Abstract
mRNA for glutathione peroxidase 1 (GPx1) is subject to cytoplasmic nonsense-mediated decay (NMD) when the UGA selenocysteine (Sec) codon is recognized as nonsense. Here, we demonstrate by moving the sole intron of the GPx1 gene that either the Sec codon or a TAA codon in its place elicits NMD when located ≥59 bp but not ≤43 bp upstream of the intron. Therefore, the exon–exon junction of GPx1 mRNA positions the boundary between nonsense codons that do and do not elicit NMD, as has been shown for the 3′-most junctions of mRNAs subject to nucleus-associated NMD. We also demonstrate by using a regulatable promoter to drive GPx1 gene expression that cytoplasmic NMD is characteristic of steady-state mRNA, in contrast to nucleus-associated NMD. These findings clarify the mechanistic relationship between cytoplasmic and nucleus-associated NMD and offer the first demonstration that nuclear introns can influence cytoplasmic NMD. Finally, by analyzing hybrid GPx1 genes, we disprove the idea that the cellular site of NMD is determined by the efficiency of translation initiation.
Keywords: introns/mRNP/nonsense-mediated mRNA decay/selenoprotein transcripts/translation
Introduction
Nonsense-mediated decay (NMD), also called mRNA surveillance, typifies all cells of all organisms that have been examined (reviewed in Peltz et al., 1994; Maquat, 1995, 1996; Ruiz-Echevarria and Peltz, 1996; Li and Wilkinson, 1998; Culbertson, 1999; Hentze and Kulozik, 1999). The pathway probably evolved to eliminate the production of nonsense-containing RNAs that are the consequence of routine abnormalities in gene expression, such as aberrant or inefficient pre-mRNA splicing and non-productive somatic cell rearrangements characteristic of the immunoglobulin and T-cell receptor genes. For reasons unknown, some mammalian mRNAs are subject to NMD exclusively in the cytoplasm, while the majority of mammalian mRNAs appear to be subject to NMD exclusively prior to release from an association with nuclei (reviewed in Maquat, 1995, 1996; Li and Wilkinson, 1998; Hentze and Kulozik, 1999; Hilleren and Parker, 1999; Sun and Maquat, 2000). Studies of nucleus-associated NMD indicate that components of the NMD pathway survey all translated mRNAs in order to distinguish those that prematurely terminate translation from those that do not. Generally, if translation terminates >50–55 nucleotides upstream of the 3′-most exon–exon junction, then the mRNA is subject to NMD (Cheng et al., 1994; Nagy and Maquat, 1998; Thermann et al., 1998; Zhang et al., 1998a,b). This rule for termination codon position within intron-containing genes explains why termination codons that allow for the production of full-length proteins usually do not elicit NMD: most reside within the final exon, and for those that reside within either the penultimate or third-to-last exon, residence is <50 nucleotides upstream of the 3′-most exon–exon junction in 98% of cases (Nagy and Maquat, 1998). The finding for T-cell receptor-β transcripts that NMD is elicited by a nonsense codon located as close as eight nucleotides upstream of an unnatural 3′-most exon–exon junction (Carter et al., 1996) may provide an exception to the rule or reflect the presence of an intron-independent destabilizing element located downstream of the junction. Models accounting for the role of introns in nucleus-associated NMD envisage the process of splicing leaving one or more proteins at or near exon–exon junctions, which, as constituents of mRNP, can influence the process of translation termination at a point when the mRNA has yet to be released from an association with nuclei into the cytoplasm (Cheng et al., 1994; Carter et al., 1996; Thermann et al., 1998; Zhang et al., 1998a,b; Le Hir et al., 2000). The finding that a fraction of nucleus-associated mRNA escapes NMD, is exported to the cytoplasm and is immune to further NMD (Takeshita et al., 1984; Urlaub et al., 1989; Baserga and Benz, 1992; Cheng and Maquat, 1993; Belgrader et al., 1993; Cheng et al., 1994; Carter et al., 1995) suggests that the resulting cytoplasmic mRNA can manifest a change in mRNP structure, translation, or both that no longer supports NMD.
The observation that NMD can take place at two distinct sites in mammalian cells raises the possibility that there are site-specific mechanistic differences. Possible differences include the influence of introns and the kinetics of decay: whether or not cytoplasmic NMD, like nucleus-associated NMD, is influenced by the position of introns or restricted to newly synthesized mRNA [i.e. mRNA during the first round(s) of translation] remains to be investigated. To date, three cellular mRNAs have been shown to be subject to cytoplasmic NMD. One is human β-globin mRNA produced in erythroid cells such as the bone marrow of patients with β°-thalassemia (Maquat et al., 1981), the bone marrow and spleen of phenylhydrazine-treated mice transgenic for various human β°-thalassemic alleles (Lim et al., 1989, 1992; Lim and Maquat, 1992) and mouse erythroleukemia (MEL) cells stably transfected with various human β°-thalassemic alleles (S.Sekularac, Y.Wang, M.Antoniou and L.E.Maquat, unpublished data). Another encodes adenine phosphoribosyltransferase (APRT) and provides the only reported example of an mRNA that is subject to cytoplasmic as well as nucleus-associated NMD (Kessler and Chasin, 1996). The last encodes glutathione peroxidase 1 (GPx1; Moriarty et al., 1997, 1998; Sun et al., 1998).
The finding that nonsense codons within the final exon of the β-globin gene result in a dominantly inherited form of the usually recessive disease β-thalassemia because of a failure to reduce mRNA abundance (Kazazian et al., 1992; Hall and Thein, 1994) is certainly consistent with a role for introns. However, the lack of experimental proof for β-globin or any other mRNA subject to cytoplasmic NMD provides an impetus for new studies. Notably, a demonstration of intron function in cytoplasmic NMD would provide the first explicit evidence for a link in mammalian cells between nuclear introns and cytoplasmic mRNA decay; while we and others have argued that nonsense codon recognition in nucleus-associated NMD is mediated by cytoplasmic ribosomes during mRNA export (Cheng et al., 1994; Thermann et al., 1998; Zhang et al., 1998a,b), the possibility that nonsense codon recognition takes place in the nucleoplasm has yet to be discounted experimentally and, in fact, is favored by a number of groups (see for example Dietz and Kendzior, 1994; Aoufouchi et al., 1996; Li and Wilkinson, 1998; Brogna, 1999; Gersappe and Pintel, 1999). Studies of erythroid cell β-globin mRNA indicate that cytoplasmic NMD (i) initiates at or near the mRNA 5′ end independently of poly(A) shortening (Lim and Maquat, 1992; S.Sekularac, Y.Wang, M.Antoniou and L.E.Maquat, unpublished data), as has been shown for NMD in Saccharomyces cerevisiae (Muhlrad and Parker, 1994), and (ii) appears to typify steady-state mRNA (Maquat et al., 1981; Lim et al., 1992). The second conclusion was based on the use of actinomycin D to block transcription in cells from either β°-thalassemic patients or transgenic mice and, if correct, specifies a very important difference between nucleus-associated and cytoplasmic NMD. Other studies of cyto plasmic β-globin mRNA produced from a rabbit β-globin gene under the control of the transiently inducible c-fos promoter demonstrated that NMD decreases mRNA half-life from >24 h to 72 min, but did not determine whether the target is steady-state mRNA (Shyu et al., 1991).
Studies of APRT mRNA have not assayed for a role of intron position in cytoplasmic NMD. While the cytoplasmic NMD of APRT was not restricted to newly synthesized (i.e. newly exported) mRNA, it was, for reasons unknown, blocked by actinomycin D (Kessler and Chasin, 1996). Considering that there would be significant differences between a decay mechanism restricted to newly synthesized mRNA and one that requires mRNA export but otherwise discounts mRNA age, additional studies become important. In view of the findings with APRT mRNA, these studies should employ a method that obviates the need to block transcription using a global effector of cellular metabolism.
