Abstract
Rhesus macaques are naturally infected with a gammaherpesvirus which is in the same lymphocryptovirus (LCV) genus as and closely related to Epstein-Barr virus (EBV). The rhesus macaque LCV (rhLCV) contains a repertoire of genes identical to that of EBV, and experimental rhLCV infection of naive rhesus macaques accurately models acute and persistent EBV infection of humans. We cloned the LCL8664 rhLCV strain as a bacterial artificial chromosome to create recombinant rhLCV for investigation in this animal model system. A recombinant rhLCV (clone 16 rhLCV) carrying a mutation in the putative immune evasion gene rhBARF1 was created along with a rescued wild-type (rWT) rhLCV in which the rhBARF1 open reading frame (ORF) was repaired. The rWT rhLCV molecular clone demonstrated viral replication and B-cell immortalization properties comparable to those of the naturally derived LCL8664 rhLCV. Qualitatively, clone 16 rhLCV carrying a mutated rhBARF1 was competent for viral replication and B-cell immortalization, but quantitative assays showed that clone 16 rhLCV immortalized B cells less efficiently than LCL8664 and rWT rhLCV. Functional studies showed that rhBARF1 could block CSF-1 cytokine signaling as well as EBV BARF1, whereas the truncated rhBARF1 from clone 16 rhLCV was a loss-of-function mutant. These recombinant rhLCV can be used in the rhesus macaque animal model system to better understand how a putative viral immune evasion gene contributes to the pathogenesis of acute and persistent EBV infection. The development of a genetic system for making recombinant rhLCV constitutes a major advance in the study of EBV pathogenesis in the rhesus macaque animal model.
Epstein-Barr virus (EBV) efficiently infects humans; nearly all humans are infected by adulthood, and once infected, humans harbor persistent virus infection for life. Tissue culture studies have revealed considerable knowledge about EBV replication, EBV immortalization of B cells, and the nature of the host immune response to EBV infection (37). However, in order to fully understand how EBV orchestrates successful infection of humans, viral gene function must also be stringently analyzed in the context of the natural host and virus-host interactions in vivo.
Rhesus macaques provide a highly accurate animal system for experimentally modeling EBV infection in humans (30, 38). The nonhuman primate host is closely related to humans, and rhesus macaques are naturally infected with a gammaherpesvirus belonging to the same lymphocryptovirus (LCV) genus as EBV. The rhesus LCV (rhLCV) genome contains a repertoire of genes identical to those of EBV, and several studies indicate that the rhLCV proteins target the same molecular pathways as their EBV homologue (2, 17, 33, 41). The biology of natural rhLCV infection in rhesus macaques is also similar to that of EBV infection in humans; e.g., rhLCV is shed in oral secretions, virtually all adult animals are infected, virus infection persists for life in peripheral blood B cells, and rhLCV-driven B-cell lymphomas can arise in immunosuppressed hosts (21, 30, 36, 38).
Rhesus macaques can be successfully infected by experimental inoculation with an rhLCV laboratory isolate (LCL8664 [35]), and experimental rhLCV infection of healthy, naïve rhesus macaques accurately models acute and persistent EBV infection in humans (30). Experimental infection of immunosuppressed macaques can result in malignant B-cell lymphoproliferation, demonstrating that LCL8664 rhLCV has tumorigenic potential (38). Thus, lymphomagenesis fails to occur after experimental infection of immunocompetent hosts not because the inoculating virus is attenuated and incapable of inducing tumors but because the host immune response controls infection. Rhesus macaques provide a unique animal model system for EBV infection by accurately reproducing the natural interaction between virus and host, resulting in life-long, asymptomatic persistent infection with tumorigenic potential.
The ability to genetically manipulate the LCL8664 rhLCV would provide a major advance in the development of the rhesus animal model. Many different herpesviruses, including EBV, have been cloned as bacterial artificial chromosomes (BACs), resulting in more rapid and widespread research utilizing viral genetics (4, 12, 27, 48). Additional challenges arise when making recombinant LCV, since it is intrinsically difficult to induce lytic LCV replication and LCV is usually recovered and propagated in latently infected, immortalized B cells. In this study, we describe the first molecular rhLCV clone. The rhLCV BAC clone was engineered to create a truncation mutant of the rhBARF1 open reading frame, a homologue for an EBV protein with putative immune evasion function (8, 45, 46). We show that rhBARF1 is capable of blocking colony stimulating factor-1 (CSF-1) cytokine signaling, similar to the EBV BARF1, and that the rhBARF1 truncation engineered into a recombinant rhLCV results in a loss of cytokine-inhibitory function. Development of recombinant rhLCV genetics opens the door for investigating the role of specific LCV genes, such as rhBARF1, in vivo after experimental infection of the natural host.
