Abstract
A novel gammaretrovirus, xenotropic murine leukemia virus-related virus (XMRV), has been identified in patients with prostate cancer and in patients with chronic fatigue syndromes. Standard Mus musculus laboratory mice lack a functional XPR1 receptor for XMRV and are therefore not a suitable model for the virus. In contrast, Gairdner's shrew-mice (Mus pahari) do express functional XPR1. To determine whether Mus pahari could serve as a model for XMRV, primary Mus pahari fibroblasts and mice were infected with cell-free XMRV. Infection of cells in vitro resulted in XMRV Gag expression and the production of XMRV virions. After intraperitoneal injection of XMRV into Mus pahari mice, XMRV proviral DNA could be detected in spleen, blood, and brain. Intravenous administration of a green fluorescent protein (GFP) vector pseudotyped with XMRV produced GFP+ CD4+ T cells and CD19+ B cells. Mice mounted adaptive immune responses against XMRV, as evidenced by the production of neutralizing and Env- and Gag-specific antibodies. Prominent G-to-A hypermutations were also found in viral genomes isolated from the spleen, suggesting intracellular restriction of XMRV infection by APOBEC3 in vivo. These data demonstrate infection of Mus pahari by XMRV, potential cell tropism of the virus, and immunological and intracellular restriction of virus infection in vivo. These data support the use of Mus pahari as a model for XMRV pathogenesis and as a platform for vaccine and drug development against this potential human pathogen.
Xenotropic murine leukemia virus-related virus (XMRV) is a gammaretrovirus originally identified in human prostate cancers (33). Small numbers of XMRV-infected cells have been observed in prostatic stromal cells but not in prostate carcinoma (33). Another study identified XMRV proviral DNA in 6 and 23% of prostate tumors when analyzed by real-time PCR and immunostaining, respectively (27). While initial studies associated XMRV almost exclusively in men who were homozygous for a variant of RNase L (R462Q), which is known to have reduced antiviral activity (33), more recent work failed to link XMRV infection and RNase L mutation (4). XMRV has also been reported in patients with chronic fatigue syndrome (CFS) (17). A total of 67% of CFS patients were positive for XMRV proviral DNA, whereas only 3.7% of healthy subjects were positive for XMRV. Subsequent testing by several other groups found no evidence of infection with XMRV in CFS patients or in healthy controls (30). In Europe, no XMRV was detected in 139 prostate cancer patients in an Irish cohort (4), while no or very few XMRV-specific DNA, RNA, or antibodies were detected in Germany or the United Kingdom cohort of CFS (7, 10, 34).
These conflicting data make it unclear to what degree XMRV infects humans and whether it plays a role in human diseases. If an etiological link is confirmed, detection and prevention of XMRV would provide novel intervention strategies for early diagnosis and treatment of both diseases. Moreover, since XMRV or XMRV-specific antibodies were detected in apparently healthy subjects, it would be critical to monitor XMRV contamination in clinical products for transfusion and transplantation.
For a better understanding of XMRV transmission, tissue tropism, and pathogenicity, studies of XMRV infection in animal models are crucial. Laboratory mice have provided important small animal model systems for many human diseases, due to their availability, size, low cost, ease of handling, and fast reproduction rate, and extensive studies have been carried out in mice to study the pathogenesis of closely related murine leukemia viruses (MLVs) (5, 11, 20, 23, 32). However, studies of XMRV pathogenesis in a mouse model have been hampered by the lack of functional receptor for XMRV in standard laboratory mice derived from Mus musculus species.
XMRV is closely related to xenotropic MLVs (X-MLVs) (33). The X-MLVs and polytropic MLVs (P-MLV) use Xpr1 as a receptor for cell entry (1, 31, 37), and so does XMRV (6, 13, 36). Xpr1 has four known variant receptor alleles in mice, Xpr1n, Xpr1sxv, Xpr1c, and Xpr1p, and each has a different susceptibility for P-MLV and/or X-MLV (35). P-MLV uses Xpr1n as receptor and most cells from Mus musculus laboratory mice express this receptor (35). Wild mice of the Eurasian genus Mus, such as Mus dunni, express the Xpr1sxv allele and are susceptible to both P-MLV and X-MLV, whereas the Asian mouse species Mus castaneus expresses Xpr1c and is susceptible only to X-MLV (19). Mus pahari is another Asian wild mouse species. This species is rooted at the base of the Mus phylogenic tree, suggesting that it may represent a Mus ancestral species. Mus pahari has the Xpr1p allele and is susceptible to X-MLV (35).
Recent data indicate that XMRV can infect Mus pahari cells (35). We therefore hypothesized that Mus pahari might be a suitable small animal model for XMRV infection. To test this, we examined the early events in XMRV infection of Mus pahari cells and Mus pahari mice in vivo.
MATERIALS AND METHODS
Mice.
The wild-derived mice, Mus pahari/EiJ, were purchased from the Jackson Laboratory. The mice were housed in the Mayo Clinic Animal Facility under the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC) guidelines with animal use protocols approved by the Mayo Clinic Animal Use and Care Committee. All animal experiments were carried out according to the provisions of the Animal Welfare Act, PHS Animal Welfare Policy, the principles of the NIH Guide for the Care and Use of Laboratory Animals, and the policies and procedures of Mayo Clinic.
Isolation of Mus pahari fibroblast cells.
Skin samples from a neonatal mouse were treated with trypsin at 37°C for 30 min. Dissociated cells were then cultured in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS) and antibiotics (penicillin and streptomycin).
Evaluation of XMRV production from the XMRV-infected Mus pahari fibroblast cells.
Supernatants of the XMRV-infected Mus pahari fibroblast cells were analyzed for production of infectious XMRV. Mus pahari cells were infected with XMRV. At 1 and 5 days after infection, culture supernatants were harvested and filtered through a 0.45-μm-pore-size filter and used to infect 2 × 105 293T cells. Total DNA was isolated from the infected 293T cells, and copy numbers of XMRV proviral DNA were quantified from 1.0 μg of total cellular DNA by using the ABI 7300 Real-Time PCR system and the following primers and probe: gag forward primer (5′-CAGTTGCTCTTAGCGGGTCT-3′), gag reverse primer (5′-TTACCTTGGCCAAATTGGTG-3′), and probe Roche library #51 (5′-FAM-GGCAGGAG-3′). Standard curves were generated with a serially diluted infectious molecular clone of XMRV VP62, pcDNA3.1(−)/VP62, where 1.0 ng of pcDNA3.1(−)/VP62 was used as 7 × 107 copies of XMRV.