We have shown recently using rat liver and cultured cell lines that GPx1 mRNA is a natural substrate for cytoplasmic NMD (Moriarty et al., 1998). Increasing the efficiency with which the selenocysteine (Sec) UGA codon is recognized as nonsense either by lowering the intracellular selenium concentration, which reduces the level of charged selenocysteyl-tRNA (Hatfield et al., 1991), or by changing the Sec codon to nonsense (UAA) increases the efficiency of NMD; in contrast, converting the Sec codon to one for cysteine (UGC) abolishes NMD (Moriarty et al., 1998). These findings indicate that half-life studies comparing the consequences of converting the UGA Sec codon to either UAA or UGC will determine whether the NMD of GPx1 mRNA is restricted to newly exported mRNA or characteristic of steady-state mRNA. An intron-less GPx1 gene fails to elicit NMD (Moriarty et al., 1998; Weiss and Sunde, 1998), indicating that at least one intron is required for cytoplasmic NMD, as has been shown for nucleus-associated NMD (Zhang et al., 1998b). However, whether or not the sole GPx1 intron functions to position the boundary between nonsense codons that do and do not elicit NMD remains to be established. The finding that moving the UGA Sec codon from within exon 1 to within exon 2 abolishes selenium-dependent regulation (Weiss and Sunde, 1998) is consistent with the intron serving as a boundary determinant. However, the recent report that not a single UGA codon generated at one of 11 positions throughout the GPx1-coding region reduced GPx1 mRNA abundance indicates that the rule established for intron position in nucleus-associated NMD does not apply to GPx1 transcripts (Wen et al., 1998). Therefore, experiments that specifically move the intron relative to the Sec codon become very important in order to investigate the mechanism of cytoplasmic NMD.
We show here that moving the sole intron of the GPx1 gene does move the boundary between nonsense codons that do and do not elicit NMD, indicating unequivocally for the first time that (i) intron position within nuclear pre-mRNA can influence translation termination directed by the resulting cytoplasmic mRNA and (ii) an exon–exon junction located >50–55 nucleotides downstream of a translation termination codon functions to elicit cytoplasmic NMD, as is the case for nucleus-associated NMD. We go on to show using a doxycycline-repressible promoter to drive GPx1 gene expression that the NMD of cytoplasmic GPx1 mRNA is not restricted to newly exported mRNA. Taken together, these data provide the first explicit evidence that an intron which is essentially confined to nuclei (Moriarty et al., 1998) can influence mRNA half-life in the cytoplasm by affecting NMD. The simplest interpretation of these data is that: (i) the process of GPx1 pre-mRNA splicing in the nucleus generates an mRNP structure at the sole exon–exon junction; (ii) at least some features of this structure are maintained during the process of export across the nuclear pore complex; and (iii) this structure influences the process of GPx1 mRNA translation termination in the cytoplasm regardless of the time after mRNA synthesis. Finally, we examine the attractive hypothesis that cytoplasmic NMD is characteristic of mRNAs that inefficiently initiate translation so that the first round of translation takes place after export to the cytoplasm, while nucleus-associated NMD is characteristic of mRNAs that efficiently initiate translation so that the first round of translation takes place before the mRNA is released from an association with nuclei into the cytoplasm. We find that (i) increasing the efficiency of GPx1 mRNA translation initiation by inserting the poly(UC) sequence from ornithine decarboxylase mRNA or (ii) replacing the entire 5′-untranslated region (UTR) and beginning coding region with corresponding sequences from an mRNA known to be subject to nucleus-associated NMD does not convert NMD from cytoplasmic to nucleus-associated. Therefore, neither the efficiency of translation initiation nor sequences confined to the 5′-UTR and beginning coding region are sufficient to dictate the cellular site of NMD.
Results
The TGA Sec codon elicits NMD when located ≥59 bp but not ≤43 bp upstream of the GPx1 intron
Since GPx1 mRNA falls into the unusual and generally uncharacterized class of mammalian mRNAs that are subject to NMD after export to the cytoplasm, we first aimed to determine whether the sole intron of the GPx1 gene functions to position the boundary between nonsense codons that do and do not elicit NMD. Normally, the rat GPx1 gene harbors a single TGA Sec codon at position 46 within exon 1, a termination codon at position 201 within exon 2, and a single intervening intron (Figure 1). Previously, we changed the TGA Sec codon to either a TAA termination codon, which is almost always recognized as nonsense, or a TGC cysteine codon, which is almost never recognized as nonsense, and demonstrated that (i) the Sec codon directs the premature termination of translation and, as a consequence, NMD some of the time regardless of the intracellular selenium concentration but less frequently at higher selenium concentrations and (ii) the TAA codon reduces mRNA abundance more efficiently than the TGA codon regardless of the selenium concentration (Moriarty et al., 1998). More specifically, the TAA codon reduces mRNA abundance ∼2- to 3-fold, while the TGA codon reduces mRNA abundance ∼2-fold under selenium-deficient conditions (Moriarty et al., 1998). We concluded that the NMD of GPx1 mRNA is inefficient relative to the NMD of many other mRNAs, and the TAA codon elicits NMD more efficiently than the TGA codon because the TGA codon is recognized as nonsense only when it fails to encode Sec.
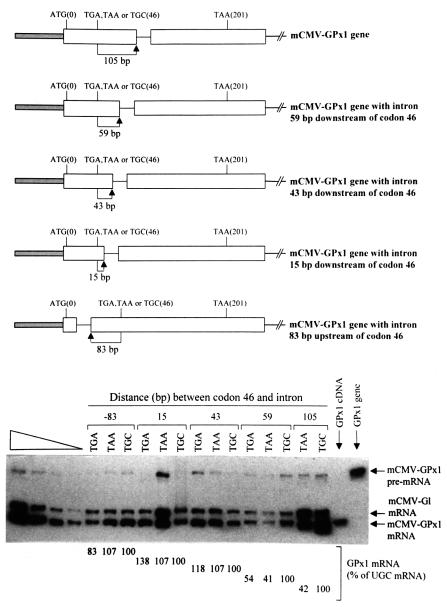
Fig. 1. Nonsense codons within the rat GPx1 gene elicit NMD when located ≥59 bp but not ≤43 bp upstream of the intron. Top: structures of the various GPx1 alleles, each of which is driven by the mCMV promoter. ATG(0) and TAA(201) specify the normal initiation and termination codons, and TGA, TAA or TGC(46) indicate the sequence at position 46, which is normally the Sec (TGA) codon. Bottom: RT–PCR analysis of the level of GPx1 mRNA as a function of intron position. NIH 3T3 cells were transiently transfected with the specified GPx1 test allele and the β-globin (Gl) reference allele. Total RNA was purified (Zhang et al., 1998b) and analyzed by RT–PCR as described (Moriarty et al., 1997, 1998). Notably, endogenous NIH 3T3 transcripts were not detected. For each intron position, which is specified as the distance in base pairs between codon 46 and the intron, the level of GPx1 mRNA was normalized to the level of Gl mRNA and is presented as a percentage of GPx1(UGC) mRNA, which was defined as 100. Results are the average of three independently performed experiments and did not differ by >8% except for the construct in which the TGA codon resided 15 bp upstream of the intron, where the maximum difference was 38% of the corresponding TGC construct. The left-most four lanes consist of serial dilutions of RNA, which were used to establish that there is a linear relationship between the amount of input RNA and the amount of each RT–PCR product. The right-most two lanes provide size standards for the products of GPx1 mRNA and GPx1 pre-mRNA.
For the present study, we generated derivatives of pmCMV-GPx1(TGA), pmCMV-GPx1(TAA) and pmCMV- GPx1(TGC) (Moriarty et al., 1997, 1998; the nature of the Sec codon at position 46 is specified in parentheses), in which each GPx1 allele is driven by the mouse cytomegalovirus (mCMV) promoter. The intron of the derivatives was moved from its usual position 105 bp downstream of the Sec codon to 59, 43 or 15 bp downstream of the Sec codon or to 83 bp upstream of the Sec codon (Figure 1). NIH 3T3 cells were transiently transfected with each of the constructs and a reference pmCMV-Gl construct (Zhang et al., 1998a), in which the β-globin (Gl) gene is driven similarly by the mCMV promoter. Total cell RNA was isolated, and RNA produced from the reference gene was used to control for variations in the efficiencies of cell transfection and RNA recovery.
