Abstract
We report the second human immunodeficiency virus (HIV) belonging to the new HIV type 1 (HIV-1) group P lineage that is closely related to the simian immunodeficiency virus found in gorillas. This virus was identified in an HIV-seropositive male hospital patient in Cameroon, confirming that the group P virus is circulating in humans. Results from screening 1,736 HIV-seropositive specimens collected in Cameroon indicate that HIV-1 group P infections are rare, accounting for only 0.06% of HIV infections. Despite its rarity, group P shows evidence of adaptation to humans.
Human immunodeficiency virus type 1 (HIV-1) is classified into groups, each of which arose from separate transmissions of simian immunodeficiency viruses (SIVs) from nonhuman primates into humans (4). Group M (major), the cause of the worldwide pandemic, and group N (non-M, non-O) viruses originated from SIV strains found in chimpanzees (SIVcpz; Pan troglodytes troglodytes), while the SIV origin of group O (outlier) viruses has not been identified (4, 5, 7, 8). Recently a new putative group, designated P, was reported to be found in a Cameroonian woman living in France (6). The group P viral sequence, RBF168, forms a distinct HIV-1 lineage that includes SIV sequences from western gorillas (SIVgor; Gorilla gorilla gorilla), suggesting that group P originated from gorillas (6, 8, 10). We report the characterization of a second HIV-1 group P strain identified in a Cameroonian hospital patient.
Diverse HIV-1 strains, including groups N and O, are circulating in Cameroon, and the SIV strains most closely related to HIV-1 were identified in chimpanzee and gorilla colonies residing within Cameroon (1, 2, 5, 9, 10). For these reasons, Cameroon is a logical site to screen for new or rare HIV variants. We implemented a variant screening strategy that utilizes serological and molecular methods (12). Specimens were collected from hospital and clinic patients and blood donors primarily in the cities of Yaoundé and Douala and were screened for HIV infection using several HIV immunoassays. Any specimen HIV seropositive by at least one assay was then screened for antibody reactivity to peptides derived from the env gp41 immunodominant region (IDR) and env gp120 V3 loop region (12). The original peptide enzyme-linked immunoassays (PEIAs) that contained peptides for HIV-1 groups M, N, and O and SIVcpz strains were modified, first to include an SIVgor IDR peptide (peptide sequence, 5′-KRLLAIETYLRDQQLLGLWGCTGKLICYTNVPW-3′) and then to include both SIVgor- and group P-specific peptides (P-IDR, 5′-LLAIETYLRDQQLLGLWGCSGQIVCYTNVPW-3′; P-V3, 5′-TRGQVQIGPMTWYNMKFYTG'-3′; and SIVgor-V3, 5′-TRGQIQVGPMTIYNSENIVG-3′). Potential HIV variants, selected based on the pattern of peptide reactivity, were subjected to molecular characterization to determine the virus present.
To estimate the prevalence of HIV-1 group P in Cameroon, a total of 1,736 HIV-seropositive specimens collected from 2006 to 2009 were screened in the PEIAs containing the SIVgor IDR peptide (n = 245) or SIVgor and group P peptides (n = 1,491). Twenty-eight specimens were selected for reverse transcription (RT)-PCR as potential group P infections: 5 from screening with the SIVgor IDR peptide and 23 from screening with SIVgor and group P peptides. Viral RNA extracted from the specimens was subjected to RT-PCR amplification using primers in regions that are relatively conserved across HIV-1 groups. A fragment of pol integrase (562 bp) was amplified using primers ppf2b and LPpoli4R, and an env gp41 fragment (362 bp) was amplified using primers JH41 and env19R (Table 1). Conditions for nucleic acid extraction, amplification, and sequencing have been described previously (13). Only specimen U14788 was confirmed as an HIV-1 group P infection; 23 were determined to be group M strains, 1 was a group O strain, and 3 were RT-PCR negative (data not shown).
TABLE 1.