Virus isolation and quantification.
Supernatants from 22Rv1 cells were purified with a 0.45-μm-pore-size filter and centrifuged through a 20% sucrose cushion at 13,000 rpm for 1 h at 4°C. The virions were washed with phosphate-buffered saline (PBS) and centrifuged at 13,000 rpm for 1 h, and the final viral pellets were resuspended in PBS. In order to obtain virus titers, the viral RNA was isolated by using a QIAamp viral RNA minikit (Qiagen). After DNase I treatment, cDNA was synthesized by using a SprintRT Complete system (Clontech). Viral genomic copy numbers were quantified by using an ABI 7300 real-time PCR system, and RNA genomic copy numbers were calculated by multiplying copy number values and each dilution factor. An infectious molecular-clone-derived XMRV, VP62, was produced by transfecting pcDNA3.1(−)/VP62 in 293T cells using FuGene 6 (Roche). Media were replaced 24 h later. At 3 days posttransfection, the supernatants were harvested, purified, and titrated as described above.
MLV GFP retroviral vector genome pseudotyped with XMRV proteins.
A Moloney MLV-based retroviral gene transfer vector, which encodes green fluorescent protein (GFP) but no viral proteins, was pseudotyped with an infectious molecular clone of XMRV (pcDNA3.1(−)/VP62) in 293T cells (24). The GFP virus was then purified as described above, and the virus titers (GFP transducing units) were determined by flow cytometry (BD FACScan) (24).
Immunostaining.
XMRV-infected Mus pahari cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.3% Triton X-100 for 15 min. Cells were blocked with 5% FBS-PBS. They were then stained with goat anti-MLV p30 (1:500) and mouse anti-β-actin (1:500) and visualized by DyLight 488-conjugated donkey anti-goat IgG antibody (1:500; Jackson Immunoresearch Laboratories) and Texas-Red-conjugated anti-mouse IgG secondary antibodies (1:500; Jackson ImmunoResearch Laboratories), respectively. Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) and analyzed by confocal microscopy (Zeiss).
Infection of mice with XMRV.
Neonatal or adult Mus pahari mice were injected intraperitoneally or intravenously with the indicated amounts of XMRV or MLV GFP vector pseudotyped with the XMRV envelope.
XMRV neutralization assay.
A GFP vector pseudotyped with XMRV was used for neutralization assay as previously reported (24). Plasma samples from XMRV-infected and uninfected Mus pahari were heat inactivated at 56°C for 30 min. Serially diluted plasma samples (1:20, 1:40, 1:80, 1:160, 1:320, and 1:640) were incubated with 2.5 × 104 infectious units (IU) of GFP-carrying XMRV at 37°C for 30 min. 293T cells (5 × 104) were then infected with the mixture of mouse plasma and XMRV. At 3 days postinfection, cells were resuspended, fixed with 4% paraformaldehyde, and analyzed for GFP-positive cell populations by flow cytometry. The percentages of GFP-positive cells were analyzed by CellQuestPro software. The detection limit for this assay is <1,000 IU/ml.
Real-time PCR.
DNA from mice tissues and blood samples were extracted by using a PureLink genomic DNA minikit (Invitrogen) according to the manufacturer's protocol. All samples were eluted in 50 μl of elution buffer, and the concentration of the DNA was determined by BioPhotometer (Eppendorf). For the real-time PCR assay, TaqMan Universal PCR Master Mix (Roche) was used with the same probe and primers as described above. The assay was analyzed by ABI 7300 real-time PCR.
Detection of XMRV DNA in blood and tissue samples.
DNA from mouse blood and tissue was extracted by using a PureLink genomic DNA minikit (Invitrogen). Nested-PCR was performed to amplify XMRV-specific sequences with platinum Taq polymerase (Invitrogen). XMRV gag was PCR amplified using the outer primers 5′-ACGAGTTCGTATTCCCGGCCGCA-3′ and 5′-CCGCCTCTTCTTCATTGTTC-3′ and the inner primers 5′-GCCCATTCTGTATCAGTTAA-3′ and 5′-AGAGGGTAAGGGCAGGGTAA-3′. For the XMRV env amplification, the outer primers 5′-TGGAAAATTACCAACCTAATGACAG-3′ and 5′-GATACAGTCTTAGTCCCCATGTT-3′ and the inner primers 5′-GATTTAGTTGGAGACAACTGGGAT-3′ and 5′-GCACCCTGGATGCTACCGGAGCC-3′ were used. The PCR products were then cloned into pGEM-T vector (Promega), and the amplified sequences were analyzed by DNADynamo (BlueTractor Software).
Immunohistochemistry.
Sections of OCT-embedded tissue specimens (5-μm thickness) on glass slides were blocked with 5% FBS-PBS for 30 min. XMRV Gag and Env were detected with goat anti-gp30/70 antibody (kindly provided by Yasu Takeuchi), followed by DyLight 488-conjugated donkey anti-goat IgG antibody (Jackson Immunoresearch). Spleen sections were also stained with the following antibodies: Alexa Fluor 647-conjugated hamster anti-mouse CD3 (1:200; Invitrogen), CD11b-FITC (1:11; Miltenyi Biotec), fluorescein isothiocyanate (FITC)-conjugated rat anti-mouse CD19 (1:200; BD Pharmingen), and FITC-conjugated anti-mouse Ly-6G (1:200; eBioscience). Nuclei were counterstained with DAPI and analyzed by confocal microscopy.
Splenocyte analysis by flow cytometry.
Spleen cells were harvested through physical dissociation. They were centrifuged (400 × g), and the pellets were lysed with red blood cell lysis buffer for 5 min at room temperature. Lysis reaction was stopped by adding RPMI medium and centrifuged at 400 × g. The pellets were washed with PBS and fixed with 4% paraformaldehyde for 5 min at room temperature. After fixation, the cells were centrifuged again and resuspended in PBS. Immunostaining was performed by using the following antibodies: phycoerythrin (PE)-conjugated rat anti-mouse CD4 (1:100; BD Pharmingen), PE-conjugated rat anti-mouse CD8a (1:100; BD Pharmingen), CD11b-PE (1:11; Miltenyi Biotec), PE-conjugated rat anti-mouse CD19 (1:100; BD Pharmingen), and PE-conjugated anti-Gr-1 (1:11; Miltenyi Biotec). Stained cells were analyzed by flow cytometry, and the data were analyzed by using CellQuestPro software.
Hematology analysis.
To provide complete blood counts (CBC), blood samples were harvested in EDTA-coated test tubes and analyzed by VetScan HMII (Abaxis). Peripheral blood smears were also fixed in methanol, and white blood cells were visualized by Wright-Giemsa staining (Volu-Sol, Inc.).
