin have a similar structural domain composition to that of hGBP1, although the sequence homology is low (Prakash et al., 2000). Whether we have to assume a similar mechnism of GTP hydrolysis via direct participation of residues from the GED/C-terminal domain is not obvious at present. It should be noted, however, that the LG domains of hGBP1 and Mx/dynamin differ. The TKxD motif is conserved in Mx and dynamins and therefore the guanine base might be bound in the conventional anti-conformation. More important is the larger distance between the P-loop and the DxxG motif in Mx/dynamin. This insertion might form a different switch I region, which could be responsible for biochemical differences within this class of GTP-binding proteins.
Conclusion
The guanine-nucleotide binding site of hGBP1 has been determined by X-ray crystallography. It shows many similarities to, but also characteristic differences from canonical GTP-binding proteins. There is a different glycosidic bond angle and the guanine base interaction is different from that of other known GTPase structures. Most importantly, the phosphate-binding site is completely closed off from solvent such that external residues are unlikely to approach the β- or γ-phosphates without major conformational changes, which might exclude the participation of either external or internal GTPase-activating domains or GEDs. Furthermore, the γ-phosphate has all its oxygens involved in tight interactions with main chain NH groups, which may be important for a better orientation of relevant reactive groups and/or better stabilization of the transition state. To obtain a more precise idea of the oligomerization-induced activation mechanism of GTP hydrolysis in GBPs and possibly the other members of the family, the structure of the hGBP1⋅GDP⋅AlFx complex has to be solved and mutational studies on residues close to the active site have to be performed. The design of such experiments is suggested by the three-dimensional structure presented here.
Materials and methods
Protein preparation
hGBP1 with an N-terminal His6 tag was expressed from pQE9 vector (Qiagen, Germany) in Escherichia coli strain BL21(DE3), as described by Praefcke et al. (1999). Cells were suspended in buffer A [50 mM Tris–HCl pH 8.0, 5 mM MgCl2, 500 mM NaCl, 10% glycerol, 20 mM imidazole, 200 µM Pefabloc (Roth GmbH + Co., Germany)], disrupted in a microfluidizer (Microfluidics Corporation, USA), bound to an Ni-NTA Superflow column (Qiagen, Germany) and eluted with buffer B (20 mM Tris–HCl pH 8.0, 5 mM MgCl2, 100 mM NaCl, 10% glycerol, 20 mM imidazole) using a 20–500 mM imidazole gradient. It was followed by gel filtration via Superdex 200 (Amersham-Pharmacia, Sweden) in buffer C (10 mM Tris–HCl pH 8.0, 2 mM MgCl2, 2 mM dithiothreitol). Fractions containing monomeric hGBP1 were pooled, concentrated to 50 mg/ml, shock-frozen in liquid nitrogen and stored at –80°C.
Crystallography
His6-tagged full-length hGBP1 (593 residues) concentrated to 97 mg/ml was incubated with a 12-fold excess of the GTP analogue, GppNHp for 3 h. This sample was used to grow crystals by the vapour diffusion method in hanging drops over a reservoir solution of 15–20% PEG3350, 150–200 mM MgCl2 at 4°C. Crystals were cryoprotected with 35% PEG3350, 200 mM MgCl2. A high-resolution native data set to 1.7 Å resolution was collected at 100 K on a MAR-Imaging Plate detector at ID2 beamline, ESRF, Grenoble, and processed with XDS (Kabsch, 1993). The space group of the crystals is C2 and the statistics of the native data are presented in Table I. The initial phases were obtained by molecular replacement with the program AmoRe, as implemented in CCP4 (version 3.5) (CCP4, 1994) using a search model based on the nucleotide-free structure of hGBP1 after removing the regions corresponding to the P-loop, switch I and switch II. Several searches using different ranges of integration radii resulted in a unique solution. Rigid body refinements of the solution thus obtained, performed with CNS (version 0.4a) (Brünger et al., 1998), resulted in an Rfree of 48.6% and an Rwork of 48%. This was followed by a round of simulated annealing as implemented in CNS, which resulted in an Rfree and Rwork of 41.5 and 36.7%, respectively; 5% of the reflections were set aside for an Rfree test before initiating any refinement. A sigma-weighted 2Fo – Fc map (CCP4 suite) computed from this model showed clear electron density for most of the omitted regions in the model, the nucleotide, GppNHp, the Mg2+ ion and the cap region, which was absent in the nucleotide-free structure. The model was built and refined through alternating cycles using the programs O (Jones et al., 1997) and CNS. The refinement statistics are presented in Table I. A typical round of refinement consisted of bulk solvent correction, positional, torsion angle-simulated annealing and B-factor refinement as implemented in CNS. Simulated annealing omit maps were used to correct or build ambiguous regions of the model. The model includes residues 7–584 of the human GBP1 sequence, GppNHp, a Mg2+ ion and 411 water molecules. There are no Ramachandran outliers. The loop comprising residues 159–166 cannot be seen in the electron density and is presumably disordered. The least-squares superpositions with the GTPases Ras and nucleotide-free hGBP1 were performed using the automatic fitting program BRAGI (Schomburg and Reichelt, 1988) and were displayed in O. Figures were generated using Molscript (Kraulis, 1991) and Raster3D (Merritt et al., 1994). The electrostatic surface potential was generated using GRASP (Nicholls et al., 1991).
