Abstract
The limited success of HIV vaccine candidates to date highlights our need to better characterize protective cell-mediated immunity (CMI). While HIV-specific CD8+ T cell responses have been defined largely by measuring gamma interferon (IFN-γ), these responses are not always protective, and it is unclear whether the same epitopes would predominate if other functional parameters were examined. Here, we assessed the epitope specificity of HIV-specific CD8+ T cell responses by multiparametric flow cytometry, measuring five CD8+ T cell functions (IFN-γ, macrophage inflammatory protein 1β [MIP-1β], tumor necrosis factor alpha [TNF-α], interleukin-2 [IL-2], and proliferative capacity) in 24 chronically HIV-infected individuals. Sixty-nine epitope-specific responses to 50 epitopes within p24 were measured. Surprisingly, most epitope-specific responses were IFN-γ negative (50/69 responses). Many responses had polyfunctional (33%) and proliferative (19%) components. An inverse association between IL-2 and proliferation responses was also observed, contrary to what was described previously. We confirm that long-term nonprogressors (LTNP) have more polyfunctional responses and also have higher-magnitude and broader p24-specific proliferation and higher levels of IL-2 and TNF-α production than do progressing controls. Together, these data suggest that the specificity of CD8+ T cell responses differs depending on the immunological readout, with a 3.5-fold increase in breadth detected by including multiple parameters. Furthermore, the identification of epitopes that elicit polyfunctional responses reinforces the need for the comprehensive evaluation of HIV vaccine candidates, and these epitopes may represent novel targets for CMI-based vaccines.
As the human immunodeficiency virus (HIV)/AIDS epidemic continues to grow, there is a desperate need for an effective vaccine. Most current HIV vaccine candidates aim to induce HIV-specific CD8+ T cell responses capable of containing viral replication and slowing disease progression. This vaccine concept is based on several lines of evidence suggesting that CD8+ T cell responses can control viral replication (7, 25, 27, 40). However, HIV-specific CD8 responses are detected in nearly all HIV-positive (HIV+) subjects regardless of disease progression (8, 20, 21, 38). Several large studies have found no correlation between HIV-specific CD8+ T cell gamma interferon (IFN-γ) secretion and viral load, and high-avidity responses to autologous virus can be measured in subjects who are progressing to AIDS (16, 26). HIV-specific CD8+ T cells are often exhausted or functionally inferior in chronic, progressive HIV-1 infection, in some cases lacking perforin expression, cytokine secretion, and proliferative capacity (30, 33). These data suggest that not all CD8+ T cell responses are effective, and responses that better correlate with protection need to be identified.
A subgroup of HIV-infected subjects, termed long-term nonprogressors (LTNP), experience slower progression to AIDS and provide a valuable model for the study of cell-mediated immunity (CMI) responses that may be capable of controlling HIV. Previous work has demonstrated that these individuals maintain stronger HIV-specific CD8+ T cell proliferation than do progressing controls (11, 30, 33). LTNP were also found to have more polyfunctional HIV-specific CD8+ T cells, as defined by the concurrent expression of the cytokines IFN-γ, interleukin-2 (IL-2), and tumor necrosis factor alpha (TNF-α); the chemokine macrophage inflammatory protein 1β (MIP-1β); and the degranulation marker CD107a (1, 5). Polyfunctional CD8+ T cell responses have been measured in humans vaccinated with the highly efficacious smallpox virus vaccine (37), while polyfunctional CD4+ T cells have been found to be protective in settings where immunity is primarily cell mediated, for example, following tuberculosis vaccination and in murine models of Leishmania major (6, 10). The detection of polyfunctional CD8+ T cells in HIV-1-exposed but -uninfected subjects potentially demonstrates that these polyfunctional responses may play a role in protection against HIV infection (17).
Most studies describing the epitope specificity of CD8+ T cell responses in HIV infection have relied extensively on IFN-γ enzyme-linked immunospot (ELISPOT) assays (44), which allow a rapid definition of positive responses. Recent studies have begun to call into question the reliability of ELISPOT assays for the assessment effective immune responses (4, 45, 47). The use of a single readout may miss many effective responses, particularly a readout that may not measure responses capable of controlling HIV infection.
For example, little is known regarding the specificity of proliferative responses despite evidence that they are associated with the control of HIV infection (11, 24, 33). Data from our laboratory examining CD8+ T cell responses to HIV Env using IFN-γ ELISPOT assays and 6-day carboxyfluorescein diacetate succinimidyl ester (CFSE) proliferation assays revealed substantial differences in the epitopes recognized between assays (31). Here we extend these observations using an unbiased epitope-mapping approach to determine the specificity of polyfunctional CD8+ T cell responses. Although proliferation and polyfunctional responses have previously been associated with LTNP, those studies utilized peptide pools or predefined epitopes to measure responses. By epitope mapping of HIV-specific CD8+ T cells with multiple parameters, we observed many readout-specific responses, including a response breadth of over 3.5 times that if IFN-γ was used alone.
MATERIALS AND METHODS
Subjects.
Study participants (n = 24) were all HIV infected and enrolled in a well-described longitudinal female sex worker cohort based in Nairobi, Kenya (18) (Table 1). Written informed consent was obtained from all study participants, and ethics review boards from the University of Manitoba and the Kenyatta National Hospital approved the study. Clinical and demographic data, including CD4 T cell counts, were collected from subjects biannually. Antiretroviral therapy (ART)-naïve subjects with CD4+ counts above 400 cells/μl for over 6 years were classified as long-term nonprogressors (LTNP). LTNP were monitored for mean and median times of 11.98 and 14.31 years, respectively (range, 6.41 to 17.43 years; n = 10). Participants who did not meet the criteria for LTNP were considered to be normal progressors (NP) (n = 9); the time to progression to AIDS has been demonstrated to be 3.5 years for this cohort (2). Additionally, five subjects on ART were included in the study.
TABLE 1.