RESULTS
Mus pahari-derived cells support XMRV entry and replication in vitro.
Yan et al. reported that Mus pahari cells express a functional receptor for XMRV (36). To confirm this, Mus p1hari fibroblasts were infected with 22Rv1-derived XMRV (13). Immunostaining with anti-MLV p30/gp70 polyclonal antibodies revealed expression of XMRV proteins in the XMRV-infected Mus pahari cells (Fig. 1A). No prominent FITC signal was observed in the uninfected Mus pahari cells. These data confirmed expression of a functional receptor for XMRV in Mus pahari cells and suggested that Mus pahari cells can support transcription and expression of XMRV genes. Filtered culture supernatants from XMRV-infected and uninfected Mus pahari cells were harvested at days 1 and 5 after infection and were used to infect 293T cells. Real-time PCR of genomic DNA from these 293T cells generated XMRV-specific amplification products (Fig. 1B), indicating that XMRV-infected Mus pahari cells produced infectious virions. Partial sequence of the env gene from the 293T cells revealed no mutations in four individual clones (data not shown). These data demonstrated that Mus pahari cells support early and late phases of XMRV replication in vitro.
FIG. 1.
Mus pahari cells express functional receptor for XMRV. (A) Detection of XMRV proteins in primary Mus pahari cells by immunostaining. XMRV Gag and Env proteins (green) were detected by goat anti-MLV-p30/gp70 polyclonal antibodies. Cells and nuclei were counterstained with anti-β-actin antibody (red) and DAPI (blue), respectively. (B) Production of infectious XMRV virions from XMRV-infected Mus pahari cells. 293T cells were infected with the supernatants harvested from XMRV-infected and uninfected Mus pahari cells at 1 and 5 days after infection. XMRV-specific proviral DNA copy numbers in 293T cells were determined by real-time PCR as described in Materials and Methods.
Infection of Mus pahari mice with XMRV.
In initial experiments, six adult and four neonatal Mus pahari mice from both sexes were injected with 22Rv1-derived XMRV. Adult mice received 2.2 × 108 genomic copies of XMRV (ca. 3.0 × 105 IU of XMRV/mouse) by the intraperitoneal route. Neonatal mice were injected subcutaneously with XMRV (5 × 107 genomic copies/mouse) on the day of birth. Two adult mice were also injected intraperitoneally with an infectious molecular clone of VP62-derived XMRV. Blood and tissue samples were isolated at various times to assay for viral biodistribution and persistence, effects on blood cell content, and immune responses against the virus.
Detection of XMRV in plasma and in blood cells.
Blood was drawn at varied times after XMRV injection and real-time PCR was performed to detect proviral DNA in blood cells. Viral DNA was detected in three of the 10 mice over 5 to 12 weeks (Table 1). This experiment was repeated in 10 adult mice to detect cell-free virus (from genomic RNA in plasma) and cell-associated virus (from proviral DNA in blood cells). No viral RNA was detected in the plasma of any of the mice 5 days after infection (data not shown). A total of 63 copies of XMRV/μl and 41 copies of XMRV/μl were detected in the plasma in 1 of 10 infected adult mice at 2 and 4 weeks after infection. This corresponded to more than 80,000 copies of XMRV viral RNA in the blood of this mouse. When cell-associated XMRV was examined, 35 to 58 copies/μg of proviral DNA were detected in three different mice. These data demonstrate detection of XMRV viral sequences in the blood of 7 of 22 mice over several weeks after infection.
TABLE 1.
Biodistribution of XMRV in Mus pahari
| Parameter | Biodistribution of XMRV in Mus paharia |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uninfected |
Adult (22Rv1 XMRV) |
Newborn (22Rv1 XMRV) |
Adult (pVP62 XMRV) |
|||||||||||
| A | B | A5 | A6 | A3 | A4 | A2 | A1 | N1 | N2 | N3 | N4 | A19 | A20 | |
| Sexb | M | F | M | M | F | F | M | M | M | M | M | M | M | M |
| wpic | NA | NA | 2 | 2 | 5 | 5 | 12 | 14 | 8 | 8 | 8 | 8 | 2 | 2 |
| Ab (NAb)d | − | − | + | + | + | − | + | + | + | + | + | + | − | + |
| Ab (WB)e | − | − | − | + | + | − | + | + | + | + | − | − | − | + |
| Isolationf | − | − | ND | ND | − | ND | − | + | − | − | + | + | − | + |
| Blood | − | − | − | − | 0.6 | − | 180 | − | − | 2.3 | − | − | − | − |
| Heart | − | − | 100 | − | − | − | − | ND | 6.3 | − | ND | ND | − | 40 |
| Liver | − | − | − | − | − | − | − | ND | − | − | ND | ND | − | − |
| Spleen | − | − | 140 | − | 62 | − | 56 | ND | 17 | − | ND | ND | 360 | − |
| Brain | − | 400 | 130 | − | − | − | 170 | ND | 62 | − | ND | ND | 100 | − |
| Prostate | − | NA | − | − | NA | NA | − | ND | − | − | ND | ND | 18 | 8.9 |
| Testis | − | NA | − | 1,100 | NA | NA | − | ND | − | − | ND | ND | − | − |
XMRV-gag genomic copy numbers in 1.0 μg of total cellular DNA are shown as numbers. NA, not available; ND, not determined. For the blood, heart, liver, spleen, brain, prostate, and testis data, a dash (−) indicates the absence of proviral DNA from blood or tissue samples. Individual mice (A, B, A5, A6, etc.) are indicated in the column headings.
M, male; F, female.
wpi, weeks postinfection.
The presence (+) or absence (−) of neutralization antibody (NAb) is indicated.
The presence (+) or absence (−) of XMRV-specific antibodies by Western blotting (WB) analysis is indicated.
The isolation (+) or not (−) of XMRV from plasma-free blood samples after cocultivation with 293T cells is indicated.
To test whether functional XMRV could be isolated from the mice, blood cells from 12 of the infected mice were cultured with 293T cells, followed by real-time PCR for viral genomes (Table 1). After three blind passages on 293T cells, XMRV sequences were detected from mice A1, A20, N3, and N4 (at 17, 1, 7, and 7 weeks, respectively, Table 1). All of the controls (three uninfected mice) were negative for XMRV. When DNA from 293T cells infected blood from mouse A1 was sequenced, a conserved G-to-A mutation was observed at the nucleotide position 790 in all XMRV gag PCR clones (six PCR clones, total of 3,480 bp). No mutations were found in the env sequence (four PCR clones, total of 1,016 bp).