MATERIALS AND METHODS
Cell culture.
The rhLCV-infected cell line LCL8664 (35) and lymphoblastoid cell lines (LCL) immortalized with natural or recombinant rhLCV were grown in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, and streptomycin at 37°C in a humidified atmosphere with 5% CO2. C33A cells (1), 293 cells (19), and BSC40 (3) were cultured in a similar manner using Dulbecco's modified Eagle medium with the same supplements. BAC1 2F5 cells (kindly provided by E. Richard Stanley) (31) were grown in alpha-minimum essential medium (alpha-MEM), supplemented with 10% fetal bovine serum, penicillin, streptomycin, 50 μM 2-mercaptoethanol, and 36 ng/ml of recombinant human CSF-1 (Cell Science).
Cloning of the rhLCV as a BAC.
A BAC vector containing the F factor sequence required for prokaryotic replication (pGS275) was kindly provided by Greg Smith and Lynn Enquist (44). This vector was modified (BACHT) by replacement of the lacZ cassette with a cytomegalovirus immediate early promoter-driven hygromycin phosphotransferase-thymidine kinase fusion gene derived from tgCMV/HyTK. The F-factor sequence, the chloramphenicol resistance gene for selection in prokaryotic cells, and the hygromycin-phosphotransferase/thymidine kinase fusion gene for positive and negative selection in eukaryotic cells were all surrounded by loxP sites for Cre-mediated removal (Fig. 1A). The BAC vector sequences were inserted into a plasmid containing the EcoRI-G rhLCV DNA fragment in order to generate a targeting plasmid for homologous recombination of the BAC vector into the rhLCV episome (RE1-BACHT). The EcoRI-G DNA fragment overlaps the right end of the BamHI-B DNA fragment (Fig. 1A; also, see Table S1 in the supplemental material) and contains 10 kb of rhLCV DNA, including all DNA from the rhBALF2 open reading frame through the rhLMP1 gene. The BAC vector was cloned into a polylinker inserted into the MluI site within the rhBARF1 open reading frame (ORF) so that approximately 5 kb of rhLCV genome sequence was flanking both sides of the inserted BAC vector to enhance homologous recombination into the rhLCV genome.
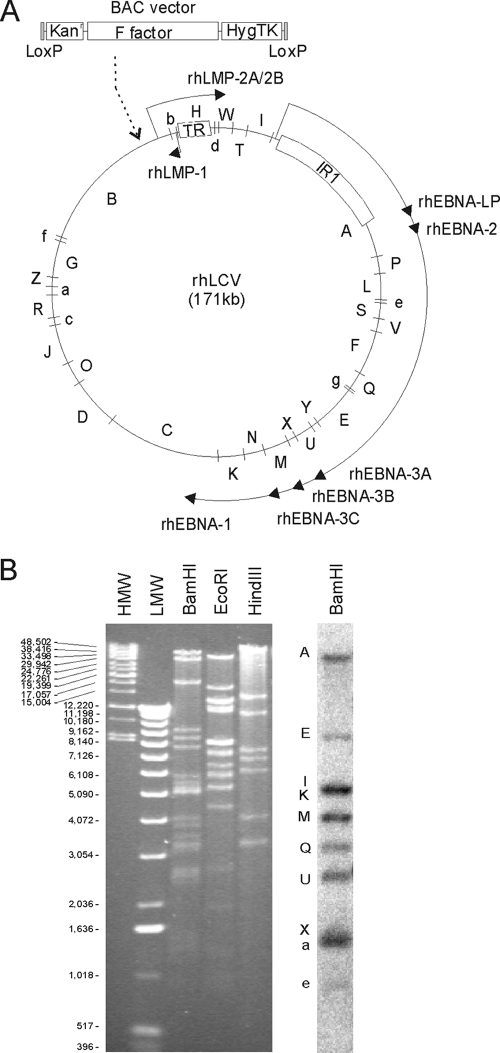
FIG. 1.