The PDB accession code for the co-ordinates is 1F5N.
Acknowledgments
Acknowledgements
We thank Drs Ilme Schlichting, Ingrid Vetter, Roman Hillig and Michael Weyand for useful discussions, Rita Schebaum for secretarial assistance, and Michael Hess for graphical presentations. We thank the staff at beamline ID2B, ESRF, Grenoble for help with data collection. The work was supported by the Deutsche Forschungsgemeinschaft (to C.H.) and by the Boehringer Ingelheim Fonds (to G.J.K.P.).
References
- Ahmadian M.R., Stege,P., Scheffzek,K. and Wittinghofer,A. (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nature Struct. Biol., 4, 686–689. [DOI] [PubMed] [Google Scholar]
- Anderson S.L., Carton,J.M., Lou,J., Xing,L. and Rubin,B.Y. (1999) Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalo myocarditis virus. Virology, 256, 8–14. [DOI] [PubMed] [Google Scholar]
- Barylko B., Binns,D., Lin,K.M., Atkinson,M.A., Jameson,D.M., Yin,H.L. and Albanesi,J.P. (1998) Synergistic activation of dynamin GTPase by Grb2 and phosphoinositides. J. Biol. Chem., 273, 3791–3797. [DOI] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova,L., Reiser,C.O., Schirmer,N.K., Sprinzl,M. and Hilgenfeld,R. (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature, 365, 126–132. [DOI] [PubMed] [Google Scholar]
- Bollag G. and McCormick,F. (1991) Differential regulation of RasGAP and neurofibromatosis gene product activities. Nature, 351, 576–579. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cheng Y.S., Colonno,R.J. and Yin,F.H. (1983) Interferon induction of fibroblast proteins with guanylate binding activity. J. Biol. Chem., 258, 7746–7750. [PubMed] [Google Scholar]
- Cheng Y.S., Patterson,C.E. and Staeheli,P. (1991) Interferon-induced guanylate-binding proteins lack an N(T)KXD consensus motif and bind GMP in addition to GDP and GTP. Mol. Cell. Biol., 11, 4717–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman D.E., Berghuis,A.M., Lee,E., Linder,M.E., Gilman,A.G. and Sprang,S.R. (1994) Structures of active conformations of Giα1 and the mechanism of GTP hydrolysis. Science, 265, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Feig L.A., Pan,B.-T., Roberts,T.M. and Cooper,G.M. (1986) Isolation of ras GTP-binding mutants using an in situ colony-binding assay. Proc. Natl Acad. Sci. USA, 83, 4607–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerstein J., Goody,R.S. and Webb,M.R. (1989) The mechanism of guanosine nucleotide hydrolysis by p21 c-Ha-Ras. The stereochemical course of the GTPase reaction. J. Biol. Chem., 264, 6188–6190. [PubMed] [Google Scholar]
- Freymann D.M., Keenan,R.J., Stroud,R.M. and Walter,P. (1999) Functional changes in the structure of the SRP GTPase on binding GDP and Mg(2+)GDP. Nature Struct. Biol., 6, 793–801. [DOI] [PubMed] [Google Scholar]
- Geyer M., Schweins,T., Herrmann,C., Prisner,T., Wittinghofer,A. and Kalbitzer,H.R. (1996) Conformational transitions in p21(Ras) and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry, 35, 10308–10320. [DOI] [PubMed] [Google Scholar]
- Gideon P., John,J., Frech,M., Lautwein,A., Clark,R., Scheffler,J.E. and Wittinghofer,A. (1992) Mutational and kinetic analysis of the GTPase-activating protein GAP–p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol. Cell. Biol., 12, 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D.L., Eccleston,J.F. and Lowe,P.N. (1999) The conserved arginine in Rho-GTPase-activating protein is essential for efficient catalysis but not for complex formation with rho GDP and aluminum fluoride. Biochemistry, 38, 985–991. [DOI] [PubMed] [Google Scholar]
- Hinshaw J.E. and Schmid,S.L. (1995) Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature, 374, 190–192. [DOI] [PubMed] [Google Scholar]