These data suggest that XMRV can infect Mus pahari blood cells in vivo. Sporadic XMRV isolation also indicates few XMRV-infected cells in circulation. These data suggest that XMRV can disseminate as cell-free and cell-associated forms and that the virus can persist in the blood and in blood cells for weeks after infection.
Biodistribution of XMRV genomes in solid tissues in Mus pahari.
Control and XMRV-infected mice were sacrificed at different times, and real-time PCR was performed on DNA from the heart, liver, spleen, brain, prostate, and testis (Table 1). In mice that were infected with 22Rv1-derived virus, XMRV sequences were detected in the heart, spleen, and brain. Interestingly, when mice were infected with an infectious molecular clone of XMRV, virus-specific sequences were also detected in the prostate. These data indicate that XMRV spreads to multiple tissue sites in Mus pahari, including tissues relevant to human XMRV associations.
Biodistribution of XMRV-infected cells in solid tissues.
Attempts to identify XMRV-infected cells in tissues by immunostaining were hampered by the lack of XMRV-specific antibodies. When sections of spleen and brain tissues from mice were immunostained with goat anti-MLV p30/gp70 polyclonal antibody, positive cells were observed in both control and infected mice (see Fig. S1 in the supplemental material).
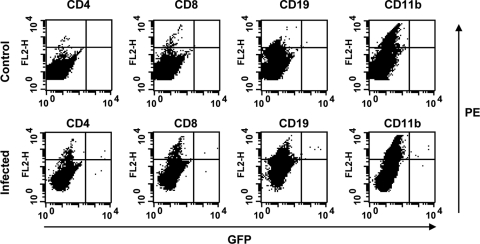
Since the anti-MLV antibody was nonspecific for XMRV, we generated a GFP vector that was pseudotyped with an infectious molecular clone of XMRV VP62 (MLV-based retroviral transfer vector genome, pseudotyped by XMRV Gag, Pol, and Env). The GFP vector pseudotyped with XMRV was injected intravenously into three adult Mus pahari mice (106 IU/mouse). At 1 week after injection, splenocytes were isolated and counterstained with PE-labeled antibodies for CD4, CD8, CD19, and CD11b to detect T cells, B cells, and monocytes/macrophages (Fig. 2 and data not shown). No GFP-positive splenocytes were observed in the control mouse or in one of the three injected animals. In a second GFP-XMRV-infected mouse, GFP-positive cells were observed in the CD4+, CD8+, CD19+, and CD11b+ cells. In the third GFP-XMRV-infected mouse, similar, but fewer GFP+/CD4+ and GFP+/CD19+ double-positive cells were also observed. However, no GFP+/CD11b+ cells were observed in this mouse. These data suggest that XMRV can infect various types of spleen cells, including CD4+ T helper cells and CD19+ B cells, in the early stage of infection. We found no notable differences in the proportions of CD4-, CD8-, CD19-, and CD11b-positive cells in XMRV-infected compared to uninfected mice.
FIG. 2.
Detection of XMRV-infected cells in spleen. Splenocytes of mice infected with a GFP-expressing XMRV were stained with PE-conjugated CD4, CD8, CD19, and CD11b, and analyzed by flow cytometry. The results of a 30,000 cell count are shown. Splenocytes of uninfected mouse were used as controls.
Influence of XMRV infection on blood and spleen cells in Mus pahari.
PCR, infection, and GFP transduction data indicated that blood cells are infected by XMRV. To assess whether XMRV infection perturbs the normal population of blood cells, blood samples from nine adult mice that received intraperitoneal injections of XMRV (2.2 × 108 genomic copies of 22Rv1-derived XMRV) were analyzed for complete blood counts (CBC) at different times after infection. The 95% reference range was calculated as (mean − 1.96 × SD) to (mean + 1.96 × SD) using CBC data from 12 uninfected adult Mus pahari (see Table S1 in the supplemental material) and used as the normal range. Modest increases in white blood cells, lymphocytes, monocytes, granulocytes, and platelets were observed in a subset of adult mice over 4 weeks after injection (Tables 2 and 3). Intriguingly, modest anemia and lymphopenia, characterized by reduced numbers of white blood cells, lymphocytes, and red blood cells, was observed in two neonatal mice injected with XMRV on the day of their birth (Table 4).
TABLE 2.
Influence of acute XMRV infection on blood parameters (2 weeks postinfection)a
| Parameterb | A7 | A8 | A9 | A10 | A11 | A12 | A13 | A15 | A16 | Normal rangec |
|---|---|---|---|---|---|---|---|---|---|---|
| WBC | 6.5 | 8.19 | 12.35 | 7.16 | 6.31 | 4.8 | 5.32 | 6.02 | 8.74 | 4.4-8.6 |
| LYM | 4.5 | 5.62 | 5.97 | 4.68 | 3.85 | 3.36 | 3.73 | 4.04 | 5.74 | 3.4-5.9 |
| MON | 0.14 | 0.32 | 0.72 | 0.46 | 0.43 | 0.16 | 0.03 | 0.31 | 0.33 | 0.01-0.32 |
| GRA | 1.83 | 2.26 | 5.66 | 2.02 | 2.03 | 1.28 | 1.56 | 1.67 | 2.68 | 0.4-2.9 |
| RBC | 10.24 | 10.93 | 10.73 | 10.1 | 10.5 | 10.31 | 10.47 | 10.84 | 10.63 | 9.1-12.1 |
| HGB | 16.6 | 17.8 | 17.1 | 16.3 | 17.1 | 15.6 | 16.9 | 16.8 | 16.2 | 14.3-19.2 |
| HCT | 41.6 | 47.57 | 44.97 | 43.94 | 46.59 | 43.32 | 46.7 | 46.79 | 44.03 | 38-52 |
| MCV | 41 | 44 | 42 | 43 | 44 | 42 | 45 | 43 | 41 | 40-45 |
| MCH | 16.2 | 16.3 | 15.9 | 16.1 | 16.3 | 15.1 | 16.2 | 15.5 | 15.2 | 14.8-16.8 |
| MCHC | 39.5 | 37.5 | 38 | 37.1 | 36.8 | 36 | 36.2 | 36 | 36.8 | 35.8-38.7 |
| PLT | 241 | 1,048 | 755 | 1,052 | 491 | 679 | 559 | 775 | 749 | 244-1,042 |
Numbers above the normal range of control mice are in boldface. Numbers below the normal range of control mice are underscored.