Demographic data for study subjects
| Subject | Disease statusa | Age (yr) | Yr of follow-up | CD4 count (cells/μl) | ART start date (day/mo/yr) | Viral load (copies/ml) | Date of viral load measurement (day/mo/yr or mo/yr) | Clade |
|---|---|---|---|---|---|---|---|---|
| 890 | LTNP | 53 | 16.8 | 570 | 144 | 5/8/2005 | A1 | |
| 1211 | LTNP | 40 | 17.43 | 437 | 8,512 | 5/8/2005 | A1 | |
| 1250 | LTNP | 49 | 6.41 | 468 | 226 | 5/2005-8/2005 | ||
| 1287 | LTNP | 36 | 6.53 | 547 | ||||
| 1424 | LTNP | 43 | 15.55 | 539 | 481 | 5/2005-8/2005 | A1 | |
| 1625 | LTNP | 43 | 14.37 | 418 | 2,400 | 19/11/2007 | A1 | |
| 1647 | LTNP | 41 | 13.55 | 731 | ||||
| 1649 | LTNP | 40 | 14.31 | 447 | 1,803 | 11/2006 | D | |
| 1654 | LTNP | 36 | 14.31 | 433 | ||||
| 1725 | LTNP | 41 | 10.79 | 452 | 1,295 | 5/2005-8/2005 | A1 | |
| 1731 | ART | 42 | 13.67 | 228 | 19/6/2007 | 50 | 5/2005-8/2005 | A1 |
| 1771 | NP | 42 | 12.74 | 186 | 6,700 | 11/2006 | ||
| 1848 | NP | 36 | 10.4 | 184 | ||||
| 1917 | ART | 37 | 7.26 | 456 | 16/12/2005 | 3,400 | 15/4/2004 | A1 |
| 1932 | ART | 36 | 6.42 | 407 | 30/5/2005 | 76 | 5/2005-8/2005 | A1 |
| 1947 | ART | 38 | 6.17 | 359 | 1/29/2007 | 479 | 5/2005-8/2005 | |
| 1971 | ART | 45 | 5.31 | 168 | 4/23/2007 | 3,000 | 28/2/2006 | |
| 1974 | NP | 44 | 5.29 | 235 | 8,100 | 6/3/2006 | D | |
| 2166 | NP | 38 | 4.67 | 233 | ||||
| 2274 | NP | 37 | 1.97 | 921 | ||||
| 2522 | NP | 27 | 0.74 | 408 | ||||
| 2531 | NP | 48 | 0.73 | 256 | ||||
| 2560 | NP | 42 | 0.7 | 446 | ||||
| 2630 | NP | 33 | 0 | 307 |
LTNP, long-term nonprogressor; NP, normal progressor; ART, subjects on antiretroviral therapy.
Flow cytometry panels.
HIV-specific responses were assessed by the simultaneous measurement of four immunological parameters in a panel that included CD3-AmCyan, CD8-allophycocyanin cynine 7 (APCCy7), IFN-γ-fluorescein isothiocyanate (FITC), MIP-1β-phycoerythrin (PE), IL-2-allophycocyanin (APC) (BD Biosciences), and TNF-α-Pacific Blue (eBiosciences). Proliferation was assessed by using a flow cytometry panel that included carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Invitrogen), CD3-AmCyan, and CD8-APCCy7 (BD Biosciences).
Peptides.
Peptides (9-mer peptides overlapping by 8 amino acids [aa]) derived from the HIV-1 p24 clade A1 ancestral sequence (Sigma-Genosys) were pooled in a matrix format using Deconvolute This!, version 1.0 (39) (courtesy of Mario Roederer, Vaccine Research Center, NIAID, NIH). Clade A1 is the predominant circulating clade in this study population (12, 35, 36). Each peptide was represented twice, resulting in 16 pools with approximately 30 peptides per pool. Peptide pools were used at 2 μg/ml/peptide, and stimulations were accompanied by 2 positive controls, cytomegalovirus (CMV), Epstein-Barr virus, and influenza virus (CEF) peptides (32 peptides/pool, 2 μg/ml/peptide; AnaSpec) and Staphylococcus aureus enterotoxin B (SEB) (0.1 μg/ml; Sigma-Aldrich), and duplicate negative controls consisting of medium alone.
Intracellular cytokine staining.
Fresh peripheral blood mononuclear cells (PBMC) were stimulated with peptide pools, CEF, SEB, or medium overnight for 14 h. The stimulation time of 14 h was chosen to optimize IL-2 expression. PBMC were incubated at 1 × 106 cells/ml in complete R-10 medium (RPMI 1640 [HyClone, Thermo Scientific] supplemented with 10% heat-inactivated fetal bovine serum [FBS] [Gibco, Invitrogen] and 1% Antimycotic Penstrep [Gibco, Invitrogen]). After 1 h of stimulation at 37°C, pretitrated amounts of monensin (Golgistop, 1 μl/ml; BD Biosciences) and brefeldin A (Golgiplug, 1 μl/ml; BD Biosciences) were added, followed by a further 13 h of incubation. Following incubation the cells were washed and stained according to the manufacturer's protocol, using a Cytofix/Cytoperm kit (BD PharMingen). Stained cells were analyzed immediately.
Proliferation assays.
PBMC were resuspended in phosphate-buffered saline (PBS) and loaded with CFSE according to the manufacturer's protocols (Molecular Probes). Cells were than stimulated with p24 peptide pools and incubated for 6 days at 37°C in R-10 medium. The same sets of negative and positive controls as those used for the overnight assays were used. On day 6, the cells were washed, stained using surface antibodies, fixed with 1% paraformaldehyde, and acquired immediately.
Data acquisition and analysis.
Cells were analyzed on an LSRII flow cytometer (BD Biosciences). Between 30,000 and 100,000 events were collected within the lymphocyte gate per sample. Data analyses were performed by using FlowJo software, version 8.7.3 (TreeStar). Boolean gates were applied to the 4 overnight functions, and the polyfunctionality of each response was assessed by using SPICE 5.1 (courtesy of Mario Roederer, Vaccine Research Center, NIAID, NIH).
Statistical analysis.