- Hirshberg M., Stockley,R.W., Dodson,G. and Webb,M.R. (1997) The crystal structure of human Rac1, a member of the Rho-family complexed with a GTP analogue. Nature Struct. Biol., 4, 147–152. [DOI] [PubMed] [Google Scholar]
- Hoffenberg S., Nikolova,L., Pan,J.Y., Daniel,D.S., Wesslingresnick,M., Knoll,B.J. and Dickey,B.F. (1995) Functional and structural interactions of the Rab5 D136N mutant with xanthine nucleotides. Biochem. Biophys. Res. Commun., 215, 241–249. [DOI] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar,N., Oswald,R.E. and Cerione,R.A. (1998) Fluoride activation of the Rho family GTP-binding protein Cdc42Hs. J. Biol. Chem., 273, 4392–4399. [DOI] [PubMed] [Google Scholar]
- Ihara K., Muraguchi,S., Kato,M., Shimizu,T., Shirakawa,M., Kuroda,S., Kaibuchi,K. and Hakoshima,T. (1998) Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem., 273, 9656–9666. [DOI] [PubMed] [Google Scholar]
- John J., Sohmen,R., Feuerstein,J., Linke,R., Wittinghofer,A. and Goody,R.S. (1990) Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry, 29, 6058–6065. [DOI] [PubMed] [Google Scholar]
- Jones T.A. and Kjeldgaard,M. (1997) Electron-density map interpretation. Methods Enzymol., 277, 173–208. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lavie A., Konrad,M., Brundiers,R., Goody,R.S., Schlichting,I. and Reinstein,J. (1998) Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5′-adenosyl) P5-(5′-thymidyl) pentaphosphate (TP5A) at 2.0 Å resolution: implications for catalysis and AZT activation. Biochemistry, 37, 3677–3686. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Montoya G., te Kaat,K., Moll,R., Schäfer,G. and Sinning,I. (2000) The crystal structure of the conserved GTPase of SRP54 from the archaeon Acidianus ambivalens and its comparison with related structures suggests a model for the SRP–SRP receptor complex. Structure Fold Des., 8, 515–525. [DOI] [PubMed] [Google Scholar]
- Moser C., Mol,O., Goody,R.S. and Sinning,I. (1997) The signal recognition particle receptor of Escherichia coli (Ftsy) has a nucleotide exchange factor built into the GTPase domain. Proc. Natl Acad. Sci. USA, 94, 11339–11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar N., Hoffman,G.R., Manor,D., Clardy,J.C. and Cerione,R.A. (1998) Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nature Struct. Biol., 5, 1047–1052. [DOI] [PubMed] [Google Scholar]
- Neun R., Richter,M.F., Staeheli,P. and Schwemmle,M. (1996) GTPase properties of the interferon-induced human guanylate-binding protein 2. FEBS Lett., 390, 69–72. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Noel J.P., Hamm,H.E. and Sigler,P.B. (1993) The 2.2 Å crystal structure of transducin-α complexed with GTPγ S. Nature, 366, 654–663. [DOI] [PubMed] [Google Scholar]
- Pai E.F., Krengel,U., Petsko,G.A., Goody,R.S., Kabsch,W. and Wittinghofer,A. (1990) Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J., 9, 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke G.J.K., Geyer,M., Schwemmle,M., Kalbitzer,H.R. and Herrmann,C. (1999) Nucleotide-binding characteristics of human guanylate-binding protein 1 (hGBP1) and identification of the third GTP-binding motif. J. Mol. Biol., 292, 321–332. [DOI] [PubMed] [Google Scholar]
- Prakash B., Praefcke,G.J.K., Renault,L., Wittinghofer,A. and Herrmann,C. (2000) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature, 403, 567–571. [DOI] [PubMed] [Google Scholar]
- Prive G.G., Milburn,M.V., Tong,L., de Vos,A.M., Yamaizumi,Z., Nishimura,S. and Kim,S.H. (1992) X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc. Natl Acad. Sci. USA, 89, 3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K., Walker,P.A., Eccleston,J.F., Smerdon,S.J. and Gamblin,S.J. (1997a) Structure at 1.65 Å of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature, 389, 758–762. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Walker,P.A., Eccleston,J.F., Nurmahomed,K., Owen,D., Laue,E., Gamblin,S.J. and Smerdon,S.J. (1997b) Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature, 388, 693–697. [DOI] [PubMed] [Google Scholar]