WBC, LYM, MON, GRA, RBC, HGB, HCT, MCV, MCH, MCHC, and PLT represent the white blood cell count (109/liter), lymphocyte count (109/liter), monocyte count (109/liter), granulocyte count (109/liter), red blood cell count (1012/liter), hemoglobin level (g/dl), hematocrit (%), mean corpuscular volume (fl), mean corpuscular hemoglobin (pg), mean corpuscular hemoglobin concentration (g/dl), and platelet count (109/liter), respectively.
The 95% reference range was calculated as (mean − 1.96 × SD) to (mean + 1.96 × SD) using CBC data from 12 uninfected Mus pahari (see Table S1 in the supplemental material) and is shown as the normal range. SD, standard deviation.
TABLE 3.
Influence of acute XMRV infection on blood parameters (4 weeks postinfection)a
| Parameterb | A7 | A8 | A9 | A11 | A12 | A13 | A14 | A15 | A16 | Normal rangec |
|---|---|---|---|---|---|---|---|---|---|---|
| SC | + | + | + | + | + | + | + | + | + | |
| WBC | 5.96 | 9.36 | 12.79 | 7.01 | 4.81 | 7.08 | 7.18 | 7.74 | 6.79 | 4.4-8.6 |
| LYM | 4.21 | 6.08 | 7.24 | 4.2 | 3.25 | 4.89 | 5.21 | 5.81 | 5.1 | 3.4-5.9 |
| MON | 0.23 | 0.06 | 0.59 | 0.04 | 0.09 | 0.04 | 0.58 | 0.35 | 0.14 | 0.01-0.32 |
| GRA | 1.52 | 3.22 | 4.97 | 2.77 | 1.48 | 2.14 | 1.39 | 1.58 | 1.54 | 0.4-2.9 |
| RBC | 9.13 | 11.08 | 9.54 | 10.43 | 10.36 | 10.76 | 9.93 | 10.37 | 11.13 | 9.1-12.1 |
| HGB | 14.3 | 18.2 | 15.7 | 17.4 | 16.3 | 17.8 | 15.6 | 16.2 | 16.3 | 14.3-19.2 |
| HCT | 39.93 | 49.23 | 41.95 | 49.01 | 44.33 | 49.15 | 40.42 | 47.249 | 44.54 | 38-52 |
| MCV | 44 | 44 | 44 | 47 | 43 | 46 | 41 | 46 | 40 | 40-45 |
| MCH | 15.7 | 16.5 | 16.4 | 16.7 | 15.7 | 16.5 | 15.7 | 15.6 | 14.6 | 14.8-16.8 |
| MCHC | 35.9 | 37 | 37.4 | 35.5 | 36.7 | 36.1 | 38.5 | 34.3 | 36.6 | 35.8-38.7 |
| PLT | 155 | 624 | 1,296 | 776 | 817 | 699 | 618 | 497 | 727 | 244-1042 |
Numbers above the normal range of control mice are in boldface. Numbers below the normal range of control mice are underscored.
WBC, LYM, MON, GRA, RBC, HGB, HCT, MCV, MCH, MCHC, and PLT represent the white blood cell count (109/liter), lymphocyte count (109/liter), monocyte count (109/liter), granulocyte count (109/liter), red blood cell count (1012/liter), hemoglobin level (g/dl), hematocrit (%), mean corpuscular volume (fl), mean corpuscular hemoglobin (pg), mean corpuscular hemoglobin concentration (g/dl), and platelet count (109/liter), respectively. XMRV seroconversion (SC) was confirmed (+) by Western blotting.
The 95% reference range was calculated as (mean − 1.96 × SD) to (mean + 1.96 × SD) using CBC data from 12 uninfected Mus pahari (see Table S1 in the supplemental material) and is shown as the normal range. SD, standard deviation.
TABLE 4.
Influence of prolonged XMRV infection on blood parametersa
| Parameterb | N1 | N2 | N3 | N3 | N3 | N4 | N4 | Normal rangec |
|---|---|---|---|---|---|---|---|---|
| wpi | 8 | 8 | 15 | 26 | 37 | 15 | 26 | |
| WBC | 3.61 | 2.31 | 5.9 | 8.24 | 5.82 | 6.32 | 7.83 | 4.4-8.6 |
| LYM | 2.57 | 1.79 | 4.11 | 5.75 | 4.25 | 4.35 | 5.64 | 3.4-5.9 |
| MON | 0.16 | 0.1 | 0.14 | 0.09 | 0.06 | 0.28 | 0.1 | 0.01-0.32 |
| GRA | 0.88 | 0.43 | 1.65 | 2.4 | 1.52 | 1.69 | 2.08 | 0.4-2.9 |
| RBC | 8.69 | 9.24 | 10.94 | 10.81 | 11.19 | 10.54 | 10.53 | 9.1-12.1 |
| HGB | 12.4 | 12.9 | 17.0 | 16.5 | 17.4 | 16.8 | 17.0 | 14.3-19.2 |
| HCT | 36.76 | 38.42 | 45.15 | 44.95 | 48.13 | 44.52 | 42.94 | 38-52 |
| MCV | 42 | 42 | 41 | 42 | 43 | 42 | 41 | 40-45 |
| MCH | 14.3 | 14.0 | 15.6 | 15.3 | 15.6 | 15.9 | 16.1 | 14.8-16.8 |
| MCHC | 33.8 | 33.7 | 37.7 | 36.8 | 36.2 | 37.7 | 39.5 | 35.8-38.7 |
| PLT | 311 | 434 | 516 | 448 | 295 | 564 | 106 | 244-1042 |
Numbers above the normal range of control mice are in boldface. Numbers below the normal range of control mice are underscored.
WBC, LYM, MON, GRA, RBC, HGB, HCT, MCV, MCH, MCHC, and PLT represent the white blood cell count (109/liter), lymphocyte count (109/liter), monocyte count (109/liter), granulocyte count (109/liter), red blood cell count (1012/liter), hemoglobin level (g/dl), hematocrit (%), mean corpuscular volume (fl), mean corpuscular hemoglobin (pg), mean corpuscular hemoglobin concentration (g/dl), and platelet count (109/liter), respectively. wpi, weeks postinfection.
The 95% reference range was calculated as (mean − 1.96 × SD) to (mean + 1.96 × SD) using CBC data from 12 uninfected Mus pahari (see Table S1 in the supplemental material) and is shown as the normal range. SD, standard deviation.