PCR primers used for nearly full-length-genome amplification of 06CMU14788
| Genome region (fragment) | Fragment size (bp)a | PCR step | Primer name | Forward or reverse primerb | Primer sequence (5′ to 3′) |
|---|---|---|---|---|---|
| LTR-gag | 1,772 | First | RE9745F-bh4 | F | CAGATYTGAGCCTGGGA |
| Opol17R | R | ACTAYTGGTCTGTCCCAAAGAG | |||
| LTR-gag (A) | 1,023 | Nested | NLTR2F | F | GAACCCACTGCTTAAGCCTCAA |
| Op24seq2R | R | TTCTATAGATGTCTCCTACTG | |||
| gag-pol | 4,387 | First | AF-2sah | F | CTAGCAGTGGCGCCCGAACAGGc |
| LPpoli4R | R | ACCTGCCATCTGTTTTCCATAATC | |||
| gag (B) | 1,407 | Nested | Ogag1F | F | GCAAATTGGATGCATGGGAA |
| Opol17R | R | ACTAYTGGTCTGTCCCAAAGAG | |||
| gag-pol (C) | 1,129 | Op24seq2F | F | CAGTAGGAGACATCTATAGAA | |
| prot4Rvh | R | TGGAGTRTTRTATGGATTTTCAGG | |||
| pol (D) | 1,375 | Opol18F | F | GAARATGGACACCAAAAATGATAGG | |
| Opol25R | R | CATTCWGGAATCCAGGTGGC | |||
| pol (E) | 1,211 | 55RT3214F | F | CAGARAGCATARTAATATGGGGAA | |
| ppr7c | R | GGTCCTTTCCAAATTGGGTCTCTGCTGTC | |||
| pol-nef | 4,739 | First | ppf2b | F | GCAGTCCATGTAGCCAGTGG |
| Onef2R | R | GAGTAAATTARCCCATCCAGTCC | |||
| pol-vif (F) | 729 | Nested | ppf10b | F | CACAATTTTAAAAGAAAAGGGGGGATTGG |
| Ham11R | R | CTTCTGTTAATYTCTGGACAC | |||
| env (H) | 985 | V3vh-1F | F | TAGGCCAGYAGTRTCAAC | |
| env17R | R | GCGAGCTCGGAGTTGTCT | |||
| pol-env | 3,856 | First | ppf2b | F | GCAGTCCATGTAGCCAGTGG |
| env19R | R | CCACAACCATTTAGTTATGTC | |||
| vif-env (G) | 2,195 | Nested | polenvPS3F | F | GGTTTTATAGACATCAYTATG |
| JH47 | R | CAATAAAAGAATTCTCCATGAC | |||
| env-LTR | 1,901 | First | JH41 | F | CAGCAGGWAGCACKATGGG |
| 9131R-2 | R | CTCYCAGGCTCARATCTGGTC | |||
| env-LTR (I) | 1,808 | Nested | Menv16F | F | GGYATAGTGCARCAGCARA |
| LTRps1-R | R | TCAAGGCAAGCTTTATTGAG |
PCR primers not included.
F, forward primer; R, reverse primer.
Primer sequence obtained from S. M. Ahumada-Ruiz, Instituto de Salud Carlos III, Madrid, Spain.
Specimen U14788, collected in 2006 from a 54-year-old male patient at the Jamot Hospital in Yaoundé, Cameroon, was seropositive for HIV in the three immunoassays used initially to screen the specimens: Murex HIV Ag/Ab Combination EIA (signal/cutoff value [s/co], 6.0; maximum possible s/co, 10; Abbott Diagnostics, Dartford, United Kingdom), Determine HIV-1/2 (positive; Inverness Medical Innovations, Japan), and HIVAB HIV-1/HIV-2 (rDNA) EIA (s/co, 13.26; maximum s/co, 14.49; Abbott Diagnostics, Abbott Park, IL). Specimen U14788 was 1 of 5 selected for RT-PCR amplification based on SIVgor IDR peptide reactivity; the ratio of SIVgor signal to group M signal was greater than 0.6 (data not shown). Subsequent retesting of the U14788 specimen using IDR and V3 PEIAs that were modified to include both group P- and SIVgor-specific peptides showed that specimen U14788 had the strongest reactivity to the group P-derived peptides (data not shown). No patient clinical data are available.