- Robinson P.J. et al. (1993) Dynamin GTPase regulated by protein kinase C phosphorylation in nerve terminals. Nature, 365, 163–166. [DOI] [PubMed] [Google Scholar]
- Scheffzek K. et al. (1997) The Ras–RasGAP complex—structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science, 277, 333–338. [DOI] [PubMed] [Google Scholar]
- Schmidt G. et al. (1996) Biochemical and biological consequences of changing the specificity of p21(Ras) from guanosine to xanthosine nucleotides. Oncogene, 12, 87–96. [PubMed] [Google Scholar]
- Schomburg D. and Reichelt,J. (1988) BRAGI: a comprehensive protein modelling program system. J. Mol. Graphics, 6, 161–165. [Google Scholar]
- Schweins T., Geyer,M., Scheffzek,K., Warshel,A., Kalbitzer,H.R. and Wittinghofer,A. (1995) Substrate-assisted catalysis as a mechanism for GTP hydrolysis of p21(Ras) and other GTP-binding proteins. Nature Struct. Biol., 2, 36–44. [DOI] [PubMed] [Google Scholar]
- Schwemmle M. and Staeheli,P. (1994) The interferon-induced 67-kDa guanylate-binding protein (hGBP1) is a GTPase that converts GTP to GMP. J. Biol. Chem., 269, 11299–11305. [PubMed] [Google Scholar]
- Schwemmle M., Richter,M.F., Herrmann,C., Nassar,N. and Staeheli,P. (1995) Unexpected structural requirements for GTPase activity of the interferon-induced MxA protein. J. Biol. Chem., 270, 13518–13523. [PubMed] [Google Scholar]
- Sever S., Muhlberg,A.B. and Schmid,S.L. (1999) Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature, 398, 481–486. [DOI] [PubMed] [Google Scholar]
- Shpetner H.S. and Vallee,R.B. (1992) Dynamin is a GTPase stimulated to high levels of activity by microtubules. Nature, 355, 733–735. [DOI] [PubMed] [Google Scholar]
- Simon I., Zerial,M. and Goody,R.S. (1996) Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J. Biol. Chem., 271, 20470–20478. [DOI] [PubMed] [Google Scholar]
- Tesmer J.J., Berman,D.M., Gilman,A.G. and Sprang,S.R. (1997) Structure of RGS4 bound to AlF4-activated G(i α1): stabilization of the transition state for GTP hydrolysis. Cell, 89, 251–261. [DOI] [PubMed] [Google Scholar]
- Tuma P.L. and Collins,C.A. (1994) Activation of dynamin GTPase is a result of positive cooperativity. J. Biol. Chem., 269, 30842–30847. [PubMed] [Google Scholar]
- Tuma P.L. and Collins,C.A. (1995) Dynamin forms polymeric complexes in the presence of lipid vesicles. Characterization of chemically cross-linked dynamin molecules. J. Biol. Chem., 270, 26707–26714. [DOI] [PubMed] [Google Scholar]
- Vetter I.R. and Wittinghofer,A. (1999) Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q. Rev. Biophys., 32, 1–56. [DOI] [PubMed] [Google Scholar]
- Warnock D.E., Hinshaw,J.E. and Schmid,S.L. (1996) Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem., 271, 22310–22314. [DOI] [PubMed] [Google Scholar]
- Warnock D.E., Baba,T. and Schmid,S.L. (1997) Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol. Biol. Cell, 8, 2553–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijland A., Parlato,G. and Parmeggiani,A. (1994) Elongation factor Tu D138N, a mutant with modified substrate specificity, as a tool to study energy consumption in protein biosynthesis. Biochemistry, 33, 10711–10717. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A. (2000) The functioning of molecular switches in three dimensions. Front. Mol. Biol., 24, 244–310. [Google Scholar]
- Yu B., Slepak,V.Z. and Simon,M.I. (1997) Characterization of a Go-α mutant that binds xanthine nucleotides. J. Biol. Chem., 272, 18015–18019. [DOI] [PubMed] [Google Scholar]
- Zhong J.-M., Chen-Hwang,M.-C. and Hwang,Y.-W. (1995) Switching nucleotide specificity of Ha-Ras p21 by a single amino acid substitution at aspartate 119. J. Biol. Chem., 270, 10002–10007. [DOI] [PubMed] [Google Scholar]