To assess whether XMRV infection could also reduce the numbers of immune cells in lymphoid tissues, we also examined the major cell populations in spleen. Although the sizes of control and infected Mus pahari spleens were notably smaller than those of commonly used laboratory mice, immunostaining for CD3 (a pan-T-cell marker), CD11b (macrophage/monocyte marker), CD19 (B cell marker), and Gr-1 (neutrophil marker) revealed no notable pathogenic changes in XMRV-infected mouse spleen (see Fig. S2 in the supplemental material), including those of the mice with pancytopenia.
Adaptive immune responses to XMRV in Mus pahari mice.
Plasma samples collected at varied times after XMRV infection had strong neutralizing antibodies in 10 of 12 infected adult and neonatal mice (Fig. 3A and Table 1). To determine the specificities of these neutralizing antibodies, the plasma samples were tested against MLV retroviral vectors (with MLV Gag and Pol) that had been pseudotyped with Env glycoproteins from vesicular stomatitis virus G, amphotropic MLV, RD114, or XMRV (25). Tenfold dilution of plasma samples from XMRV-infected Mus pahari completely neutralized infection of the vector pseudotyped with XMRV Env (>97% neutralization). In contrast, <5% neutralization was observed at a 1:10 dilution of mouse serum against vectors pseudotyped with other viral glycoproteins, confirming the specificity of neutralizing activity (data not shown). It is worth noting that no cross-neutralizing activity was observed against a closely related amphotropic MLV Env-pseudotyped vector. These data demonstrated the rapid induction of high levels of XMRV-specific neutralizing antibodies in the infected mice.
FIG. 3.
Induction of XMRV-specific antibodies in XMRV-injected mice. (A) Serially diluted plasma (1:20, 1:40, 1:80, 1:160, 1:320, and 1:640) isolated from XMRV-infected and uninfected control mice were incubated with GFP-expressing XMRV. Three days after infection, GFP-positive cell populations were analyzed by flow cytometry. Representative data with plasma dilutions at 1:20 and 1:640 were shown. (B) Detection of XMRV Env and Gag proteins by Western blotting with plasma samples of XMRV-infected mice. Cell lysates from PC3 cells (XMRV antigen negative, which is indicated as “−”), and cell lysates from XMRV-infected PC3 cells (XMRV antigen positive; “+”) were used as antigens. Plasma samples isolated at various time points (weeks postinfection, indicated under each blot) were used. Cell lysates were also analyzed by anti-MLV p30/gp70 antibody as a positive control.
When plasma samples were tested against XMRV structural proteins from cell lysates of XMRV-infected and uninfected PC3 prostate carcinoma cells, Env-specific antibodies were observed as early as 2 weeks after XMRV injection in some of the mice (Fig. 3B). Western blotting analysis with plasma samples obtained at 12 and 14 weeks after infection showed stronger XMRV Env signals and additional signals for XMRV precursor Gag and capsid proteins. These kinetic antibody responses were in sharp contrast to responses in Mus musculus BALB/c mice, which developed no detectable levels of XMRV-specific antibodies after multiple intraperitoneal injection of XMRV (data not shown). Western blotting with the mouse A4 plasma, which showed no detectable XMRV-neutralizing antibodies, did not detect XMRV-specific proteins. Since we failed to detect XMRV sequences in the mouse A4, we speculate that this mouse was not infected by XMRV. In addition, despite their high levels of XMRV-specific neutralizing antibodies, plasma samples from an adult mouse A5 and two mice N3 and N4, which received XMRV on the day of birth, repeatedly failed to show XMRV Env and Gag proteins. It is likely that the neutralizing antibodies in these mice recognized the epitopes that were sensitive to sodium dodecyl sulfate or thermal denaturation. XMRV Env-specific immune responses were also observed viral molecular clone injected mouse A19 by the neutralization assays (data not shown) and Western blotting (Fig. 3B). No XMRV-specific antibodies were detected in the mouse A20 at 2 weeks postinfection, perhaps due to less robust immune responses at this early time point.
Potential intracellular restriction of XMRV in Mus pahari mice.
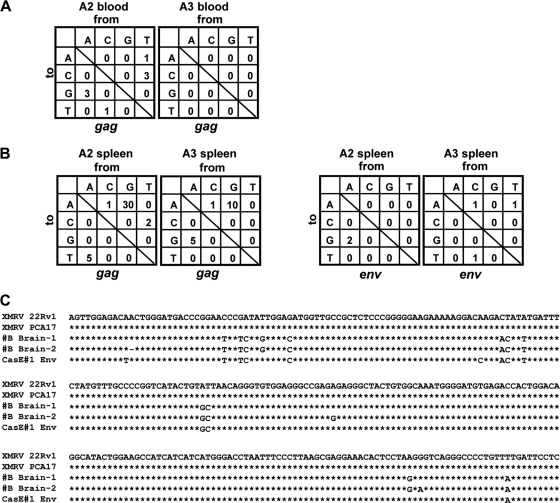
Since XMRV is sensitive to the human APOBEC3G and -3F-mediated hypermutation (8, 22), we analyzed XMRV sequences recovered from infected Mus pahari mice. Partial gag and env sequences were amplified from blood cell DNA from mice A2 and A3 (Table 1), and these were compared to the reference XMRV sequence (GenBank accession no. NC_007815). Eight nucleotide substitutions and one nucleotide deletion were found in the 5,830 bp of mouse A2 blood-derived gag sequences (10 PCR clones, no identical mutations found) (Fig. 4A). No mutations were observed in A2 blood-derived env sequences (12 PCR clones, total of 3,060 bp) and in the mouse A3 blood-derived gag sequences (3 PCR clones, total of 1,740 bp). When the XMRV sequences in the spleen of infected mice were determined, high levels of G-to-A hypermutations were found in the gag sequences, but not in the env sequences, of mouse A2 and A3 spleen-derived XMRV sequences (Fig. 3B). No notable mutations were observed in XMRV sequences cloned from the brain tissues of mice A2 and A3 (data not shown). These data suggest the existence of APOBEC3-like intracellular immunity, which restricts XMRV replication through G-to-A hypermutation, in Mus pahari spleen cells.
FIG. 4.
Sequence analysis of partial XMRV gag and env genes recovered from infected Mus pahari. (A) The numbers of mutations in the gag gene from blood samples of mice A2 (total of 5,830 bp sequenced) and A3 (total of 1,740 bp sequenced) were summarized. (B) Numbers of mutations in the gag and env genes from spleen samples of mice A2 and A5 (in 5,830 bp for both gag sequenced and 1,785 bp for both env sequenced) were summarized. (C) Alignment of MLV-like sequences (#B Brain-1 and -2) identified in the brain of uninfected mouse #B was shown with corresponding env sequences of XMRV (22Rv1 and PCA17) and MLV CasE#1.