Relative to constructs harboring the TGC cysteine codon, both the TGA Sec codon and the TAA nonsense codon failed to elicit NMD when the intron was moved either 83 bp upstream or as close as 15 or 43 bp downstream (Figure 1). However, both codons elicited NMD when the intron was moved 59 bp downstream (Figure 1). Notably, the extent of NMD was comparable with the extent when the intron was located at its natural site, i.e. 105 bp downstream of the codons (Figure 1; data not shown for TGA), and the cellular site of NMD remained cytoplasmic (data not shown). Therefore, GPx1 mRNA is not an exception to the rule for termination codon position within intron-containing genes, which states that nonsense codons located >50–55 nucleotides upstream of the 3′-most exon–exon junction elicit NMD. These results are important because they define an intron as a cis-acting sequence that determines which nonsense codons elicit cytoplasmic NMD. Therefore, the role of introns in nucleus-associated and cytoplasmic NMD can now be viewed as similar. Additionally, given the controversy over the cellular site of nonsense codon recognition in nucleus-associated NMD, these studies of cytoplasmic NMD are the first to link the presence of an intron, which is detected by RT–PCR essentially exclusively in nuclei (Moriarty et al., 1998), to a mechanism that undoubtedly involves translation by cytoplasmic ribosomes. In so doing, they provide the first explicit evidence that nuclear events influence cytoplasmic events in NMD.
The NMD of GPx1 mRNA is not restricted to newly exported cytoplasmic mRNA
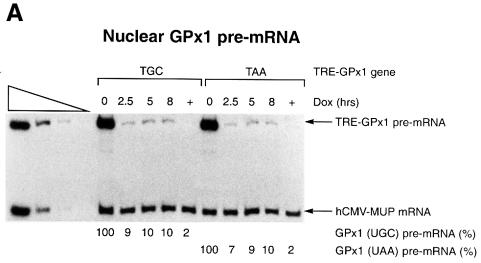
In order to determine whether the cytoplasmic NMD of GPx1 mRNA is restricted to newly exported mRNA, possibly analogously to the restriction of nucleus- associated NMD to newly synthesized mRNA, the mCMV promoter of the mCMV-GPx1 (TGC) and mCMV-GPx1(TAA) alleles was substituted with the regulatable TRE promoter (Clontech). The alleles were then transiently introduced into HeLa Tet-Off cells (Clontech), which support TRE-GPx1 gene transcription in the absence of doxycycline. Initially, HeLa Tet-Off cells were transiently transfected with a pTRE-GPx1 test plasmid and a phCMV-MUP reference plasmid in the absence of doxycycline so that the TRE-GPx1 plasmids produced GPx1 RNA. After 24 h, doxycycline was added to 1 µg/ml in order to shut off GPx1 RNA production, and nuclear and cytoplasmic RNAs were harvested at varying times thereafter. RT–PCR analysis of nuclear RNA indicated that the TRE-GPx1(TGC) and TRE- GPx1(TAA) alleles produced comparable amounts of nuclear pre-mRNA at each time point (Figure 2A), as expected given that NMD affects only cytoplasmic GPx1 mRNA (Moriarty et al., 1998). Transfections performed in the continual presence of 1 µg/ml doxycycline generated only a barely detectable level of TRE-GPx1 pre-mRNA, indicating that the block in transcription by doxycycline was nearly complete (Figure 2A, lanes marked ‘+’). Accordingly, a decrease in the level of nuclear TRE-GPx1 pre-mRNA to 7–9% the level at the time (0 h) of doxycycline addition was evident for both alleles by the earliest (2.5 h) time assayed after doxycycline addition (Figure 2A). All of these findings indicate that the rate and extent of the doxycycline-induced block in TRE-GPx1 gene transcription were sufficiently high to allow for measurements of cytoplasmic TRE-GPx1 mRNA half-life.
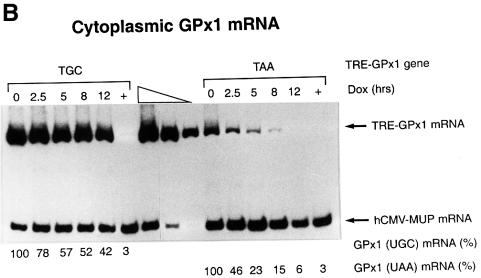
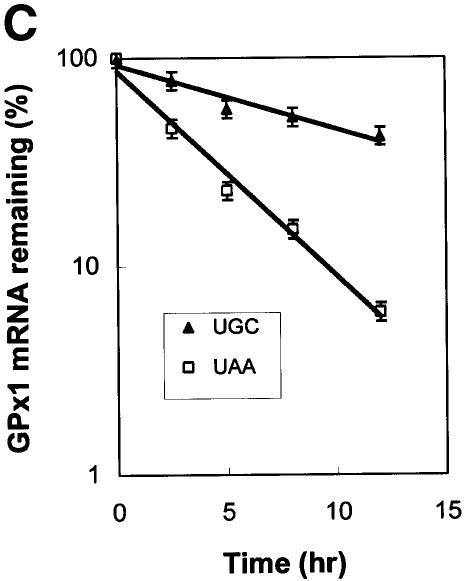
Fig. 2. NMD of cytoplasmic GPx1 mRNA is not restricted to newly exported mRNA. HeLa Tet-Off cells were transiently transfected with two plasmids: pTRE-GPx1(TGC) or pTRE-GPx1(TAA) test plasmid, in which the GPx1 gene harbors either a cysteine codon or a nonsense codon in place of the Sec codon and is under the control of the doxycycline-repressible TRE promoter, and the phCMV-MUP reference plasmid. After 24 h, doxycycline was added to shut off GPx1 gene transcription, and nuclear (A) and cytoplasmic (B) RNA was harvested at the specified times. Lanes labeled ‘+’ indicate that doxycycline was present throughout the experiment and provide a measurement of the amount of leaky transcription. Quantitations of nuclear GPx1 pre-mRNA demonstrate that the shut-off in transcription was evident by 2.5 h after doxycycline addition. For each lane, the level of GPx1 mRNA was normalized to the level of MUP mRNA and is presented as a percentage of the normalized level of either UGC- or UAA-containing mRNA, which was defined as 100. Half-lives are calculated to be 8–10 h for GPx1(UGC) mRNA and 2.5 h for GPx1(UAA) mRNA (C).
RT–PCR analysis of cytoplasmic RNA indicated that the level of mRNA from the TRE-GPx1(TAA) allele was reduced relative to that of mRNA from the TRE-GPx1(TGC) allele at the time (0 h) of doxycycline addition (Figure 2B), consistent with the TAA-induced NMD observed previously (Moriarty et al., 1998). Cytoplasmic mRNA from the TRE-GPx1(TGC) allele was degraded with a half-life of 8–10 h (Figure 2B and C), in keeping with previous estimates obtained using a block in transcription with actinomycin D (Bermano et al., 1996). In contrast, cytoplasmic mRNA from the TRE-GPx1(TAA) allele was degraded with a half-life of only 2.5 h and was degraded to completion by 12 h after the addition of doxycycline (Figure 2B and C). We conclude that the NMD of GPx1(UAA) mRNA (i) is not restricted to newly exported cytoplasmic mRNA and (ii) targets all GPx1(UAA) mRNA molecules with sufficient time. These findings contrast with those found for the NMD of nucleus-associated mRNA, which is restricted to newly synthesized mRNA and is characteristic of only a fraction of mRNA, the rest of which is resistant to NMD and exported to the cytoplasm where it manifests a normal half-life.