Schema of the rhLCV genome and restriction fragment analysis of the clone 16 rhLCV BAC. (A) BamHI restriction map of the LCL8664 rhLCV genome. The relative size and position of each rhLCV BamHI DNA fragment are shown in the LCL8664 rhLCV episome. BAC vector sequences were inserted into the BamHI-B fragment by homologous recombination in LCL8664 cells in order to recover the clone 16 rhLCV BAC. The terminal repeats (TR) and major internal repeat (IR1) are represented by boxes. The major transcriptional units for the latent infection genes are represented by the solid arrows and shown for orientation. (B) Restriction digestion analysis of clone 16 rhLCV BAC. Clone 16 rhLCV BAC DNA digested with BamHI, EcoRI, or HindIII and separated by gel electrophoresis was visualized by ethidium bromide staining and UV illumination (left). The predicted sizes of the BamHI, EcoRI, and HindIII fragments in the LCL8664 rhLCV genome and clone 16 rhLCV BAC are shown in Table S1 in the supplemental material. The relative migration of high-molecular-weight markers (HMW) and low-molecular-weight markers (LMW) is shown. BamHI-digested clone 16 rhLCV BAC DNA was transferred to a nylon membrane and hybridized with a pool of radiolabeled oligonucleotide probes in order to identify the BamHI-A, -E, -I, -K, -M, -Q, -U, -X, -a, and -e DNA fragments (right).
The RE1-BACHT targeting vector was electroporated into LCL8664 cells, and hygromycin-resistant clones were selected. Clones with the BAC vector sequences recombined into the rhLCV episome were identified by Gardella gel electrophoretic separation of viral episomes (18) and hybridization of Southern blots with a radiolabeled probe derived from BAC vector DNA as previously described (28). Hirt DNA (22) was isolated from positive, hygromycin-resistant LCL8664 clones and electroporated into DH10B cells, and BAC DNA was recovered by selection for chloramphenicol-resistant Escherichia coli.
Restriction enzyme digests were performed on BAC DNA extracted from recombinant E. coli with a R.E.A.L. Prep 96 plasmid kit (Qiagen). In order to identify specific rhLCV DNA fragments, Southern blots were hybridized with 32P end-labeled oligonucleotides from the following rhLCV BamHI DNA fragments: BamHI-A, TGGAAGGAGAATGCTTTTATGA; BamHI-E, TCGGGAGGTCGGGCGTA; BamHI-I, TTATTATAGCGCTTGCACCTG; BamHI-K, CCGTTTCTGTGCTAGGTAGGA; BamHI-M, GGGAGGCATCACAATCACAC; BamHI-Q, CATCTGAGGCTGAAGTTACC; BamHI-U, TACCCCTACACACATCCGGT; BamHI-X, GTGGTATCTGGGCGATGAAT; BamHI-a, CTGGCCAGACTGGACGCCTGG; and BamHI-e, CTGGCCAGACTGGACGCCTGG.
Construction of a rescued wild-type (rWT) rhLCV BAC.
The antibiotic selection marker in the original rhLCV BAC clone was changed from chloramphenicol to kanamycin resistance using a plasmid (pKD119)-based system for lambda recombinase-mediated homologous recombination (10) as previously described (7). This change facilitated transfer to chloramphenicol-resistant BM2710 cells used to introduce the BAC into eukaryotic cells (20). The kanamycin-resistant rhLCV BAC clone derived with the RE1-BACHT targeting vector was designated clone 16 rhLCV BAC.
The rWT rhLCV BAC was generated by restoring the rhBARF1 ORF in the clone 16 rhLCV BAC using positive and negative selection with the galactokinase (GalK) recombineering system (49). In clone 16 rhLCV BAC, the BAC vector sequences interrupted the rhBARF1 ORF so that the amino-terminal two-thirds of the ORF was on the left side of the vector insertion and the carboxy-terminal third of the ORF was on the right side of the vector insertion. The overall approach involved two steps employing positive and then negative selection for GalK. First, GalK was targeted to the rhLCV BAC by homologous recombination so that the GalK insertion deleted the 3′ end of the rhBARF1 ORF on the right side of the BAC insertion. Recombinants were identified by positive selection for growth on galactose as the carbon source. The right-hand border of the BAC insertion was then fused to rhLCV sequence starting 86 nucleotides (nt) downstream of the rhBARF1 termination codon by homologous recombination of a synthetic oligonucleotide with the fusion sequence, resulting in GalK gene removal and survival under negative selection with 2-deoxy-galactose, which forms a toxic intermediate when phosphorylated by GalK. In the second step, GalK was reinserted immediately adjacent to the left-hand border of the BAC insertion, and then GalK was replaced by a DNA fragment so that a fully intact rhBARF1 ORF along with 85 nt downstream of the termination codon was restored on the left side of the BAC vector. All manipulations of the BAC were monitored by restriction fragment analysis, PCR amplification, and nucleotide sequencing. This strategy essentially moved the BAC vector insertion downstream of an intact rhBARF1 ORF.