Exogenous MLV-like sequences identified in a control mouse.
Unexpectedly, a brain sample from a control mouse B gave a positive signal by real-time PCR (Table 1). Sequencing and BLAST analysis of the amplified XMRV-like sequences identified an MLV sequence, which is most closely related to a previously reported, wild-mouse-derived MLV strain, CasE#1 (35) (Fig. 4C). This observation suggests that CasE#1-like exogenous MLV strain may circulate in commercially available Mus pahari colonies.
DISCUSSION
In this study, we examined the permissivity of Mus pahari to XMRV infection. We demonstrated in vitro that Mus pahari cells support the early and late phases of XMRV replication. In vivo, we demonstrated persistent detection of viral RNA, proviral DNA, and infectious virus in the mice for prolonged periods after single injection. XMRV appeared to infect immune cells, including CD4+ T cells and CD19-positive B cells.
These data indicate that Mus pahari mice hold promise as a model for XMRV infection. Once it is optimized to increase the frequency of detectable infection, the mice should be a useful platform for screening XMRV diagnostics and XMRV therapeutics. Single XMRV injection induced strong immune responses against viral Env and Gag proteins. Therefore, Mus pahari would also appear to be a viable model for testing vaccine strategies against this virus should it be determined that it poses legitimate risks to humans.
Modest anemia and lymphopenia was observed in two neonatally XMRV-injected mice. These two mice showed continual minor nose bleeding and were smaller than their two litter mates (N3 and N4) that showed no notable clinical signs. Although the precise mechanism remains to be determined, the observed modest anemia and lymphopenia were likely due to the continual bleeding. Alternatively, XMRV infection in newborn mice may induce bone marrow suppression. Since these two mice were littermates, it also remains possible that these clinical signs were not associated with XMRV infection. Additional studies are needed to determine whether these are XMRV-related phenomena; however, these experiments are difficult since virus-injected neonates are frequently cannibalized by the mother.
This XMRV-associated pancytopenia is of interest considering recently identified gammaretroviral Env-mediated immunosuppression (28). It is plausible that persistent XMRV infection in immune cells can induce immunological dysregulation. If this is confirmed, induction of immunosuppression could play a role in XMRV-associated diseases, perhaps including prostate cancer and CFS. Loss of immune control of transformed cells is a classic event in tumorigenesis. Likewise, aberrant cellular immunity has been observed in patients with CFS (2, 12). Further long-term studies will determine whether XMRV infection in Mus pahari induces transient or persistent immunosuppression, followed by malignancy and neurological or immunological disorders.
Although our results demonstrated that Mus pahari can be infected with XMRV and used for studying the virus infection, we experienced several Mus pahari-specific issues. First of all, handling of these mice requires special precautions. Mus pahari mice are faster and more aggressive than inbred laboratory mice. In addition, they are more susceptible to loss of skin when they are held by their tail. Other husbandry issues include lower successful mating rates (ca. 50%), small litter size (typically two to four pups), and parental cannibalism. Finally, given its potential role in human disease, it is crucial to contain these XMRV-infected mice in biosafety level 2 facilities.
In contrast to commonly used Mus musculus laboratory mice, Mus pahari has no endogenous xenotropic and mink cell focus-forming virus env sequences in its genome (14, 15). Indeed, except for the one brain sample that harbored an exogenous CasE#1-like MLV sequence, our XMRV gag- and env-specific real-time PCRs and standard XMRV gag and env sequence-specific PCR primers did not amplify any MLV-related sequences from uninfected Mus pahari DNA samples. These data confirmed no MLV-like endogenous retrovirus in Mus pahari, and we concluded that the sequences amplified by our PCR primers were of XMRV origin, rather than related endogenous retroviruses in Mus pahari. BLAST analysis of the amplified sequences recovered from XMRV-infected mice were most closely related to previously reported XMRV sequences. Nevertheless, these data do not rule out the possible existence of endogenous retroviruses in Mus pahari. Indeed, when we used PCR primers that were previously used to detect XMRV pol in human cells (24), all DNA samples from infected and uninfected Mus pahari tissues gave positive signals. This suggests that Mus pahari cells may have an endogenous retrovirus or embedded pol gene with some homology to XMRV.
We also identified an MLV-related sequence in an uninfected Mus pahari brain. BLAST analysis identified a wild mouse-derived exogenous MLV strain, CasE#1, as the closest relative of this sequence. The same sequences were not detected in other Mus pahari DNA samples, indicating that this mouse had an exogenous CasE#1-like MLV. Although CasE#1 was originally isolated from a wild mouse trapped in the area of Lake Casitas, California (3), a subsequent study implied that this wild mouse population may have acquired mouse gammaretroviruses from Asian mice (15). We speculate that the CasE#1-related MLV is naturally circulating in wild-mouse populations in Thailand, which was then introduced into commercially available Mus pahari populations when colonies were established using the Thai wild mice.
In addition to well-characterized conventional innate and adaptive immunity, intrinsic antiviral mechanisms play critical roles in restricting retrovirus replication (9, 21, 29). In mice, the mutagenic cytidine deaminase activity of APOBEC3 has been linked with the nucleotide sequence diversity among endogenous and exogenous gammaretroviruses; APOBEC3 restricts AKV MLV in AKR mice through introduction of G-to-A hypermutations (16), while APOBEC3 knockout mice show enhanced replication and pathogenesis of Moloney MLV (18). Moreover, mouse APOBEC3 has been under positive selection throughout Mus evolution, suggesting the essential role of APOBEC3 in controlling retroviral replication in vivo (26). Recent studies have demonstrated that XMRV restriction by human APOBEC3 proteins and mouse APOBEC3 in vitro (8, 22). Consistent with these observations, we found prominent G-to-A mutations in the XMRV gag sequences recovered from infected Mus pahari spleen, suggesting the possible involvement of APOBEC3-mediated hypermutations in controlling XMRV replication in vivo in Mus pahari. No notable G-to-A hypermutations were found in the XMRV sequences amplified from blood or brain samples, suggesting that APOBEC3 is expressed to higher levels in spleen than in blood or brain cells. It is also possible that XMRV replicates more actively in spleen than in blood or brain. Further studies are necessary to address the involvement of APOBEC3 in controlling XMRV replication in vivo.