Neither the 5′-UTR nor the context of the translation initiation codon influences the cellular site of GPx1 NMD
An important as yet unresolved issue pertaining to mammalian NMD focuses on how some mRNAs are degraded in association with nuclei, either in the nucleoplasm or during transit through the nuclear pore complex, while other mRNAs are degraded after export to the cytoplasm is complete. According to one simplistic theory, mRNAs subject to nucleus-associated NMD are translated more efficiently than mRNAs subject to cytoplasmic NMD so as to have access to ribosomes, either nuclear or cytoplasmic, before release into the cytoplasm. Support for the possibility of translation during export is provided by the very large (35–40 kb) Balbiani ring granule of the dipteran Chironomus tentans, which invariably exits the nucleus and enters the cytoplasm 5′ end first and becomes associated with ribosomes while the 3′ end is still in the nucleoplasm (Mehlin et al., 1992). According to this theory, the NMD of GPx1 mRNA would be converted from cytoplasmic to nucleus-associated with a sufficiently large increase in the efficiency of translation initiation so that some of the mRNA is first translated while nucleus-associated. The validity of this theory could be assessed in two ways: (i) by inserting a sequence into GPx1 mRNA that increases the efficiency of translation initiation; and (ii) by substituting sequences of GPx1 mRNA that determine translation initiation efficiency with corresponding sequences of TPI mRNA, which is subject to nucleus-associated NMD.
Two sets of constructs were generated in order to test this theory. For the first set, a TC-rich translational enhancer from the 5′-UTR of the ornithine decarboxylase (ODC) gene (Grens and Scheffler, 1990) was utilized. When inserted into the 5′-UTR of a chloramphenicol acetyltransferase (CAT) gene, this enhancer increased the translation initiation efficiency of CAT mRNA 3-fold in both mammalian cells transfected with the gene and reticulocyte lysates programmed with transcripts from the gene (Pyronnet et al., 2000; S.Pyronnet and N.Sonenberg, personal communication). Notably, an effect of this magnitude is significantly larger than that which typically derives from altering the immediate sequence context of the AUG initiation codon (see for example Dasso et al., 1990; polysome-association data for mCMV-GPx1 variant transcripts are not shown), indicating that the degree to which initiation codon contexts match the Kozak consensus sequence (GCCGCCPuCCAUGG; reviewed in Kozak, 1999) cannot be used alone to predict translation initiation efficiencies in either intact cells or cell extracts. Therefore, mRNA sequence information alone cannot be used to confirm or eliminate the possibility that translation initiation efficiencies correlate with the cellular site of NMD.
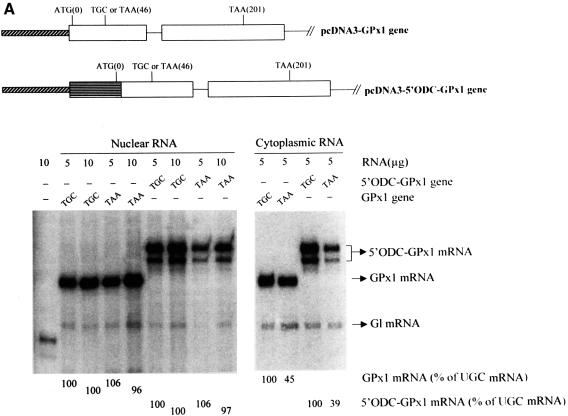
Initially, the ODC TC-rich sequence and first four codons of the CAT gene (referred to simply as the ODC sequence) were used to replace the 5′-UTR and the first four codons of the GPx1(TGC) and GPx1(TAA) genes to generate pcDNA-5′ ODC-GPx1(TGC) and pcDNA-5′ ODC-GPx1(TAA), respectively. In order to measure the effect on GPx1 mRNA translation initiation in reticulocyte lysates, intron-less derivatives were then made. Consistent with studies of CAT mRNA translation, GPx1(UGC) mRNA harboring the ODC sequence was translated in reticulocyte lysates ∼2-fold more efficiently than mRNA lacking the ODC sequence (Figure 3). Next, Cos cells were transiently transfected with the intron-containing pcDNA-5′ ODC-GPx1 constructs. According to the results of RNase mapping, the ODC sequence was of little consequence to either the extent or cellular site of NMD: in either the presence or absence of the sequence, the level of nuclear GPx1 (UAA) mRNA was on average 100% that of nuclear GPx1 (UGC) mRNA, and the level of cytoplasmic GPx1 (UAA) mRNA was on average 42% that of cytoplasmic GPx1 (UGC) mRNA (Figure 4A).
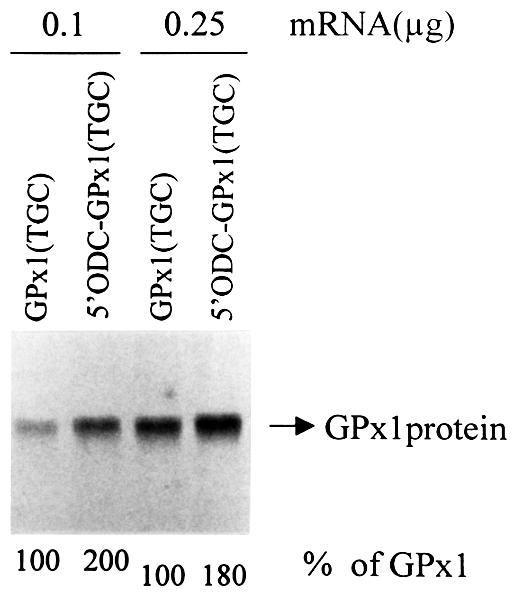
Fig. 3. Replacing the GPx1 5′-UTR and beginning of the coding region with the ODC TC-rich translational enhancer and beginning of the CAT coding region increases the efficiency of GPx1 mRNA translation initiation in reticulocyte lysates ∼2-fold. Intron-less versions of pcDNA3-GPx1(TGC) and pcDNA-5′ ODC-GPx1(TGC) were transcribed in vitro using T7 RNA polymerase, and 0.1 or 0.25 µg of each transcript were translated in vitro in the presence of [35S]methionine. Protein was electrophoresed in an SDS–polyacrylamide gel. For each amount of transcript analyzed, the level of 5′ODC-GPx1(UGC) protein is presented as a percentage of GPx1(UGC) protein, which was defined as 100.
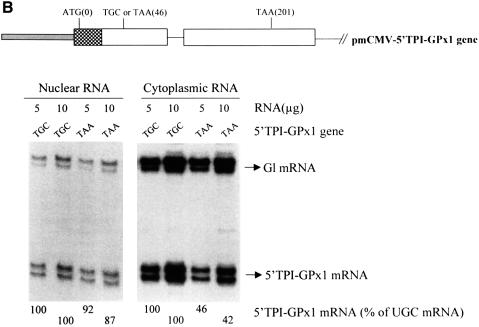
Fig. 4. Neither the extent nor cellular site of GPx1 NMD is affected by substituting the GPx1 5′-UTR and beginning of the coding region within the ODC translational enhancer and beginning of the CAT gene or the corresponding regions of the TPI gene. (A) Structures of the pcDNA3-GPx1 and pcDNA3-5′ODC-GPx1 genes. The diagonally striped bar represents the CMV promoter of pcDNA3 and the horizontally striped exonic region represents part of the 5′-UTR of ODC and the first four codons of the CAT gene. Cos cells were transiently transfected with the specified GPx1 test allele and the Gl reference allele. Nuclear and cytoplasmic RNA was purified (Zhang et al., 1998b), and 5 or 10 µg were analyzed by RNase mapping using uniformly 32P-labeled antisense RNAs (see Materials and methods). Cytoplasmic RNA was exposed to X-ray film for a shorter time than nuclear RNA. The analysis of untransfected cells demonstrated that RNase-resistant 32P measured either test or reference RNAs. The finding that 10 µg of RNA protected ∼2-fold more 32P than 5 µg of RNA demonstrated that the analysis was quantitative. The level of each GPx1 and 5′ODC-GPx1 mRNA was normalized to the level of Gl mRNA and is presented as a percentage of the level of the corresponding UGC-containing mRNA, which was defined as 100. Results are the average of three independently performed experiments and did not differ by >7%. (B) Structure of the 5′TPI-GPx1 gene. The gray bar/attached checkered exonic region represent, respectively, the mCMV promoter/5′-UTR and first 22 codons of the TPI gene. RNase mapping and quantitation of the level of 5′TPI-GPx1 mRNA were as described for (A). For both (A) and (B), antisense RNAs that appear as RNase-resistant doublets reflect incomplete or excessive RNase digestion rather than structural heterogeneity of the mRNA under analysis.