Replication of rhLCV BACs in eukaryotic cells and recovery of recombinant viruses in immortalized B cells.
In order to generate viruses from the BAC clones, an rhLCV BAC was transferred to E. coli BM2710 cells, which can mediate transfer of recombinant DNA into mammalian cells after simple coincubation due to coexpression of the invasin gene from Yersinia pseudotuberculosis and the listeriolysin O gene from Listeria monocytogenes (20). C33A cells were exposed to BM2710 cells carrying a rhLCV BAC, and stable transfectants were selected by culture in medium containing 800 μg/ml of hygromycin. Hygromycin-resistant C33A clones were induced for lytic replication by exposure to 20 ng/ml phorbol 12-myristate 13-acetate, 3 mM butyrate, and recombinant adenoviruses expressing BZLF1 or BRLF1. The amount of lytic replication was measured by immunoblotting with EBV-immune human sera to detect rhLCV lytic infection protein expression. Inducible clones were expanded, and cell-free supernatants from induced cells were used to infect rhesus macaque lymphocytes. Infected cells were cultured for 6 to 8 weeks in RPMI complete medium containing 0.5 μg/ml cyclosporine, and wells containing immortalized LCL were expanded in RPMI complete medium containing 400 μg/ml hygromycin. The BAC vector sequence was excised by electroporation of a Cre expression plasmid (pGS212, kindly provided by Greg Smith and Lynn Enquist) into the LCL established with BAC-derived rhLCV, and transfected cells were selected by culture in RPMI complete medium containing 100 μM ganciclovir.
Southern blotting and PCR analysis of LCL containing BAC-derived rhLCV.
Genomic DNA was isolated from LCL, digested with EcoRI, separated by gel electrophoresis, transferred by Southern blotting, and hybridized with a 32P-labeled rhLCV EcoRI-G DNA probe. PCR analysis of the BAC insertion site from LCL8664 and clone 16 rhLCV was performed with primers RE1-MLU-F0 (TTAGCGTGGTGAAGCCCCTGAC) and RE1-4692 (GTCCCCAGCCCACAGCAAACTAA), and PCR analysis of the BAC insertion site from rWT rhLCV was performed with primers RE1-MLU-F0 and Right check R (CAGGGAGCCTATCCACCGTGGAGCCGGTCT). PCR products were detected by gel electrophoresis, Southern blot transfer, and hybridization with a 32P end-labeled rhBARF1 (GGTACACCACAACAGAG) or loxP (ATAACTTCGTATAGCATACATTATACGAAGTTAT) oligonucleotide probe.
Large-scale virus preparations, titration for transforming units, and measurement of viral DNA units.
Large volumes of LCL8664, clone 16, or rWT rhLCV LCL were grown in RPMI complete medium, harvested, and resuspended in RPMI complete medium with 20 ng/ml phorbol 12-myristate 13-acetate and 3 mM butyrate at 3 × 106 cells/ml. After an overnight incubation, cells were removed from the induction medium and resuspended in RPMI complete medium only. Cell supernatants were harvested after 5 days of culture and passed through a 0.45-μm filter. Virus was pelleted by high-speed centrifugation at 10,000 × g for 2 h at 4°C and resuspended in 1 ml of RPMI complete medium.
Transformation titration assays were performed by making 10-fold serial dilutions of virus from 1:200 to 1:20,000,000 and testing at least five replicates at each dilution with rhesus macaque lymphocytes (200,000/well in a 96-well microtiter plate) cultured with RPMI complete medium and 0.5 μg/ml cyclosporine. Wells with clearly growing LCL were scored microscopically after at least 8 weeks of culture. Transforming units were defined as the reciprocal virus dilution required to achieve a 50% endpoint, i.e., wells containing immortalized cells, by the Reed-Muench calculation.