In summary, our data demonstrated that Mus pahari mice support XMRV replication in vitro and in vivo, while they can control its replication through innate and adaptive immune responses. They can be used to study the modes of XMRV transmission or to evaluate anti-XMRV strategies such as anti-XMRV vaccines in vivo. Long-term XMRV infection study in large number of Mus pahari would also reveal possible pathogenesis caused by XMRV in vivo. Thus, Mus pahari would provide the animal model testing to in part satisfy Koch's postulate regarding XMRV. Since we found that AZT strongly blocks XMRV replication in vitro (24), we are currently evaluating its preventive and therapeutic effects on XMRV infection and replication in vivo.
Supplementary Material
Acknowledgments
We thank Robert H. Silverman and Yasuhiro Takeuchi for the XMRV infectious clone, pVP62/pcDNA3.1(−), and the goat anti-MLV p30/gp70 antibody, respectively. We thank Ianko Iankov, Rae M. Myers, and Suzanne M. Greiner for excellent technical supports.
This study was supported by the National Institutes of Health (R56AI074363), Mayo Clinic Career Development Project in Prostate SPORE grant CA91956-080013, and the Mayo Foundation (to Y.I.).
Footnotes
Published ahead of print on 17 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Battini, J. L., J. E. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. U. S. A. 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caligiuri, M., et al. 1987. Phenotypic and functional deficiency of natural killer cells in patients with chronic fatigue syndrome. J. Immunol. 139:3306-3313. [PubMed] [Google Scholar]
- 3.Cloyd, M. W., M. M. Thompson, and J. W. Hartley. 1985. Host range of mink cell focus-inducing viruses. Virology 140:239-248. [DOI] [PubMed] [Google Scholar]
- 4.D'Arcy, F., et al. 2008. No evidence of XMRV in Irish prostate cancer patients with the R462Q mutation. Eur. Urol. Suppl. 7:271. [Google Scholar]
- 5.DesGroseillers, L., and P. Jolicoeur. 1984. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J. Virol. 52:448-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, B., et al. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. U. S. A. 104:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groom, H. C., et al. 2010. Absence of xenotropic murine leukaemia virus-related virus in UK patients with chronic fatigue syndrome. Retrovirology. 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groom, H. C., M. W. Yap, R. P. Galao, S. J. Neil, and K. N. Bishop. 2010. Susceptibility of xenotropic murine leukemia virus-related virus (XMRV) to retroviral restriction factors. Proc. Natl. Acad. Sci. U. S. A. 107:5166-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 10.Hohn, O., et al. 2009. Lack of evidence for xenotropic murine leukemia virus-related virus (XMRV) in German prostate cancer patients. Retrovirology 6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolicoeur, P., N. Nicolaiew, L. DesGroseillers, and E. Rassart. 1983. Molecular cloning of infectious viral DNA from ecotropic neurotropic wild mouse retrovirus. J. Virol. 45:1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klimas, N. G., F. R. Salvato, R. Morgan, and M. A. Fletcher. 1990. Immunologic abnormalities in chronic fatigue syndrome. J. Clin. Microbiol. 28:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knouf, E. C., et al. 2009. Multiple integrated copies and high-level production of the human retrovirus XMRV (xenotropic murine leukemia virus-related virus) from 22Rv1 prostate carcinoma cells. J. Virol. 83:7353-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak, C. A. 1985. Susceptibility of wild mouse cells to exogenous infection with xenotropic leukemia viruses: control by a single dominant locus on chromosome 1. J. Virol. 55:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozak, C. A., and R. R. O'Neill. 1987. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J. Virol. 61:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langlois, M. A., K. Kemmerich, C. Rada, and M. S. Neuberger. 2009. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J. Virol. 83:11550-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lombardi, V. C., et al. 2009. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science 326:585-589. [DOI] [PubMed] [Google Scholar]
- 18.Low, A., C. M. Okeoma, N. Lovsin, M. de las Heras, T. H. Taylor, B. M. Peterlin, S. R. Ross, and H. Fan. 2009. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology 385:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyu, M. S., and C. A. Kozak. 1996. Genetic basis for resistance to polytropic murine leukemia viruses in the wild mouse species Mus castaneus. J. Virol. 70:830-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mosier, D. E., R. A. Yetter, and H. C. Morse III. 1987. Functional T lymphocytes are required for a murine retrovirus-induced immunodeficiency disease (MAIDS). J. Exp. Med. 165:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 22.Paprotka, T., et al. 2010. Inhibition of xenotropic murine leukemia virus-related virus by APOBEC3 proteins and antiviral drugs J. Virol. 84:5719-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruscetti, S., L. Davis, J. Field, and A. Oliff. 1981. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J. Exp. Med. 154:907-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakuma, R., T. Sakuma, S. Ohmine, R. H. Silverman, and Y. Ikeda. 2010. Xenotropic murine leukemia virus-related virus is susceptible to AZT. Virology 397:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakuma, T., et al. 2010. Characterization of retroviral and lentiviral vectors pseudotyped with XMRV envelope glycoprotein. Hum. Gene Ther. 12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanville, B., et al. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog. 6:e1000974. [DOI] [PMC free article] [PubMed]
- 27.Schlaberg, R., D. J. Choe, K. R. Brown, H. M. Thaker, and I. R. Singh. 2009. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc. Natl. Acad. Sci. U. S. A. 106:16351-16356. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Schlecht-Louf, G., et al. 2010. Retroviral infection in vivo requires an immune escape virulence factor encrypted in the envelope protein of oncoretroviruses. Proc. Natl. Acad. Sci. U. S. A. 107:3782-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stremlau, M., et al. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 30.Switzer, W. M., et al. 2010. Absence of evidence of xenotropic murine leukemia virus-related virus infection in persons with chronic fatigue syndrome and healthy controls in the United States. Retrovirology 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tailor, C. S., A. Nouri, C. G. Lee, C. Kozak, and D. Kabat. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. U. S. A. 96:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, C. Y., R. Khiroya, R. S. Schwartz, and J. M. Coffin. 1984. Role of recombinant ecotropic and polytropic viruses in the development of spontaneous thymic lymphomas in HRS/J. mice. J. Virol. 50:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urisman, A., et al. 2006. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.van Kuppeveld, F. J., et al. 2010. Prevalence of xenotropic murine leukaemia virus-related virus in patients with chronic fatigue syndrome in the Netherlands: retrospective analysis of samples from an established cohort. BMJ 340:c1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan, Y., R. C. Knoper, and C. A. Kozak. 2007. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruses and their XPR1 receptors elucidate receptor determinants of virus entry. J. Virol. 81:10550-10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan, Y., Q. Liu, and C. A. Kozak. 2009. Six host-range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 6:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, Y. L., et al. 1999. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 21:216-219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.