Since a 2- to 3-fold increase in the efficiency of translation initiation may not be adequate to convert the NMD of GPx1 mRNA from cytoplasmic to nucleus-associated, a second set of GPx1 constructs was generated utilizing sequences from the 5′ end of the TPI gene, which produces mRNA subject to nucleus-associated NMD. The 5′-UTR and first 17 codons of mCMV-GPx1(TGC) and pmCMV-GPx1(TAA) were replaced with the 5′-UTR and first 22 codons of the TPI gene to generate pmCMV-5′ TPI-GPx1(TGC) and pmCMV-5′ TPI-GPx1(TAA), respectively. The results of RNase mapping indicated that, again, there was no appreciable effect on either the extent or cellular site of NMD: the TPI sequence had no appreciable effect on the relative nuclear levels of UGC- and UAA-containing mRNAs and did not alter the degree of cytoplasmic NMD (Figure 4B). Therefore, sequences that determine the translation initiation efficiency of TPI mRNA, when used to replace the corresponding sequences of GPx1 mRNA, do not convert the cellular site of GPx1 NMD from cytoplasmic to nucleus-associated. We conclude that neither translation initiation efficiency nor some other feature of the 5′ end of mRNA determines the cellular site of NMD.
Discussion
Prior to these studies, little was known about the mechanistic relationship between nucleus-associated and cytoplasmic NMD in mammalian cells. The lack of knowledge was largely attributable to the paucity of cellular mRNAs known to be subject to cytoplasmic NMD. The discovery that GPx1 mRNA is a natural substrate of cytoplasmic NMD allowed for the demonstration that a dominant-negative version of hUpf1p abrogates the NMD of both GPx1 mRNA, which is strictly cytoplasmic, and β-globin mRNA expressed in non-erythroid cells, which is strictly nucleus-associated, indicating that this group 1 RNA helicase functions in NMD at both cellular sites (Sun et al., 1998). The finding that substituting the TPI or GPx1 promoters with the mCMV promoter did not interfere with the nucleus-associated NMD of TPI mRNA (Cheng et al., 1994) or the cytoplasmic NMD of GPx1 mRNA (Moriarty et al., 1998) indicated that the cellular site of NMD for these mRNAs was not dictated by the promoter. In search of additional comparisons between nucleus-associated and cytoplasmic NMD, we demonstrate here that the cytoplasmic NMD of GPx1 mRNA is dependent on intron position within the GPx1 gene (Figure 1). We find that a nonsense codon elicits NMD when located ≥59 but not ≤43 bp upstream of the sole intron of the GPx1 gene (Figures 1 and 5), consistent with the rule established in studies of nucleus-associated NMD that a nonsense codon must reside >50–55 bp upstream of the 3′-most intron in order to elicit NMD (Cheng et al., 1994; Nagy and Maquat, 1998; Thermann et al., 1998; Zhang et al., 1998a,b). However, we find that the cytoplasmic NMD of GPx1 mRNA is not restricted to newly synthesized mRNA, i.e. mRNA that has just entered the cytoplasm (Figure 2), in contrast to data indicating that nucleus-associated NMD is restricted to newly synthesized mRNA, i.e. mRNA prior to release from an association with nuclei into the cytoplasm (reviewed in Maquat, 1995; Li and Wilkinson, 1998; Hentze and Kulozik, 1999). Our finding that cytoplasmic NMD is not restricted to newly exported mRNA is consistent with previous studies of β-globin mRNA produced in erythroid cells (Lim et al., 1992), APRT mRNA (Kessler and Chasin, 1996), unspliced RNA of Rous sarcoma virus (Barker and Beemon, 1991) and a variety of transcripts that derive from naturally intron-less genes in S.cerevisiae (Zhang et al., 1997).
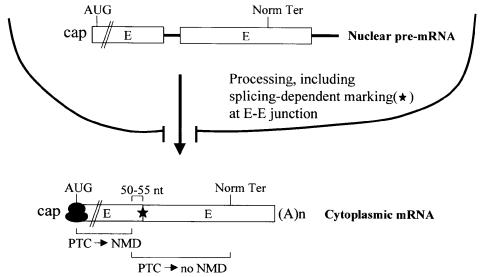
Fig. 5. A model for intron function in the NMD of cytoplasmic mRNA. Nuclear pre-mRNA that consists of at least two exons (E) and a single intervening intron (thick line) undergoes splicing and 3′ end formation, not necessarily in that order. The process of splicing leaves behind a mark of several proteins (star) at or near each E–E junction of product mRNA (Le Hir et al., 2000). If the resulting mRNP prematurely terminates translation >50–55 nucleotides upstream of the 3′-most E–E junction, then the mRNP will be subject to NMD. NMD takes place only after export to the cytoplasm is complete, probably because the first round of translation takes place only after export, the mRNP is first conducive to NMD only after export, or both.
As has been proposed for mRNAs subject to nucleus-associated NMD (reviewed in Maquat, 1995; Li and Wilkinson, 1998; Nagy and Maquat, 1998; Hentze and Kulozik, 1999; Hilleren and Parker, 1999; Le Hir et al., 2000), data presented here for an mRNA subject to cytoplasmic NMD suggest that the process of splicing influences mRNP structure in a way that influences NMD. In support of this idea, pre-mRNA splicing in HeLa cell nuclear extracts has been shown to result in the formation of a stable complex of proteins at or near the exon–exon junctions of spliced products that remain after spliceosome dissociation (Le Hir et al., 2000), and pre-mRNA splicing in Xenopus laevis oocytes has been shown to result in a change in mRNP structure that correlates with an increase in the efficiency of mRNA export to the cytoplasm (Luo and Reed, 1999). Despite similarity in the role of introns in nucleus-associated and cytoplasmic NMD, differences in the kinetics of NMD at the two cellular sites indicate that there must be differences in the kinetics of the splicing-mediated influence on mRNP structure at each site. In the case of nucleus-associated NMD, some type of mRNP remodeling that precludes further NMD is envisaged for the fraction of mRNA that is released into the cytoplasm (reviewed in Maquat, 1995), since release is accompanied by an association with polysomes and, therefore, a mechanism for nonsense codon recognition (Kessler and Chasin, 1996; Stephenson and Maquat, 1996). In contrast, cytoplasmic NMD may simply (i) fail to be translated until completely cytoplasmic, (ii) acquire a structure that supports NMD only after export to the cytoplasm, or (iii) both.
The cellular location of cytoplasmic NMD provides a strong indication that nonsense codon recognition involves cytoplasmic ribosomes. A comparably strong case for the involvement of cytoplasmic ribosomes in nucleus-associated NMD is difficult to make: arguments based on the findings that nucleus-associated NMD is abrogated by a variety of translational effectors consisting of (i) ribosome-binding inhibitors of translation (Qian et al., 1993; Menon and Neufeld, 1994; Carter et al., 1995), (ii) suppressor tRNA (Belgrader et al., 1993; Li et al., 1997), (iii) secondary structure in the 5′-UTR that blocks translation initiation (Belgrader et al., 1993; Thermann et al., 1998), (iv) poliovirus infection, which inactivates cap-dependent translation (Carter et al., 1995), and (v) translation reinitiation downstream of and in frame with a nonsense codon (Zhang and Maquat, 1997) are significantly confounded by the cellular location of NMD. Since the possibility that nonsense codon recognition in nucleus-associated NMD involves nuclear ribosomes cannot be discounted (for a review see Li and Wilkinson, 1998), data presented provide the strongest suggestion that introns can influence the termination of cytoplasmic translation. Notably, recent studies of S.cerevisiae indicating that NMD requires mRNP marking by the hnRNP-like protein Hrp1/Nab4 illustrate a different way in which nuclear processes can influence cytoplasmic mRNA half-life by affecting NMD (González et al., 2000).