Real-time DNA PCR was performed using SYBR green (Applied Biosystems) and rhEBER32F (GGAGGAGATGAGTGTGACTTAAATCA) and rhEBER148R (TGAACCGAAGAGAGCAGAAACC) primers. The virus preparations (50, 5, and 0.5 nl of each) were added into replicate PCR mixtures and amplified for 40 cycles (15 s at 95°C, 30 s at 60°C, and 30 s at 72°C). A plasmid containing the rhLCV EBERs (CC1-C) was used as a standard, and plasmid standards from 105 to 101 copies were based on spectrophotometric quantitation of plasmid DNA. All values used for comparison were run in the same assay and averaged, and relative values were expressed as arbitrary DNA units.
Semiquantitative endpoint reverse transcription-PCR for rhLMP2A and GAPDH.
Total RNA was isolated (Trizol) from LCL8664-, rWT-, or clone 16 rhLCV-infected LCL, and 1 μg of RNA was reverse transcribed using Super Script II reverse transcriptase (Invitrogen) and gene-specific primers (rhLMP2A-E2R [GTAACAATGCCGACGAGGAT] and GAPDH-R [CCAGTGGACTCCACGACGTA]) for 50 min at 42°C and 15 min at 72°C. Tenfold serial dilutions of the cDNA (2 μl to 200 pl) were PCR amplified for 35 cycles (15 s at 95°C, 30 s at 55°C, and 30 s at 72°C) using primers specific for exons 1 and 2 of rhLMP2A (rhLMP2A-E1A [GGAATCCACCTCCTTACG] and rhLMP2A-E2R) or GAPDH (GAPDH-F [GCGAGATCCCTCCAAAATCA] and GAPDH-R). PCR products were separated by gel electrophoresis and visualized after ethidium bromide staining.
Recombinant rhLCV protein expression, affinity purification, and CSF-1-dependent growth assay.
The EBV BARF1 ORF was carboxy-terminally tagged with a Flag epitope and cloned into pcDNANeo (pcDNA EBV BARF1 c-Flag). The rhBARF1 ORF from rWT and clone 16 rhLCV was amplified with a carboxy-terminal Flag epitope and cloned into the pSG5 plasmid (pSG5 rWT rhBARF1 or pSG5 clone 16 rhBARF1, respectively). All expression vectors were sequenced to confirm the identity and accuracy of the representative ORF. Recombinant rhBZLF1 and rhBVRF2 were expressed from recombinant vaccinia viruses. The rhBZLF1 and rhBVRF2 ORFs were amino-terminally tagged with a Flag epitope and cloned into the pABT4587 vector. Recombinant vaccinia viruses were generated as previously described (16, 29).
Recombinant EBV BARF1 and rhBARF1 proteins were expressed by transient transfection of expression plasmids into 293 cells with Effectine (Qiagen). After 36 h, supernatants and cell lysates were collected and analyzed by immunoblotting with an anti-Flag monoclonal antibody (M2; Sigma). Recombinant rhBZLF1 and rhBVRF2 were expressed by infection of BSC40 cells with recombinant vaccinia viruses at a multiplicity of infection (MOI) of 10. After 12 h, cell lysates were collected and analyzed by immunoblotting with an anti-Flag monoclonal antibody.
EBV BARF1 and rWT rhBARF1 were affinity purified from cell supernatants. rWT rhBARF1, clone 16 rhBARF1, rhBZLF1, and rhBVDF2 were affinity purified from transfected cells lysed in 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2 mM EDTA, 1% NP-40, and protease inhibitors (Roche). Recombinant proteins were immunoprecipitated with agarose beads coupled to anti-Flag monoclonal antibody and eluted with 3 × Flag peptide (Sigma) in accordance with the manufacturer's instructions. Affinity-purified proteins were quantified using albumin standards after acrylamide gel electrophoresis and silver staining (Bio-Rad).
Assays for CSF-1-dependent growth were performed in duplicate wells using 96-well plates with 5,000 BAC1 2F5 cells/well, 100 μl of alpha-MEM medium with 9 ng/ml CSF-1, and various amounts of recombinant protein. After 3 days, cell growth was measured using a tetrazolium-based colorimetric assay (CCK8; Dojindo Molecular Technologies, Inc.). All assays were correlated with visual microscopic examination of all wells and manual counting of selected wells. The average background value from cells cultured with no CSF-1 was subtracted from all test values. The growth of cells cultured with 9 ng/ml CSF-1 in the absence of recombinant LCV protein represented 100% growth.
RESULTS
Cloning of rhLCV as a BAC.