HIV-specific CD8+ T cell responses were considered positive for a given parameter if they were ≥2-fold higher than the mean of their respective negative controls. Data reported are values after background subtraction. Polyfunctional responses were also background subtracted, and a lower threshold corresponding to the 90th percentile of distribution of negative values was built for each cytokine pattern, and values below this threshold were set to 0. For some participants limited cell numbers prevented the confirmation of all putative epitopes; in these cases the cutoff for a positive pool response was raised to 3-fold over background, and therefore, only the strongest peptides were confirmed. Statistical analyses were performed by using Graph Pad Prism 5.0 and SPICE 5.1. All correlations were determined by using Spearman's rank correlation. Breadth and magnitude comparisons between subject groups were determined by using Mann-Whitney tests.
RESULTS
Epitope mapping of p24 using parallel measurements of four CD8+ T cell functions.
HIV-specific CD8+ T cell responses to p24 epitopes were mapped by simultaneously measuring IFN-γ, MIP-1β, IL-2, and TNF-α production following overnight stimulations with 16 peptide pools representing HIV-1 clade A1 p24. Representative intracellular and proliferation responses are shown in Fig. 1a. For this particular participant, CD8+ T cell responses to a single peptide were detectable for all four effector functions, while proliferation was detected for another peptide. The full complexity of the short-term response to all peptide pools was then examined by using Boolean gating, yielding 16 unique response combinations for the four individual readouts measured (Fig. 1b).
FIG. 1.
Representative polyfunctional intracellular cytokine staining and 6-day CFSE dilution. (A) Initial gating of forward-scatter area (FSC-A) versus forward-scatter height (FSC-H) used to eliminated doublets. The side-scatter area (SSC-A) versus the FSC-A was used to identify lymphocytes. After gating on CD3+ CD8+ cells, further gates were made for each of the respective functions (IFN-γ, MIP-1β, TNF-α, IL-2, and proliferation). Shown are representative data from subject ML2630, including overnight responses to the p24 peptide QGQMVHQSL (aa 7 to 14) and the day 6 response to the peptide SDIAGTTST (aa 102 to 110). (B) Boolean gates were applied to the 4 overnight functions. The 16 possible CD8+ T cell response combinations are shown for LTNP, ART, and NP. IFN-γ, MIP-1β, IL-2, and/or TNF-α responses are shown as positive (+) and negative (−). (C) Comparison of the mean responses to p24 peptide pools between LTNP, ART, and NP. For simplicity, responses are grouped by number of functions, matched to the colored bars shown in B. LTNP and ART have a higher frequency of polyfunctional responses than do NP (P < 0.001).
LTNP maintain a higher degree of HIV-specific CD8+ T cell functionality and stronger proliferation, IL-2, and TNF-α responses.
Previous work suggested that LTNP maintain a higher degree of polyfunctional HIV-specific CD8+ T cell responses than do NP (5, 48). However, this was examined mostly using predefined epitopes or peptide pools and not a comprehensive epitope-screening approach. After Boolean gating and stratification by disease status and ART use, we confirmed that a higher degree of CD8+ T cell polyfunctionality in LTNP was observed for our study (Fig. 1c). HIV-specific CD8+ T cell responses from LTNP and ART subjects (Fig. 1b, blue and red bars, respectively) displayed a higher functional profile than did those from NP (P < 0.0001) (Fig. 1b, green bars, and c). While responses that included all four functions were nearly absent from NP, they were observed at a low frequency in LTNP and at a relatively high frequency in subjects on ART, presumably due to reconstituted immune responses in the latter.
Although their CD8+ T cell responses were more polyfunctional, LTNP did not have a higher breadth of response than did NP, regardless of the parameter taken into account (Table 2). Pool-specific responses in LTNP were typically of a higher magnitude than those observed for NP (proliferation, P < 0.0001; IL-2, P = 0.0310; TNF-α, P = 0.0052 [by Mann-Whitney test]) (Table 2). The total breadth of responses, measured by the total number of responding pools regardless of parameter or disease profile, was not associated with either viral load or CD4 count. However, CD4 counts were associated with the breadth of both pool-specific proliferation and TNF-α responses (r = 0.41 and P = 0.0453, and r = 0.41 and P = 0.0461, respectively, by Spearman rank correlation) (Table 3). These associations suggest that polyfunctional p24 responses are an important correlate of LTNP in this cohort.
TABLE 2.
Comparison of breadths and magnitudes of responses in LTNP and NP
| Comparison |
P valuea |
|||||
|---|---|---|---|---|---|---|
| Total | Proliferation | IFN-γ | MIP-1β | IL-2 | TNF-α | |
| Breadth for LTNP vs NP | 0.2372 | 0.1629 | 0.4542 | 0.3435 | 0.4246 | 0.1517 |
| Magnitude for LTNP vs NP | <0.0001 (LTNP > NP) | 0.2913 | 0.7295 | 0.0310 (LTNP > NP) | 0.0052 (LTNP > NP) | |
Boldface type indicates a significant association.
TABLE 3.
Comparison of breadths of responses with CD4 counts and viral loads
| Comparison |
r value, P valuea |
|||||
|---|---|---|---|---|---|---|
| Total | Proliferation | IFN-γ | MIP-1β | IL-2 | TNF-α | |
| CD4 count | 0.33, 0.1143 | 0.41, 0.0453 | −0.12, 0.5781 | −0.39, 0.0596 | −0.002, 0.9934 | 0.41, 0.0461 |
| Viral load | 0.23, 0.4354 | 0.43, 0.1439 | 0.15, 0.6329 | 0.24, 0.4332 | 0.39, 0.1836 | −0.18, 0.5577 |
Boldface type indicates a significant association.
Different functional readouts define divergent specificities of CD8+ T cell responses.
We hypothesized that regardless of the progression status, the specificity of responses to p24 pools would differ depending on the functional readout of the HIV-specific CD8+ T cell response that was measured. In a proportion of participants (7/24 participants), pool-specific IFN-γ responses were observed in the absence of any other functional parameters; conversely, the majority of participants (20/24) displayed one or more of MIP-1β, IL-2, and/or TNF-α in the absence of IFN-γ (Table 4). These data suggest that many CD8 T cell responses are missed if IFN-γ was the only parameter measured.