Studies presented here rule out the possibility that either the translation initiation efficiency or some other feature of an mRNA 5′ end dictates the cellular site of NMD (Figure 4): the NMD of GPx1 mRNA remained cytoplasmic when the 5′-UTR and first 17 codons were replaced with the corresponding region of TPI mRNA, which is subject to nucleus-associated NMD. Along similar lines, we have found that a decrease in the translation initiation efficiency of TPI mRNA that was mediated by insertion of a hairpin into the 5′-UTR of the mRNA was of no consequence to the cellular site of NMD (Belgrader et al., 1993). Notably, however, the decrease in efficiency severely reduced the extent of NMD, consistent with the restriction of NMD to the fraction of time the mRNA was nucleus-associated (Belgrader et al., 1993). Future studies will aim to understand why there are two mammalian cell sites for NMD.
Materials and methods
Plasmid constructions
Moving the GPx1 intron. A 721 bp SacII–BsaI fragment comprising 584 bp of the GPx1 coding region and 137 bp of the GPx1 3′-UTR was purified from pmCMV-GPx1(TGA) intron-less (Moriarty et al., 1997) and inserted into the SacII and SmaI sites of pBluescript-KS–. Prior to insertion, the BsaI end was made blunt with Klenow and dNTPs. The pBluescript-GPx1 construct was mutagenized (Kunkel et al., 1987) using one of two mutagenic antisense primers, each of which creates an NcoI site. An NcoI site was generated downstream of GPx1 codon 46 using the antisense primer 5′-CCCAGACGCCCATGGAGATCATTC-3′ (where underlined nucleotides specify the NcoI site and italicized nucleotides are mutagenic) or the antisense primer 5′-CACCAGGCCCCATGGCCCCAGACG-3′. Next, the 252 bp XmaI–DraIII fragment extending from GPx1 exon 1 to GPx1 exon 2 of each mutagenized plasmid was used to replace the corresponding fragment of pmCMV-GPx1(TGA) (Moriarty et al., 1997, 1998). The 216 bp intron of GPx1 was then inserted into one of the NcoI sites, the unique XmaI site or the unique BssHII site in order to generate intron insertions located, respectively, 59, 43 or 15 bp downstream of codon 46 or 83 bp upstream of codon 46. Each site maintains a consensus intron–exon junction when Klenow filled and ligated to the 216 bp intron, and the insertion process resulted in the addition of six extra nucleotides (four from the filled insertion site and two from the ends of the PCR products). The 216 bp intron was generated by amplifying pmCMV-GPx1 using the sense primer 5′-GGTATGTGAGACGGGATGG-3′, the antisense primer 5′-CCTAGGGAAAGCAAAGAC-3′ and Vent polymerase (New England Biolabs). The blunt ends of the amplification product were then phosphorylated. In order to generate the intron insertion 59, 43 and 15 bp downstream of codon 46 within either pmCMV-GPx1(TAA) intron-less or pmCMV-GPx1(TGC) intron-less, the 468 bp XmaI–DraIII fragment that harbors the appropriately positioned intron was used to replace the corresponding fragment of pmCMV-GPx1(TAA) or pmCMV-GPx1(TCG). In order to generate the intron insertion 83 bp upstream of codon 46 within either pmCMV-GPx1(TAA) intron-less or pmCMV-GPx1(TGC) intron-less, the 248 bp SacII–BssHII fragment that harbors the intron at the BssHII site (which is regenerated after intron insertion) was used to replace the corresponding fragment of either pmCMV-GPx1(TAA) intron-less or pmCMV-GPx1(TGC) intron-less.
Construction of pTRE-GPx1(TAA or TGC). Two fragments, consisting of the 680 bp KpnI–DraIII fragment from pmCMV-GPx1(TGC) or pmCMV-GPx1(TAA) that extends from the 5′-UTR into exon 2 and was Klenow treated at the KpnI site, and the 420 bp DraIII–EcoRI fragment that extends from exon 2 into 3′-flanking DNA, were generated and inserted into the EcoRI- and Klenow-treated SacII sites of pTRE (Clontech) so that GPx1 gene expression was driven by the Tet-responsive (Tet-Off™) promoter.
Construction of pcDNA3-GPx1(TGC or TAA) and intron-less counterparts. The GPx1 gene was amplified as a 1.1 kbp fragment using the sense primer 5′-CCAAAAGCTTCTACAGTATGTCTGCTGC-3′ (where underlined nucleotides specify a HindIII site) and the antisense primer 5′-CCGTTCTAGAGGAAGTTTAGACTCG-3′ (where underlined nucleotides specify an XbaI site). The resulting PCR product, which extends from the 5′-UTR into 3′-flanking DNA, was cleaved with HindIII and XbaI and inserted into the HindIII and XbaI sites of pcDNA3 (Invitrogen). Subsequently, the 240 bp XbaI–BbsI fragment of vector DNA that includes the bovine growth hormone polyadenylation sequences was deleted. The 865 bp SacII–XbaI fragment extending from exon 1 into 3′-flanking DNA of pmCMV-GPx1(TGC) intron-less (Moriarty et al., 1997) was used to replace the corresponding fragment of pcDNA3-GPx1(TGC) in order to generate the intron-less counterpart.
Construction of pcDNA3-5′ODC-GPx1 (TGC or TAA) and intron-less counterparts. The GPx1 gene was amplified as a 1.07 kbp fragment using the sense primer 5′-GCTGGGATCCTCTCCGCGGTGGCACAG-3′ (where underlined nucleotides specify a BamHI site) and the same antisense primer as in the previous section. The resulting PCR product, which extends from codon 5 into 3′-flanking DNA, was cleaved with BamHI and XbaI and ligated to XbaI- and HindIII-cleaved pcDNA3 together with the 120 bp HindIII–Sau3AI fragment from the 5′-UTR through codon 4 of pcDNA3-5′ODC-CAT (a gift from S.Pyronnet and N.Sonenberg). The 240 bp XbaI–BbsI fragment of vector DNA that includes the bovine growth hormone polyadenylation sequences was then deleted. The intron-less counterpart was generated as described in the previous section.
Construction of pmCMV-5′TPI-GPx1 (TGC or TAA). The 650 bp BamHI–SacI fragment that includes the entire mCMV promoter and first 22 codons of TPI exon 1 was inserted in place of the 620 bp XbaI–BssHII fragment of pmCMV-(TGC) or (TAA) that includes the entire mCMV promoter and first 17 codons of the GPx1 gene. Prior to insertion, the 650 bp fragment and vectors were Klenow treated.
Cell transfections, tetracycline repression studies and RNA isolation
In experiments that did not involve repression, NIH 3T3 and Cos cells were maintained in minimal essential medium (Life Technologies) containing 10% fetal bovine serum (FBS) (Hyclone). NIH 3T3 cells (15 cm dish; 40–50% confluency) were transiently transfected with a pmCMV-GPx1 test plasmid (30 µg) and the pmCMV-Gl reference plasmid (10 µg) using calcium phosphate and harvested 48 h later as described (Moriarty et al., 1998). Cos cells (15 cm dish; 40–50% confluency) were transiently transfected with a pcDNA3-GPx1, pcDNA3-5′ODC-GPx1 or pmCMV-5′TPI-GPx1 test plasmid (30 µg) and the pmCMV-Gl reference plasmid (12 µg) and harvested as described for NIH 3T3 cells.