LCL8664 is a naturally occurring rhLCV strain derived from a B-cell lymphoma arising in a simian immunodeficiency virus-infected macaque (35). The rhLCV genome sequence was determined from LCL8664 rhLCV (40), and LCL8664 rhLCV has been used as the source of rhLCV for experimental animal infections (30). Therefore, we cloned the LCL8664 rhLCV strain as a bacterial artificial chromosome (BAC).
The BAC vector was inserted into the BamHI B DNA fragment of the rhLCV episome by homologous recombination after transfection of a targeting plasmid into the LCL8664 cell line (Fig. 1A). The targeting plasmid contained 10 kb of rhLCV DNA fragment with the BAC vector cloned into the middle of the viral DNA sequence. This targeting plasmid was used because (i) this 10-kb DNA fragment was the first large fragment of rhLCV DNA cloned, (ii) there was a convenient MluI site in the middle of the rhLCV DNA fragment, i.e., there would be equal amounts of viral DNA on both sides of the BAC insertion to facilitate homologous recombination, (iii) the BAC insertion into the MluI site disrupted the rhBARF1 ORF, and (iv) EBV deleted for BARF1 was known to be replication and B cell immortalization competent in vitro (8).
BAC DNA was recovered in E. coli from cell clone 16, in which the BAC vector had recombined into the rhLCV episome. As shown in Fig. 1B, the clone 16 rhLCV BAC showed expected rhLCV DNA fragments by restriction fragment analysis with BamHI, EcoRI, and HindIII (see Table S1 in the supplemental material). Radioactively labeled oligonucleotides with sequences from 10 different rhLCV BamHI DNA fragments (A, E, I, K, M, Q, U, X, a, and e) also hybridized to the expected restriction fragments (Fig. 1B). Of note, the rhLCV BamHI-A oligonucleotide hybridized to the second largest, and not the largest, BamHI DNA fragment in the clone 16 rhLCV BAC. This was consistent with homologous recombination and insertion of the 10-kb BAC vector into the rhLCV BamHI-B fragment, making it larger than the BamHI-A fragment (see Table S1). These studies suggested that the complete rhLCV genome was represented in the clone 16 rhLCV BAC.
BRLF1-dependent replication of the rhLCV BAC in C33A cells.
We transferred the clone 16 rhLCV BAC into 293 cells, since these cells have been used to successfully replicate the EBV BAC into virus (7, 12). However, all attempts to recover virus from 293 cells transfected with clone 16 rhLCV BAC were unsuccessful. Therefore, several other cell types were tested for their ability to replicate the rhLCV BAC. Transforming virus was successfully recovered by introducing the clone 16 rhLCV BAC into the epithelial cell line C33A and inducing cells for lytic replication.
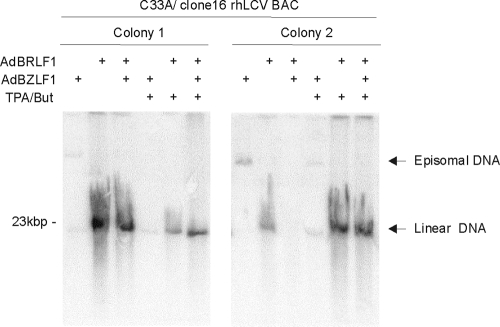
Lytic replication of clone 16 rhLCV BAC in C33A cells was dependent on BRLF1 overexpression. As shown in Fig. 2, lytic replication and accumulation of linear rhLCV DNA was induced by infection with recombinant adenoviruses expressing BRLF1 but not BZLF1. The addition of phorbol ester and butyrate enhanced BRLF1-induced lytic replication in some, but not most, clones (Fig. 2 and data not shown). Thus, lytic replication of the rhLCV BAC was dependent on BRLF1 overexpression in C33A cells, whereas our experience with a B95-8 EBV BAC showed that virus was readily induced in 293 cells with BZLF1 expression and/or phorbol-butyrate treatment (7). LF2, which is deleted in the B95-8 EBV (23, 32, 34), has been shown to inhibit BRLF1 transactivation (5). The LCL8664 rhLCV has a full-length genome and encodes an rhLF2 (40). Thus, clone 16 rhLCV BAC can express rhLF2, LF2 can inhibit BRLF1-induced transactivation, and BRLF1 overexpression may overcome the natural rhLF2 inhibition of lytic replication in epithelial cells.
FIG. 2.