TABLE 4.
p24 pool responses showing disconnect between IFN-γ and other short-term parameters
| Subject | Total no. of IFN-γ-positive responses | No. of IFN-γ responses in the absence of other short-term parameters | Total no. of MIP-1β, IL-2, and/or TNF-α responses in the absence of IFN-γ |
|---|---|---|---|
| 890 | 0 | 0 | 7 |
| 1211 | 2 | 0 | 0 |
| 1250 | 1 | 0 | 5 |
| 1287 | 0 | 0 | 1 |
| 1424 | 9 | 1 | 5 |
| 1625 | 1 | 0 | 0 |
| 1647 | 8 | 1 | 6 |
| 1649 | 2 | 2 | 3 |
| 1654 | 6 | 2 | 2 |
| 1725 | 3 | 0 | 2 |
| 1731 | 1 | 0 | 2 |
| 1771 | 10 | 0 | 4 |
| 1848 | 0 | 0 | 2 |
| 1917 | 0 | 0 | 8 |
| 1932 | 2 | 0 | 2 |
| 1947 | 0 | 0 | 0 |
| 1971 | 5 | 1 | 2 |
| 1974 | 1 | 0 | 5 |
| 2166 | 2 | 2 | 2 |
| 2274 | 1 | 0 | 5 |
| 2522 | 0 | 0 | 0 |
| 2531 | 2 | 2 | 1 |
| 2560 | 0 | 0 | 2 |
| 2630 | 3 | 0 | 3 |
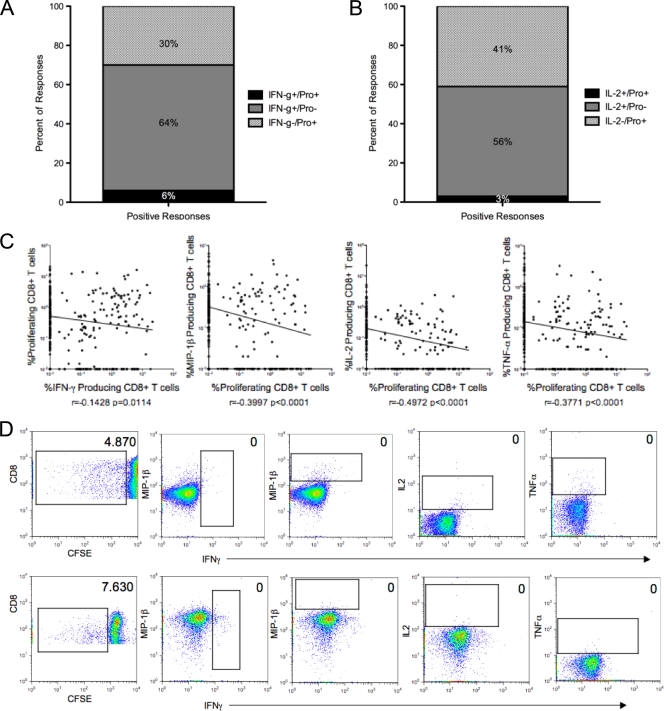
For the majority of the subjects (67%; 16/24), at least one p24 pool was IFN-γ positive (IFN-γ+) in the absence of a proliferative response. Similar data were obtained by examining proliferation responses: 50% of subjects (12/24) had at least one proliferation-positive pool that was negative for IFN-γ. In total, the majority of IFN-γ responses did not have an accompanying proliferative response (91%; 54/59), and 25/30 (83%) of proliferation-positive pools lacked a corresponding IFN-γ response. Only 6% of responding pools were positive in both assays, all of which occurred in the LTNP, while 64% were IFN-γ+ and proliferation negative, and 30% were IFN-γ negative (IFN-γ−) and proliferation positive (Fig. 2a). These data suggest that specific epitopes within these p24 pools are resulting in either an IFN-γ or a proliferative response but rarely both in the same subject.
FIG. 2.
Disconnect between overnight and proliferation assays. (A) Percentage of CD8+ T cells that were IFN-γ positive in the overnight assay and proliferation negative (Pro−) in the day 6 assay, those that were IFN-γ negative and proliferation positive, and those that were positive in both assays. (B) Percentage of CD8+ that were IL-2 positive in the overnight assay and proliferation negative in the day 6 assay, those that were IL-2 negative proliferation positive, and those that were positive in both assays. (C) Inverse correlation between pool-specific proliferative responses and all four overnight readouts (Spearman's rank correlation). Shown are percentages of day 6 and overnight responses for all subjects and all responses. (D) Background-adjusted CD8+ T cell responses in subjects ML2531 (NP) (top) and ML1647 (LTNP) (bottom). In each of these instances, the only response to the respective pool was proliferation (pools 14 and 8, respectively).
To better understand the relationship between specific effectors and proliferative responses, we first determined the association between short-term pool-specific responses in LTNP and NP subjects. This approach allows the evaluation of both the concurrent expression of multiple cytokines, where one cell is responsible for the secretion of both effectors, and the coexpression of multiple cytokines, where the same sample is positive for two cytokines but is expressed by different cells. Interestingly, HIV-specific CD8+ T cell responses from LTNP and NP differed in many associations, principally those involving TNF-α expression (Table 5). In general, LTNP exhibited more consistent associations between overnight parameters than did NP. This supports the observation that LTNP were more likely to have a concurrent expression of multiple parameters and have a higher degree of polyfunctionality.
TABLE 5.
Correlations between short-term parameters stratified by disease status
Boldface type indicates a significant association. Shading indicates associations that differ between LTNP and NP.
We next examined the correlation between overnight and proliferation responses. Across the entire data set, the magnitude of IFN-γ-positive responses correlated inversely with the magnitude of the proliferation response to the corresponding pool (r = −0.14 and P = 0.0114 by Spearman's rank correlation) (Fig. 2c). This is similar to what has previously been described, where substantial differences were observed in the epitope specificities between IFN-γ ELISPOT and 6-day proliferation assays (31). In the present study, proliferation responses were inversely correlated with the magnitude of all overnight readouts, including MIP-1β, IL-2, and TNF-α (all P < 0.0001) (Fig. 2c and d). When stratified by disease profile, IFN-γ and proliferation responses did not correlate in either LTNP or NP (Table 6). In addition, proliferation also correlated inversely with the all other effector functions in both groups (P < 0.05). These data again demonstrate substantial differences in the antigen specificities of CD8+ T cells measured by proliferative assays compared to any readout in overnight assays.