In experiments that involved repression in order to study the decay of GPx1(UAA) and GPx1(UGC) mRNAs, HeLa Tet-Off cells (Clontech) were maintained in α-minimal essential medium (MEM) (Life Technologies) supplemented with 10% FBS that lacked tetracycline (Clontech) and contained 100 µg/ml of G418. For transient transfection, a confluent 15 cm plate was split (1:5) 20 h prior to transfection using antibiotic-free α-MEM plus tetracycline-free 10% FBS. Cells were co-transfected for 5 h with phCMV-MUP (1.75 µg; Belgrader and Maquat, 1994) and the appropriate pTRE-GPx1 (5.25 µg) by using Lipofectamine Plus reagent (Life Technologies). After an additional 19 h, fresh α-MEM plus 10% FBS containing 1 µg/ml doxycycline (Sigma) was added in order to shut off GPx1 gene transcription. Cells were harvested at the specified times. Nuclear and cytoplasmic RNA were isolated as described (Zhang et al., 1998a).
RT–PCR analyses
For all RT–PCRs (Cheng and Maquat, 1993; Moriarty et al., 1998; Sun et al., 1998), cDNA was synthesized using 0.04–5.0 µg of total, nuclear or cytoplasmic RNA that had been treated with RQ1 DNase (1 U per 5 µg of RNA for 1 h at 37°C; Promega Corp) using reverse transcriptase (Superscript, Gibco) and random hexamers (Promega). Each PCR contained one-fifth of the reverse transcriptase reaction mixture, 0.12 mM each of the four deoxynucleotides, 4 µCi of [α-32P]dATP (3000 Ci/mmol; Amersham), 0.5 µM each of two primers and 2.5 U of Taq DNA polymerase (Promega). cDNAs of mCMV-GPx1 pre-mRNA and mCMV-GPx1 and mCMV-Gl mRNAs were amplified using a sense primer 5′-ACCACCGTAGAACGCAGATCG-3′, which corresponds to the common mCMV promoter region. cDNA of mCMV-GPx1 pre-mRNA was amplified using an antisense primer 5′-CCCGATGAGTCACCAGGAAG-3′, which corresponds to the GPx1 intron, and cDNA of mCMV-GPx1 mRNA was amplified using the antisense primer 5′-CTTCTCACCATTCACCTCGCACTT-3′, which corresponds to GPx1 exon 2. The antisense primer used to amplify cDNA of mCMV-Gl mRNA, 5′-CGGGGTGAAGCTCCTTGCCAAG-3′, corresponds to exon 3 of the mouse β-globin gene (Cheng and Maquat, 1993). Notably, GPx1 cDNA that derived from the endogenous NIH 3T3 cell gene was not amplified. Primers for the amplification of hCMV-MUP cDNA, which in some experiments was used in place of amplifying mCMV-Gl cDNA, were as previously described (Sun et al., 1998). One-tenth of each PCR was electrophoresed in a 4% denaturing polyacrylamide gel, and RT–PCR products were quantified using a PhosphorImager and ImageQuant software (Molecular Dynamics).
In vitro transcription and translation
pcDNA3-GPx1 (TGC) intron-less and pcDNA3-5′ ODC-GPx1 (TGC) intron-less DNAs were linearized with XbaI. Capped transcripts were generated in vitro using T7 RNA polymerase (Promega) and quantitated by measuring the OD260 and staining with ethidium bromide after electrophoresis in agarose. Varying amounts of each transcript (0.1 and 0.25 µg) were translated in vitro using rabbit reticulocyte lysates (Promega) as specified by the manufacturer in the presence of [35S]methionine (1000 Ci/mmol; Amersham). Translation products were electrophoresed in SDS–10% acrylamide, the gel was dried and protein was quantitated by phosphoimaging and autoradiography.
In vitro transcription and RNase protection assays
To generate a template for the synthesis of antisense Gl RNA, the Klenow-filled 370 bp BbsI–XmnI fragment that extends from intron 1 to intron 2 of the pmCMV-Gl gene was inserted into the HincII site of pGEM-3Z (Promega). The resulting plasmid was subsequently linearized with AccI (Figure 4A) or HindIII (Figure 4B). To generate a template for the synthesis of antisense ODC-GPx1 RNA, the 400 bp MscI–EcoRI fragment that extends from the middle of the ODC poly(TC) sequence into GPx1 intron 1 of the ODC-GPx1 gene was inserted into the EcoRI and HincII sites of pGEM-3Z. The resulting plasmid was then linearized with NcoI. In vitro transcription (Ambion) was performed in 20 µl using 1 µg of linearized plasmid DNA, 40 µCi of [32P]UTP (3000 µCi/mmol; Amersham), 2 µl of 100 µM UTP, 1 µl each of 10 mM ATP, CTP and GTP, and 20 U of RNA polymerase.
RNase protection assays were performed with 5 or 10 µg of nuclear or cytoplasmic RNA using the RPAIII kit (Ambion) as specified by the manufacturer. Briefly, cellular RNA was hybridized to Gl and GPx1 in vitro-synthesized antisense RNAs (∼8 × 105 c.p.m.; at least a 5-fold molar excess relative to the complementary cellular RNA) in 10 µl overnight at 42°C and subsequently treated with RNase A (50 U/ml) and RNase T1 (200 U/ml) for 1 h at 37°C. Protected fragments were precipitated, electrophoresed in 5% denaturing polyacrylamide and quantitated by phosphoimaging and autoradiography after the gel was dried.
Acknowledgments
Acknowledgements
The authors thank Xiaojie Li for technical assistance, members of the Maquat laboratory for helpful discussions, Stéphane Pyronnet and Nahum Sonenberg for ODC sequences and communicating unpublished data, and Yasuhito Ishigaki for comments on the manuscript. P.M.M. is grateful to C. Channa Reddy for salary support while working in the Maquat laboratory. This work was supported by Public Health Service Research grants GM52822 and GM59614 to L.E.M.