BRLF1-dependent lytic replication of clone 16 rhLCV BAC in C33A cells. Two individual colonies of C33A cells stably carrying clone 16 rhLCV BAC were induced for lytic viral replication, and viral DNA separated by Gardella gel electrophoresis of induced cells was detected on Southern blots. Cells were induced for lytic replication by various combinations of adenoviruses expressing BZLF1 (AdBZLF1) or BRLF1 (AdBRLF1) and chemical treatment with phorbol ester and butyrate (TPA/But). Migration of the 23-kb DNA marker is shown on the left.
Recovery of BAC-derived virus in immortalized B cells and excision of BAC vector sequences.
Virus produced from clone 16 rhLCV BAC in C33A cells was recovered by infection of rhesus macaque lymphocytes with cell-free supernatants and establishment of immortalized LCL. These LCL were hygromycin resistant (data not shown), as expected for cells immortalized with BAC-derived virus. As shown in Fig. 3A, the BAC vector insertion introduces several new EcoRI sites splitting the original 10-kb EcoRI-G rhLCV DNA fragment. Southern blot analysis showed that a 10-kb fragment was detected when EcoRI-digested genomic DNA from an LCL infected with LCL8664 rhLCV was hybridized with a radiolabeled EcoRI-G DNA probe (Fig. 3B). However, in the LCL immortalized with virus derived from the clone 16 rhLCV BAC, the EcoRI-G DNA probe detected two smaller fragments around 5 kb and no 10-kb DNA fragment. These data were consistent with the presence of BAC-derived virus, and the absence of wild type rhLCV, in the LCL infected with virus derived from the clone 16 rhLCV BAC.
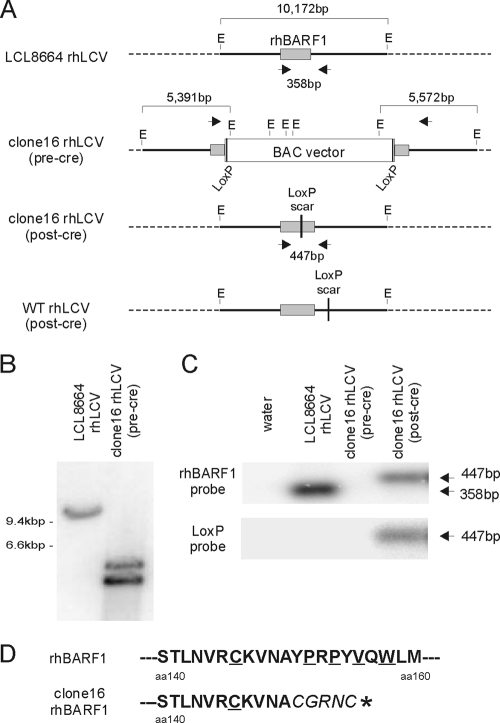
FIG. 3.
Recovery of BAC-derived clone 16 rhLCV in an LCL, Cre-mediated excision of BAC vector sequences from the viral episome, and predicted amino acid sequence for the truncated rhBARF1 ORF. (A) Schematic of the EcoRI-G rhLCV DNA fragment containing the rhBARF1 ORF in LCL8664 rhLCV, BAC-derived clone 16 rhLCV before (pre-cre) and after (post-cre) Cre-mediated excision, and post-Cre, BAC-derived rWT rhLCV constructed by repairing the rhBARF1 ORF from clone 16 rhLCV BAC. EcoRI digestion sites (E) and locations of PCR primers (arrowheads) are shown. (B) Confirmation of recombinant rhLCV DNA in an LCL infected with clone 16 rhLCV (pre-Cre). Genomic DNA from an LCL immortalized with LCL8664 rhLCV or pre-Cre clone 16 rhLCV was digested with EcoRI, and Southern blots were hybridized with a radiolabeled EcoRI-G rhLCV DNA fragment to detect restriction fragment polymorphisms associated with the insertion of the BAC vector in the viral DNA. (C) PCR confirmation of successful Cre-mediated excision of the BAC vector in the rhLCV clone 16 LCL. Genomic DNA from LCL immortalized with LCL8664 rhLCV, pre-Cre clone 16 rhLCV, or post-Cre clone 16 rhLCV was amplified with the primers shown in panel A. PCR products were hybridized with a radiolabeled oligonucleotide probe derived from the rhBARF1 ORF (top) or the loxP sequence (bottom). (D) Effects of the loxP scar sequence on the rhBARF1 amino acid sequence from clone 16 rhLCV. PCR amplified DNA from LCL infected with clone 16 rhLCV was sequenced, and amino acid sequences from residues 140 to 160 are shown. The loxP scar sequence results in an in-frame insertion of five new amino acids (italicized in the bottom sequence) and a premature termination codon (*). rhBARF1 amino acid residues highly conserved in CSF-1R and other c-Fms family members are underlined.