TABLE 6.
Correlations between short-term parameters and proliferation stratified by disease status
| Proliferation in subject type |
r value, P valuea |
|||
|---|---|---|---|---|
| IFN-γ | MIP-1β | IL-2 | TNF-α | |
| LTNP | −0.10, 0.2828 | −0.41, <0.0001 | −0.51, <0.0001 | −0.30, 0.0010 |
| NP | −0.07, 0.4207 | −0.32, 0.0007 | −0.41, <0.0001 | −0.32, 0.0003 |
Boldface type indicates a significant association.
HIV-1-specific IL-2+ and proliferating CD8+ T cell responses were previously observed to correlate strongly (48). Therefore, the inverse relationship between IL-2-secreting HIV-specific CD8+ T cells and proliferation responses (r = −0.49; P < 0.0001) (Fig. 2c) observed in our study was unexpected. In the present study, 70% (17/24) of subjects had at least one pool that was IL-2+ but did not have an accompanying proliferative response. Similarly, 58% of subjects (14/24) had at least one proliferation-positive pool that was negative for IL-2. In total, only 5% (2/41) of IL-2 responses had an accompanying proliferative response, and only 7% (2/30) of the proliferation responses had a corresponding IL-2 response (Fig. 2b). While only 3% of pools were IL-2+ and proliferation positive, 56% were IL-2+ and proliferation negative, and 41% were IL-2− and proliferation positive (Fig. 2b). The data suggest an even greater disparity between proliferation and IL-2 than what was observed for IFN-γ and suggest that IL-2 might not always be a reliable predictor of proliferative capacity.
The epitope specificities of CD8+ T cell responses differ based on functional readout.
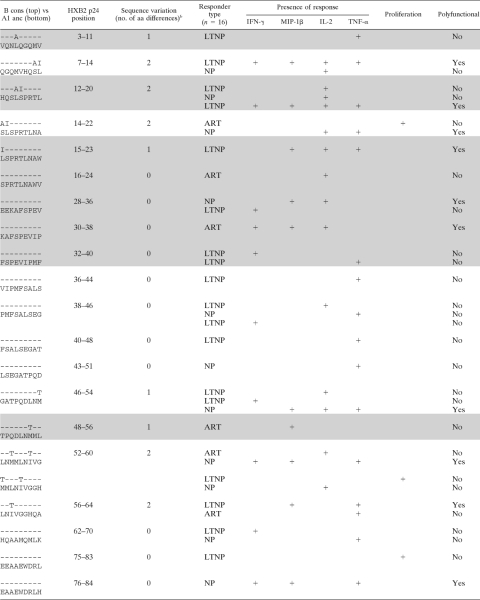
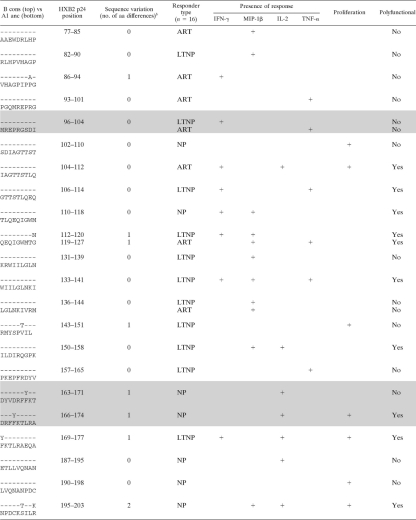
To further dissect differences between functional readouts at the epitope level, we confirmed the epitope specificities of pool responses. Putative responding peptides were identified by using the pooling matrix, and these were subsequently confirmed at later time points for 16/24 subjects (126 putative epitopes were tested with a range of 4 to 21 peptides tested/subject). Similar numbers of epitopes were tested between patient groups (data not shown). All 16 subjects responded to at least one of their putative epitopes, making a total of 69 responses to 50 epitopes (Table 7). There was no difference in the breadths of the response to the p24 epitopes between LTNP and NP (P = 0.9423; mean numbers of peptides of 4.7 for LTNP and 4.5 for NP) (data not shown).
TABLE 7.
Fifty identified epitopes and their associated response profilesa
Shown is a comparison of B consensus (B cons) sequences with recognized peptides from the A1 uncestral (A1 anc) library used in the current study. Shading indicates epitopes that are considered best-defined epitopes (BDE) or can be found within a longer, 10-aa to 15-aa BDE (23).
Number of amino acid differences between the sequences noted.
In agreement with the pool data, non-IFN-γ responses were observed for a high proportion (50/69; 73%) of peptide-specific responses. These data clearly show that a substantial portion of immune responses would remain undetected had we measured IFN-γ alone. Proliferation was a component of 13/69 (18%) responses, and only one response was proliferation positive and IFN-γ+. Consistent with pool-specific responses, the percentages of epitope-specific IFN-γ-responding cells were inversely correlated with the percentages of proliferating cells (r = −0.66; P < 0.0001) (data not shown). Furthermore, the majority (9/13; 69%) of proliferative responses detected did not have any corresponding responses measured in the overnight assay. Where a corresponding overnight function was present, it was often IL-2 (3/4 [75%] [1 LTNP and 2 NP]). However, the majority (22/25) of the IL-2 responses did not have a corresponding proliferation response, and the percentage of proliferating cells was inversely correlated with the magnitude of the IL-2 response (r = −0.72; P < 0.0001) (data not shown).
To determine if the addition of extra parameters would lead to the definition of new epitopes, we compared epitopes identified in our study with previously characterized epitopes. Of the 50 epitopes identified in this study, 12 (24%) are considered best-defined epitopes (BDE) or can be found within a longer BDE (10 to 15 aa) (19), while the remaining epitopes have not been fully characterized (Table 7). To elucidate possible sequence variations that may have led to the detection of novel epitopes, we compared the clade A1 ancestral sequence used in this study and the clade B consensus sequence used in previous epitope-mapping studies (Table 7) (19). Many of the previously uncharacterized epitopes (13/38; 33%) had ≥1 amino acid variation between the study sequence and the clade B consensus sequence, possibly accounting for their lack of recognition in previous studies. However, 100% of the 12 recognized BDE also had ≥1 amino acid variation between the two sequences. Therefore, sequence differences are likely not the reason why these new epitopes have not been previously defined, and the addition of functional readouts could increase the number of HIV epitopes that are defined.