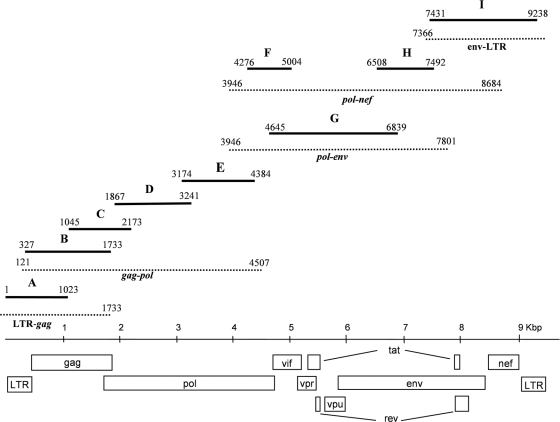
The nearly complete viral genome, designated 06CMU14788, was amplified and sequenced using proviral DNA extracted from a blood specimen of U14788. Nine overlapping fragments were generated using a combination of long-range- and nested-PCR procedures (Fig. 1 and Table 1). First-round long-range-PCR amplifications for fragments LTR-gag, gag-pol, pol-nef, and env-LTR were performed using 2.5 units of Pfu Turbo polymerase (Stratagene, La Jolla, CA) and 2.5 units of Taq polymerase (Bioline USA, Inc., Taunton, MA) in 1× Pfu Turbo buffer, 0.2 mM each deoxynucleoside triphosphate (dNTP), 20 pmol each primer, and 2 to 5 μl of DNA template in a 50-μl reaction mixture. The PCR conditions consisted of preincubation at 94°C for 2 min and then 40 cycles at 94°C for 15 s, 50°C or 55°C for 30 s, and 72°C for 6 min (gag-pol and pol-nef) or 2 min (LTR fragments), followed by 72°C for 10 min and holding at 4°C. The pol-env fragment was amplified using Qiagen reagents (Qiagen GmbH). Nested PCR was performed with AmpliTaq DNA polymerase (Applied Biosystems, Carlsbad, CA). Prior to sequencing, PCR fragment H, which spans the highly variable env gp120 V3 loop, was cloned using a Qiagen PCR cloning plus kit (Qiagen GmbH, Hilden, Germany). Amplified products were sequenced directly (13). Sequencing primers are available upon request. The sequence data were assembled and edited using Sequencher version 4.8 (Gene Codes Corporation, Ann Arbor, MI). One clone of fragment H that was most representative of the consensus sequence for three clonal sequences was selected for the final genome contig. The assembled genome sequence is 9,238 nucleotides in length and encodes open reading frames for all the HIV-1 structural and regulatory genes.
FIG. 1.
PCR amplification of the 06CMU14788 genome. First-round-PCR products are shown as dotted lines, and nested-PCR products are shown as solid lines. The nested products are labeled A to I, with numbers indicating the 5′- and 3′-nucleotide positions within the assembled 06CMU14788 genome sequence.
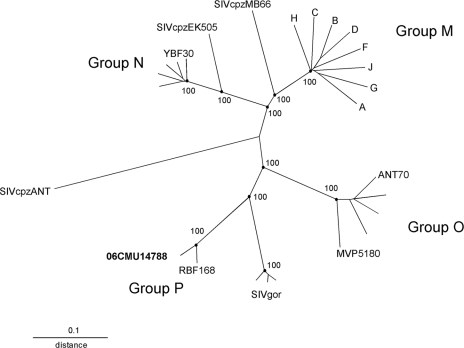
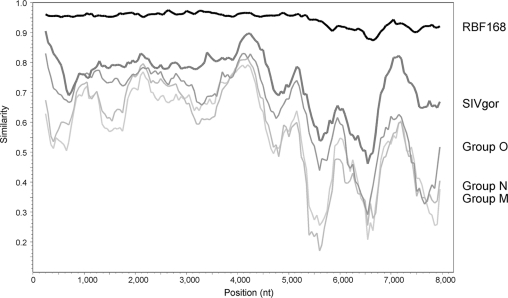
Phylogenetic analysis of the complete genome sequence confirmed the designation of 06CMU14788 as HIV-1 group P. The genome sequences for 06CMU14788 and group P strain RBF168 were manually aligned, using Lasergene MegAlign (version 7.2.1; DNAStar, Madison, WI), to an HIV-1 genome alignment from the HIV sequence database (http://www.hiv.lanl.gov) that was modified to limit the number of reference strains. Phylogenetic analysis was performed using PHYLIP software (version 3.5c; J. Felsenstein, University of Washington, Seattle, WA); evolutionary distances were estimated using DNADIST, and phylogenetic relationships were determined using Neighbor with SIVcpzANT as the outgroup. The tree was constructed using TreeView (R. D. M. Page, University of Glasgow, United Kingdom), and branch reproducibility was done using SEQBOOT (100 replicates) and CONSENSE. The phylogenetic tree derived from the genome alignment shows that 06CMU14788 and RBF168 cluster closely together on a branch shared with the SIVgor strains (Fig. 2). A similarity plot of a genome alignment restricted to the sequences of 06CMU14788, RBF168, group M strain HXB2, group N strain YBF30, group O strain ANT70, and SIVgor strain CP2139 confirms this close relationship across the length of the genome. The similarity plot was obtained using SimPlot (version 3.5.1; S. Ray, Johns Hopkins University, Baltimore, MD) with 06CMU14788 as the query sequence with a window of 500 nucleotides and a step of 50 nucleotides. The 06CMU14788 genome has greater than 87% similarity to RBF168 at the DNA level and has greater similarity to SIVgor than to HIV-1 groups M, N, and O (Fig. 3).
FIG. 2.
Phylogenetic tree derived from nucleotide alignment of genome sequences. HIV-1 group M is represented by single sequences for each subtype A through H; group N and group O are each represented by 5 sequences, with isolates YBF30, ANT70, and MVP5180 indicated, and SIVgor is represented by 3 sequences, CP684, CP2135, and CP2139. The alignment consisted of 7,509 nucleotides after gaps were stripped. Reproducibilities of key nodes are shown as percentages.