References
- Aoufouchi S., Yélamos,J. and Milstein,C. (1996) Nonsense mutations inhibit RNA splicing in a cell free system: recognition of mutant codon is independent of protein synthesis. Cell, 85, 415–422. [DOI] [PubMed] [Google Scholar]
- Barker G.F. and Beemon,K. (1991) Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol. Cell. Biol., 11, 2760–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S.J. and Benz,E.J.,Jr (1992) β-Globin nonsense mutation: deficient accumulation of mRNA occurs despite normal cytoplasmic stability. Proc. Natl Acad. Sci. USA, 89, 2935–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P. and Maquat,L.E. (1994) Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol. Cell. Biol., 14, 6326–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Cheng,J. and Maquat,L.E. (1993) Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc. Natl Acad. Sci. USA, 90, 482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermano G., Arthur,J.R. and Hesketh,J.E. (1996) Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett., 387, 157–160. [DOI] [PubMed] [Google Scholar]
- Brogna S. (1999) Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA, 5, 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M.S., Doskow,J., Morris,P., Li,S., Nhim,R.P., Sandstedt,S. and Wilkinson,M.F. (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem., 270, 28995–29003. [DOI] [PubMed] [Google Scholar]
- Carter M.S., Li.S. and Wilkinson,M.F. (1996) A splicing-dependent regulatory mechanism that detects translation signals. EMBO J., 15, 5965–5975. [PMC free article] [PubMed] [Google Scholar]
- Cheng J. and Maquat,L.E. (1993) Nonsense codons can reduce the abundance of nuclear mRNA without affecting the abundance of pre-mRNA or half-life of cytoplasmic mRNA. Mol. Cell. Biol., 13, 1892–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Belgrader,P., Zhou,X. and Maquat,L.E. (1994) Introns are cis effectors of the nonsense-codon-mediated reduction in nuclear mRNA abundance. Mol. Cell. Biol., 14, 6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M.R. (1999) RNA surveillance: unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet., 15, 74–80. [DOI] [PubMed] [Google Scholar]
- Dasso M.C., Milburn,S.C., Hershey,J.W.B. and Jackson,R.J. (1990) Selection of the 5′-proximal translation initiation site is influenced by mRNA and eIF-2 concentrations. Eur. J. Biochem., 187, 361–371. [DOI] [PubMed] [Google Scholar]
- Dietz H.C. and Kendzior,R.J.,Jr (1994) Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nature Genet., 8, 183–188. [DOI] [PubMed] [Google Scholar]
- Gersappe A. and Pintel,D.J. (1999) A premature termination codon interferes with the nuclear function of an exon splicing enhancer in an open reading frame-dependent manner. Mol. Cell. Biol., 19, 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González C.I., Ruiz-Echevarría,M.J., Vasudevan,S., Henry,M.F. and Peltz,S.W. (2000) The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol. Cell, 5, 489–499. [DOI] [PubMed] [Google Scholar]
- Grens A. and Scheffler,I.E. (1990) The 5′- and 3′-untranslated regions of ornithine decarboxylase mRNA affect the translational efficiency. J. Biol. Chem., 265, 11810–11816. [PubMed] [Google Scholar]
- Hall G.W. and Thein,S. (1994) Nonsense codon mutations in the terminal exon of the β-globin gene are not associated with a reduction in β-mRNA accumulation: a mechanism for the phenotype of dominant β-thalassemia. Blood, 83, 2031–2037. [PubMed] [Google Scholar]
- Hatfield D., Lee,B.J., Hampton,L. and Diamond,A.M. (1991) Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res., 19, 939–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. and Kulozik,A.E. (1999) A perfect mRNA: RNA surveillance and nonsense-mediated decay. Cell, 96, 307–310. [DOI] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H.H. Jr, Dowling,C.E., Hurwitz,R.L., Coleman,M., Stopeck,A. and Adams,J.G.,III (1992) Dominant thalassemia-like phenotypes associated with mutations in exon 3 of the β-globin gene. Blood, 79, 3014–3018. [PubMed] [Google Scholar]
- Kessler O. and Chasin,L.A. (1996) Effects of nonsense mutations on nuclear and cytoplasmic adenine phosphoribosyltransferase RNA. Mol. Cell. Biol., 16, 4426–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A., Roberts,J.D. and Zakour,R.A. (1987) Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol., 154, 367–382. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Moore,M.M. and Maquat,L.E. (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon–exon junctions. Genes Dev., 14, 1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Li S. and Wilkinson,M.F. (1998) Nonsense surveillance in lymphocytes? Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- Li S., Leonard,D. and Wilkinson,M.F. (1997) T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: a nuclear-associated translation-like mechanism. J. Exp. Med., 185, 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.-K. and Maquat,L.E. (1992) Human β-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a 5′ cap-like structure at the 5′ termini. EMBO J., 11, 3271–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.-K., Mullins,J.J., Chen,C.M., Gross,K.W. and Maquat,L.E. (1989) Novel metabolism of several β°-thalassemic β-globin RNAs in the erythroid tissues of transgenic mice. EMBO J., 8, 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S.-K., Sigmund,C., Gross,K.W. and Maquat,L.E. (1992) Nonsense codons in human β-globin mRNA result in the production of mRNA degradation products. Mol. Cell. Biol., 12, 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou M.J. and Reed,R. (1999) Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA, 96, 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. (1995) When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA, 1, 453–465. [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E. (1996) Defects in RNA splicing and the consequence of shortened translational reading frames. Am. J. Hum. Genet., 59, 279–286. [PMC free article] [PubMed] [Google Scholar]
- Maquat L.E., Kinniburgh,A.J., Rachmilewitz,E.A. and Ross,J. (1981) Unstable β-globin mRNA in mRNA-deficient β°-thalassemia. Cell, 27, 543–553. [DOI] [PubMed] [Google Scholar]
- Mehlin H., Daneholt,B. and Skoglund,U. (1992) Translocation of a specific pre-messenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell, 69, 605–613. [DOI] [PubMed] [Google Scholar]
- Menon K.P. and Neufeld,E.F. (1994) Evidence for degradation of mRNA encoding α-l-iduronidase in Hurler fibroblasts with premature termination alleles. Cell Mol. Biol., 40, 999–1005. [PubMed] [Google Scholar]
- Moriarty P.M., Reddy,C.C. and Maquat,L.E. (1997) The presence of an intron within the rat gene for selenium-dependent glutathione peroxidase 1 is not required to protect nuclear RNA from UGA-mediated decay. RNA, 3, 1369–1373. [PMC free article] [PubMed] [Google Scholar]
- Moriarty P.M., Reddy,C.C. and Maquat,L.E. (1998) Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol., 18, 2932–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D. and Parker,R. (1994) Premature translational termination triggers mRNA decapping. Nature, 370, 578–581. [DOI] [PubMed] [Google Scholar]
- Nagy E. and Maquat,L.E. (1998) A rule for termination-codon position within intron-containing genes: when nonsense affects mRNA abundance. Trends Biochem. Sci., 23, 198–199. [DOI] [PubMed] [Google Scholar]
- Peltz S.W., Feng,H., Welch,E. and Jacobson,A. (1994) Nonsense-mediated decay in yeast. Prog. Nucleic Acid Res. Mol. Biol., 47, 271–298. [DOI] [PubMed] [Google Scholar]
- Pyronnet S., Pradayrol,L. and Sonenberg,N. (2000) A cell cycle-dependent internal ribosome entry site. Mol. Cell, 5, 607–616. [DOI] [PubMed] [Google Scholar]
- Qian L.L., Theodor,L., Carter,M., Vu,N., Sasaki,A.W. and Wilkinson,M.F. (1993) T cell receptor-β mRNA splicing: regulation of unusual splicing intermediates. Mol. Cell. Biol., 13, 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria M.J., Czaplinski,K. and Peltz,S.W. (1996) Making sense of nonsense in yeast. Trends Biochem. Sci., 21, 433–438. [DOI] [PubMed] [Google Scholar]
- Shyu A.-B., Belasco,J.G. and Greenberg,M.E. (1991) Two distinct destabilizing elements in the c-fos message trigger deadenylation as the first step in rapid mRNA decay. Genes Dev., 5, 221–231. [DOI] [PubMed] [Google Scholar]
- Stephenson L. and Maquat,L.E. (1996) Cytoplasmic mRNA for human triosephosphate isomerase is immune to nonsense-mediated mRNA decay despite forming polysomes. Biochimie, 78, 1043–1047. [DOI] [PubMed] [Google Scholar]
- Sun X. and Maquat,L.E. (2000) mRNA surveillance in mammalian cells: the relationship between introns and translation termination. RNA, 6, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Perlick,H.A., Dietz,H.C. and Maquat,L.E. (1998) A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 10009–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita K., Forget,B.G., Scarpa,A. and Benz,E.J.,Jr (1984) Intranuclear defect in β-globin mRNA accumulation due to a premature translation termination codon. Blood, 64, 13–22. [PubMed] [Google Scholar]
- Thermann R., Neu-Yilik,G., Deters,A., Frede,U., Wehr,K., Hagenmeier,C., Hentze,M.W. and Kulozik,A.E. (1998) Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J., 17, 3484–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell,P.J., Cuidad,C.J. and Chasin,L.A. (1989) Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol. Cell. Biol., 9, 2868–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.L. and Sunde,R.A. (1998) Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA, 4, 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Weiss,S.L. and Sunde,R.A. (1998) UGA codon position affects the efficiency of selenocysteine incorporation into glutathione peroxidase-1. J. Biol. Chem., 273, 28533–28541. [DOI] [PubMed] [Google Scholar]
- Zhang J. and Maquat,L.E. (1996) Evidence that the decay of nucleus-associated nonsense mRNA for human triosephosphate isomerase involves nonsense codon recognition after splicing. RNA, 2, 235–243. [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y. and Maquat,L.E. (1998a) Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA, 4, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sun,X., Qian,Y., LaDuca,J.P. and Maquat,L.E. (1998b) At least one intron is required for nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol., 18, 5272–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Welch,E.M., Hogan,K., Brown,A.H., Peltz,S.W. and Jacobson,A.J. (1997) Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA, 3, 234–244. [PMC free article] [PubMed] [Google Scholar]