Interestingly, polyfunctionality (defined as ≥2+ responses) comprised 23/69 (33%) of the responses (Table 7). Both BDE and the newly described epitopes frequently stimulated polyfunctional responses (5/17 and 18/52, respectively; P = 0.59) and responses in the absence of IFN-γ (11/17 and 39/52, respectively; P = 0.35). Interestingly, the uncharacterized epitopes were solely responsible for the proliferation responses that were observed (n = 13). These data suggest not only that it is important to measure multiple functions but also that the measurement of responses at multiple time points is critical.
This polyfunctional epitope-mapping approach revealed a further disconnect between IFN-γ and proliferative responses. As one might predict from the pool data, individual epitopes often preferentially elicited IFN-γ and not proliferation, and visa versa. This is shown for subject ML1211 for pool 3 and pools 7 and 10, respectively (Fig. 3a and b) and for the deconvoluted peptides (Fig. 3b), with peptide 86 eliciting a polyfunctional 4+ response, while two peptides from this subject (peptides 66 and 186) were positive for proliferation but none of the short-term parameters.
FIG. 3.
HIV-specific CD8+ T cell specificity differs depending on the effector readout, as shown for a representative subject. (A) Pool-specific responses for ML1211. Proliferation responses were measured at day 6, and four different readouts were measured after overnight stimulation. This subject has divergent long-term and short-term responses, including proliferation only (pool 10) and 4+ without proliferation (pool 3). Pool 15 is an example of a 4+ polyfunctional response with accompanying proliferation. (B) Deconvoluted peptide responses for subject ML1211. Peptide 86 elicited a polyfunctional 4+ response, while two peptides, peptides 66 and 186, preferentially elicited a proliferation-only response. (C) Pool-specific responses for subject ML1932. Many responses were 1+, with an MIP-1β response to pool 14 and an IL-2 response to pools 5 and 11. Polyfunctional responses to pools 7 and 16 were seen with the concurrent expression of IFN-γ and MIP-1β. (D) Deconvoluted peptide responses for subject ML1932. Responses to peptides 202, 203, and 208 were 1+, with an MIP-1β response and a TNF-α response, respectively. Peptides 209 and 210 elicited a 2+ response, with an IFN-γ+ IL-2+ response and an MIP-1β+ TNF-α+ response, respectively.
In addition, we observed numerous examples of individual epitopes selectively expressing one or more overnight parameters. Data for subject ML1932 (Fig. 3c and d) demonstrate the functional disparity of CD8 overnight responses at both the pool and epitope levels. This subject responded with monofunctional responses to 3/5 responsive pools (MIP-1β responses to pool 14 and IL-2 responses to pools 5 and 11) but also had polyfunctional responses to pools 7 and 16, with the coexpression of IFN-γ and MIP-1β. This participant showed similar selectivenesses in expression at the peptide level, with responses to peptides 202, 203, and 208 being monofunctional and independent from all other parameters (MIP-1β+ and TNF-α, respectively). Several polyfunctional responses were also seen: peptide 209 was IFN-γ+ IL-2+, while peptide 210 was MIP-1β+ TNF-α+. These data further show that the functional readout used to define CD8+ T cell activity can play a major role in determining the epitope specificity of HIV-specific CD8+ T cells.
DISCUSSION
Recent advances in immunology have allowed the opportunity to redefine the breadth and specificity of the large, complex antiviral CD8+ T cell responses, expanding our understanding of protective immunity. For example, acute CD8+ T cell responses to smallpox and yellow fever virus vaccinations, measured by cellular activation, intracellular cytokine staining, and tetramer frequencies, revealed that these responses are of a much higher magnitude than was previously appreciated (34). While assessments of HIV-specific responses have traditionally relied on assays that measure IFN-γ secretion, this cytokine does not always correlate with protective immunity, nor does it accurately describe the full breadth of HIV-specific CD8+ T cell responses (15, 26, 45). Other studies have demonstrated that HIV+ LTNP maintain a more functional response than do those who progress to AIDS (1, 5), but whether these responses differ in specificity remains largely unexplored. Here we have mapped CD8+ T cell epitopes more comprehensively than has been previously described, using multiple overnight parameters and long-term proliferation assays. Critically, over two-thirds of the CD8 responses detected in our study were detected in the absence of a parallel IFN-γ response (50/69 responses). These data suggest that previous epitope-mapping studies may have missed out on important immune responses by focusing solely on IFN-γ. Our data further reveal that CD8+ T cell responses are complex and that particular epitopes within a given individual can be associated with different functional profiles. Identifying and understanding epitopes that elicit polyfunctional and proliferative responses will serve as a valuable resource for vaccine design and ensure that wider verities of protective responses are obtained.
By epitope mapping using multiple parameters, we identified 50 epitopes in HIV-1 p24, 38 of which have not yet been fully characterized according to the Los Alamos HIV Immunology Database. A number of factors could explain the identification of these novel epitopes. For one, we used shorter peptides than most prior studies (9-mer peptides compared to 15-mer peptides), which have an increased sensitivity to detect responses (3, 13, 14, 41). Second, the database of optimal CD8+ T cell HIV epitopes may be biased toward clade B, while this epitope-mapping study was conducted in Kenya, where clade A1 predominates (12, 32, 35, 36). Although the use of the clade A1 ancestral sequence may be a contributing factor leading to the identification of novel epitopes, we also found that the 12 previously defined epitopes could also be recognized between clades, despite sequence differences. A third, and perhaps more important, factor is the inclusion of a greater number of functional parameters as readouts to define responses. The measurement of four functional parameters (IFN-γ, MIP-1β, IL-2, and TNF-α) plus proliferation expanded the breadths of the detected responses by over 3.5-fold. The majority of novel epitopes identified in this study did not include IFN-γ (39/52 responses), 13 of which elicited proliferation responses. Interestingly, a number of these previously unidentified epitopes induced polyfunctional (≥2+) responses (18/52) but not necessarily IFN-γ. One caveat of our study, however, was that the longer incubation time was not optimized to detect IL-2 and not IFN-γ. It is possible that we would have detected more epitope-specific responses producing IFN-γ, had we used a standard 6-h assay. However, together, these data support the idea that the breadth of HIV-specific immunity may be even greater than previously appreciated.