FIG. 3.
Similarity plot of nucleotide genome alignment using 06CMU14788 as the query sequence. The alignment consisted of 8,218 nucleotides (nt) after gaps were stripped and was evaluated using a window of 500 nucleotides and a step of 50 nucleotides. The thick black line represents group P isolate RBF168, and the thick dark-gray line represents SIVgor CP2139. The thinner gray lines represent group O ANT70 (medium gray), group N YBF30 (light gray), and group M HXB2 (pale gray).
Based on the screening strategy utilized in this study, the prevalence of group P in HIV-1-infected individuals in Cameroon is low (1 of 1,736 infections; 0.06%). In comparison, HIV-1 group O accounted for 2.2% and group N for 0.1% of HIV infections determined by using this strategy in similar Cameroonian study populations (1, 9). Group P infections may not be efficiently detected by current HIV screening tests due to the absence of group P-specific reagents for antibody detection. Thus, group P infections may have been missed because of our strategy of evaluating only HIV-seropositive specimens. In addition, the PEIAs may have failed to identify group P infections; reasons may include high cross-reactivity to non-P peptides, leading to misclassification; masking of group P reactivity due to coinfection with group M or O; and/or lack of antibody reactivity to the specific group P and SIVgor peptide sequences utilized in the assays. Alternatively, it is possible that group P may be restricted to specific regions of Cameroon that were not represented in our study population or originated elsewhere in West Central Africa with isolated introduction into the urban center of Yaoundé. Both group P-infected individuals had a connection to Yaoundé; U14788 was a hospital patient there, and RBF168 lived near Yaoundé prior to moving to France (6).
These data document the identification of a second HIV-1 group P infection in a Cameroonian individual, confirming that this new HIV lineage is circulating in humans. Despite the low prevalence of group P, that 06CMU14788 has the signature human-specific adaptation mutation, lysine at position 30 in the gag protein, and that RBF168 (methionine at gag position 30) replicates to high levels in human cell culture provide evidence that group P has adapted to humans and is capable of spreading between humans (6, 11).
Currently we know that the SIV strains that gave rise to HIV-1 have been introduced into the human population at least four times (groups M, N, O, and P) (4, 5, 6, 8, 10). These include only those zoonotic transmissions that were successful, i.e., that established an infection with subsequent transmission within a human population. The SIV strains that gave rise to HIV-2 have been introduced at least eight times (groups A to H). However, only HIV-2 groups A and B appear to be successful (3). The resulting AIDS pandemic shows the consequences of these viruses spreading and becoming well established in the human population before they were recognized as a public health issue. Thus, it is important to monitor for emergence of novel HIV strains to limit their spread.
Nucleotide sequence accession numbers.
The GenBank accession number of the 06CMU14788 genome sequence from proviral DNA is HQ179987, and those for the clonal sequences of the env V3 loop region not included in the genome sequence are HQ179988 and HQ179989. The pol and env sequences obtained from viral RNA are HQ179990 and HQ179991, respectively.
Acknowledgments
We thank David Robertson for helpful discussions and review of the manuscript.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Brennan, C. A., et al. 2008. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J. Acquir. Immun. Defic. Syndr. 49:432-439. [DOI] [PubMed] [Google Scholar]
- 2.Corbet, S., et al. 2000. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Damond, F., et al. 2004. Identification of a highly divergent HIV type 2 and proposal for a change in HIV type 2 classification. AIDS Res. Hum. Retroviruses 20:666-672. [DOI] [PubMed] [Google Scholar]
- 4.Gao, F., et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 5.Keele, B. F., et al. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plantier, J.-C., et al. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871-872. [DOI] [PubMed] [Google Scholar]
- 7.Simon, F., et al. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032-1037. [DOI] [PubMed] [Google Scholar]
- 8.Takehisa, J., et al. 2009. Origins and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallari, A., et al. 2010. Four new HIV-1 group N isolates from Cameroon: prevalence continues to be low. AIDS Res. Hum. Retroviruses 26:109-115. [DOI] [PubMed] [Google Scholar]
- 10.Van Heuverswyn, F., et al. 2006. SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 11.Wain, L. V., et al. 2007. Adaptation of HIV-1 to its human host. Mol. Biol. Evol. 24:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi, J., et al. 2004. HIV infections in northwestern Cameroon: identification of HIV type 1 group O and dual HIV type 1 group M and group O infections. AIDS Res. Hum. Retroviruses 20:944-957. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi, J., et al. 2010. Near full-length sequence of HIV type 1 subtype J strain 04CMU11421 from Cameroon. AIDS Res. Hum. Retroviruses 26:693-697. [DOI] [PubMed] [Google Scholar]