There was a striking discordance between responses measured in overnight assays and those measured in proliferation assays, with CD8+ T cells responding to epitopes by either secreting a cytokine or proliferating but rarely both. This suggests that a given epitope in a subject may preferentially induce either effector memory CD8+ T cell (TEM)-like (IFN-γ) or central memory CD8+ T cell (TCM)-like (proliferative) responses. This observation was not limited to IFN-γ but was apparent for all short-term readouts, as proliferation responses were inversely correlated with IFN-γ, MIP-1β, and TNF-α, all associated with TEM responses. This was even more pronounced at the epitope level, where the vast majority of proliferation responses (9/13) were completely independent of overnight parameters. While an explanation for these data remains unclear, one hypothesis is that the cells that respond in an overnight assay are not equipped to survive for the length of a proliferation assay, which is more likely to identify cells that need time to become activated in vitro. CD8+ T cell survival may be an important precursor of proliferation, whereas the short-term expression of cytokines, particularly IL-2, may not require survival to the same extent. Some studies suggested that survival capabilities, measured by exhaustion markers, might be the key attribute of a TCM cell (9).
Contrary to what was observed in a previous study (48), our data consistently showed an inverse correlation between IL-2 and proliferation, which rarely corresponded. A possible explanation for these differences is that previous studies stimulated cells for 6 h to measure IL-2, whereas we measured this cytokine in 14-h assays. However, another important difference between our data and those of Zimmerli et al. (48) is that our correlations were based on an epitope-screening approach that used IL-2 and proliferation to define CD8+ T cell specificity, compared to measuring IL-2+ CD8+ T cells that were also IFN-γ+. Since IL-2 and proliferation were both inversely correlated with IFN-γ in our cohort, it is possible that by focusing solely on IFN-γ responses, IL-2 and proliferation responses independent of IFN-γ may have been missed in previous studies. A lack of an association between proliferation and IL-2 in the bulk of Gag-specific CD8+ T cells was recently observed for a Chinese cohort, supporting our findings (29).
Virus-specific CD8+ T cells in chronic infections such as HIV experience substantial functional exhaustion. It has been suggested that this occurs in a hierarchical manner as the infection persists, where IL-2, cytolysis, and proliferation are the first to be lost, followed by the loss of TNF-α and, finally, by the loss of IFN-γ and anergy/deletion (28, 42, 43, 46). Based on this model, the reliance on IFN-γ to screen immune responses may overestimate the effectiveness of the immune response by measuring a significant proportion of CD8+ T cells that may be in the final stages of exhaustion. However, our data also challenge this model by finding many CD8+ T cell responses that are independent of IFN-γ, including the display of IL-2 and proliferation in isolation from other functions. Many functional divisions in epitope specificity were seen among the overnight parameters. An examination of all responses clearly shows that epitopes can stimulate many combinations of responses, often in the absence of IFN-γ. These data suggest that in a chronic viral infection, CD8+ T cell responses can be complex and heterogeneous.
In agreement with data from previous work, our study found that LTNP possess a more polyfunctional HIV-specific CD8+ T cell response than do NP (5). However, our polyfunctional profile is slightly different from those reported by previous studies, due to a longer incubation time, which likely underestimates MIP-1β responses that are optimally detected at 6 h. LTNP also responded with stronger p24-specific proliferation, IL-2, and TNF-α responses than did NP. These data are consistent with data from other studies which found the level of proliferation to be significantly lower in progressive chronic infection than primary infection or LTNP (30) and that responses to p24 may be an important target for the control of HIV replication (22). In addition, LTNP demonstrated more consistent associations between functional parameters than did progressors, including the rarely observed coexpression of proliferation and IFN-γ responses. This may be important for HIV vaccines, based on a recent study with nonhuman primates which showed that macaques vaccinated with a CMV vector expressing simian immunodeficiency virus (SIV) had robust TEM responses, characterized by polyfunctionality, and were less likely to become infected following low-dose challenge (23). However, the authors of that study stressed that the parallel induction of TCM responses, similar to what occurs in many LTNP, may provide a second line of defense in case infection becomes established.
In summary, we show that within a given HIV-infected subject, it is possible to identify novel epitopes that induce qualitatively distinct CD8+ T cell responses. Screening for responses by the measurement of five CD8+ T cell functions revealed a greater complexity of CD8+ T cell immunology and described a greater breadth of response than would have been derived from the measurement of any one response on its own. We have shown that epitope specificity differs between short- and long-term assays as well as within the short-term assay. A multiparametric approach to defining epitope specificity is one that could be used in vaccine evaluations in order to determine the true immunogenicity of a given vaccine candidate. Moreover, because polyfunctionality and proliferative responses have been associated with an attenuated HIV-1 disease course, these data and similarly comprehensive epitope-mapping studies could identify a number of additional epitopes that may serve as useful targets for successful HIV-1 vaccines that will presumably induce a wider range of epitope-specific responses.
Acknowledgments
We thank the study participants and the staff at the Majengo clinic (1985 to present) for their dedication and perseverance. We also thank Mario Roederer at the Vaccine Research Center, NIAID, NIH, for providing Deconvolute This! and SPICE software. F.P. is a Tier I Canada Research Chair in susceptibility and resistance to infection.
M.R. is supported by the MHRC, CIHR, and CIHR IID and GH training program. This work was supported by Canadian Institute of Health Research grant HOP 86192. We declare that we have no conflicts of interest.
M.R., L.R.M., S.A.K.K., and T.B.B. conceived and designed the experiments. M.R. and L.R.M. performed the experiments. M.R. and L.R.M. analyzed the data. M.R., L.R.M., S.A.K.K., and T.B.B. wrote the paper. J.K., C.W., M.K., F.A.P., and T.B.B. performed clinical work and cohort maintenance.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Almeida, J. R., et al. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anzala, O. A., et al. 1995. Rapid progression to disease in African sex workers with human immunodeficiency virus type 1 infection. J. Infect. Dis. 171:686-689. [DOI] [PubMed] [Google Scholar]
- 3.Beattie, T., et al. 2004. Screening for HIV-specific T-cell responses using overlapping 15-mer peptide pools or optimized epitopes. AIDS 18:1595-1598. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, M. S., H. L. Ng, A. Ali, and O. O. Yang. 2008. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. J. Infect. Dis. 197:390-397. [DOI] [PubMed] [Google Scholar]
- 5.Betts, M. R., et al. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beveridge, N. E., et al. 2007. Immunisation with BCG and recombinant MVA85A induces long-lasting, polyfunctional Mycobacterium tuberculosis-specific CD4+ memory T lymphocyte populations. Eur. J. Immunol. 37:3089-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borrow, P., et al. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 9.Chomont, N., et al. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darrah, P. A., et al. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13:843-850. [DOI] [PubMed] [Google Scholar]
- 11.Day, C. L., et al. 2007. Proliferative capacity of epitope-specific CD8 T-cell responses is inversely related to viral load in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowling, W. E., et al. 2002. Forty-one near full-length HIV-1 sequences from Kenya reveal an epidemic of subtype A and A-containing recombinants. AIDS 16:1809-1820. [DOI] [PubMed] [Google Scholar]
- 13.Draenert, R., et al. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275:19-29. [DOI] [PubMed] [Google Scholar]
- 14.Draenert, R., et al. 2004. Impact of intrapeptide epitope location on CD8 T cell recognition: implications for design of overlapping peptide panels. AIDS 18:871-876. [DOI] [PubMed] [Google Scholar]
- 15.Draenert, R., et al. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Draenert, R., et al. 2004. Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J. Virol. 78:630-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson, A. L., et al. 2008. Potentially exposed but uninfected individuals produce cytotoxic and polyfunctional human immunodeficiency virus type 1-specific CD8(+) T-cell responses which can be defined to the epitope level. Clin. Vaccine Immunol. 15:1745-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fowke, K. R., et al. 1996. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 348:1347-1351. [DOI] [PubMed] [Google Scholar]
- 19.Frahm, N., B. Baker, and C. Brander. 2008. Identification and optimal definition of HIV-derived cytotoxic T lymphocyte (CTL) epitopes for the study of CTL escape, functional avidity and viral evolution, p. 3-24. In B. T. M. Korber, et al. (ed.), HIV molecular immunology 2008. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM.
- 20.Gea-Banacloche, J. C., et al. 2000. Maintenance of large numbers of virus-specific CD8+ T cells in HIV-infected progressors and long-term nonprogressors. J. Immunol. 165:1082-1092. [DOI] [PubMed] [Google Scholar]
- 21.Goulder, P. J., et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 22.Goulder, P. J., and D. I. Watkins. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, S. G., et al. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton, H., et al. 2006. Preservation of T cell proliferation restricted by protective HLA alleles is critical for immune control of HIV-1 infection. J. Immunol. 177:7406-7415. [DOI] [PubMed] [Google Scholar]
- 25.Jin, X., et al. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klenerman, P., Y. Wu, and R. Phillips. 2002. HIV: current opinion in escapology. Curr. Opin. Microbiol. 5:408-413. [DOI] [PubMed] [Google Scholar]
- 27.Koup, R. A., et al. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristensen, N. N., J. P. Christensen, and A. R. Thomsen. 2002. High numbers of IL-2-producing CD8+ T cells during viral infection: correlation with stable memory development. J. Gen. Virol. 83:2123-2133. [DOI] [PubMed] [Google Scholar]
- 29.Li, H., et al. 2009. Proliferation, but not interleukin 2 production, of Gag-specific CD8+ T cells is associated with low HIV viremia and high CD4 counts in HIV-1-infected Chinese individuals. J. Acquir. Immune Defic. Syndr. 52:1-8. [DOI] [PubMed]
- 30.Lichterfeld, M., et al. 2004. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J. Exp. Med. 200:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKinnon, L. R., et al. 2008. Substantial intrapatient differences in the breadth and specificity of HIV-specific CD8+ T-cell interferon-gamma and proliferation responses. J. Acquir. Immune Defic. Syndr. 49:123-127. [DOI] [PubMed] [Google Scholar]
- 32.McKinnon, L. R., et al. 2009. Epitope mapping of HIV-specific CD8+ T cells in a cohort dominated by clade A1 infection. PLoS One 4:e6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Migueles, S. A., et al. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. D., et al. 2008. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity 28:710-722. [DOI] [PubMed] [Google Scholar]
- 35.Neilson, J. R., et al. 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 73:4393-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peters, H. O., et al. 2008. An integrative bioinformatic approach for studying escape mutations in human immunodeficiency virus type 1 gag in the Pumwani sex worker cohort. J. Virol. 82:1980-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Precopio, M. L., et al. 2007. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J. Exp. Med. 204:1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price, D. A., et al. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. U. S. A. 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roederer, M., and R. A. Koup. 2003. Optimized determination of T cell epitope responses. J. Immunol. Methods 274:221-228. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz, J. E., et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 41.Sekaly, R. P. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 205:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin, H., and E. J. Wherry. 2007. CD8 T cell dysfunction during chronic viral infection. Curr. Opin. Immunol. 19:408-415. [DOI] [PubMed] [Google Scholar]
- 43.Streeck, H., et al. 2008. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med. 5:e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Streeck, H., N. Frahm, and B. D. Walker. 2009. The role of IFN-gamma Elispot assay in HIV vaccine research. Nat. Protoc. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 45.Valentine, L. E., et al. 2008. Recognition of escape variants in ELISPOT does not always predict CD8+ T-cell recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J. Virol. 82:575-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wherry, E. J., J. N. Blattman, K. Murali-Krishna, R. van der Most, and R. Ahmed. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, O. O. 2003. Will we be able to ‘spot’ an effective HIV-1 vaccine? Trends Immunol. 24:67-72. [DOI] [PubMed] [Google Scholar]
- 48.Zimmerli, S. C., et al. 2005. HIV-1-specific IFN-gamma/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc. Natl. Acad. Sci. U. S. A. 102:7239-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]