Leaving the BAC vector in other herpesviruses can have adverse effects when the recombinant virus is used to infect the natural host (6). Therefore, we removed the BAC vector from the BAC-derived rhLCV molecular clone by transfecting and expressing Cre recombinase in the LCL. The Cre recombinase excised the BAC vector DNA flanked by loxP sites, and successful Cre-mediated recombination was selected for by cell growth in ganciclovir-containing medium, since the BAC vector contained a hygromycin-thymidine kinase fusion gene.
Cre-transfected, ganciclovir-resistant cells were analyzed by PCR to confirm removal of the BAC vector. PCR primers located on either side of the BAC vector insertion site (Fig. 3A) amplified 358 bp of viral DNA from LCL8664 rhLCV that hybridized with a rhBARF1 but not a loxP probe (Fig. 3C). The same primers failed to amplify a visible product of any size from the LCL immortalized with clone 16 rhLCV BAC before transfection with a Cre expression plasmid (Fig. 3C and data not shown). This was likely due to the large target size created by the BAC vector insertion (>10 kbp), resulting in less efficient PCR amplification. However, in the Cre-transfected, ganciclovir-resistant LCL, a PCR product that hybridized with both rhBARF1 and loxP probes was detected (Fig. 3C). As expected, this PCR fragment was slightly larger than the LCL8664 rhLCV product due to the loxP scar sequence and polylinker predicted to remain after Cre-mediated excision. Sequencing of the PCR products confirmed successful excision of BAC vector sequences and insertion of loxP scar and polylinker sequences, resulting in a frameshift after amino acid 150 in rhBARF1, the introduction of five new amino acids, and a premature termination codon (Fig. 3D). Thus, the BAC-derived, Cre-excised rhLCV molecular clone (now referred to as clone 16 rhLCV) should express a mutant rhBARF1 deleted for the domain homologous to the CSF-1 receptor (CSF-1R; conserved residues in the homology domain underlined in Fig. 3D) and 70 carboxy-terminal residues out of 220 total amino acids.
Construction of a rescued wild-type rhLCV BAC by repair of the clone 16 rhLCV BAC.
In order to construct a rescued wild-type (rWT) rhLCV BAC representing the LCL8664 strain, the rhBARF1 ORF was repaired in the clone 16 rhLCV BAC. In brief, this was accomplished by first removing the carboxy-terminal portion of the rhBARF1 ORF on the right side of the BAC vector insertion and then restoring a complete rhBARF1 ORF on the left side of the BAC vector insertion. Techniques employing positive and negative selection for galactokinase (49) were used to repair the rhBARF1 ORF and essentially move the BAC vector 85 nt downstream of an intact rhBARF1 ORF.
The rWT rhLCV BAC was transferred to C33A cells, where lytic replication was induced, and BAC-derived virus was recovered by peripheral blood mononuclear cell (PBMC) infection and B-cell immortalization. LCL were transfected with the Cre expression plasmid, and rWT rhLCV with the BAC vector excised was recovered in ganciclovir-resistant LCL. The rWT rhLCV contains the same loxP scar sequence that differs only in its location, being within the rhBARF1 ORF and 958 nt upstream of the rhLMP2A initiator codon in clone 16 versus 3′ to the rhBARF1 ORF and 654 nt upstream of the rhLMP2A initiator codon in the rWT virus (Fig. 3A). The viral genome from the beginning of rhBARF1 to rhLMP2A was sequenced from both recombinant viruses and was identical to LCL8664 rhLCV, except for the presence of the scar sequence in the appropriate locations. Thus, the rWT rhLCV represents a full-length LCL8664 molecular clone, as well as a wild-type rescue clone for the rhBARF1 mutation in clone 16 rhLCV.
Efficiency of B-cell immortalization and lytic replication by the rWT and clone 16 rhLCV.
rWT and clone 16 rhLCV were competent for viral replication and B cell immortalization based on the successful recovery from BAC-transfected C33A cells. More quantitative experiments were performed to determine whether the rWT rhLCV was comparable to the natural LCL8664 rhLCV in terms of viral replication and B-cell-immortalizing properties and to evaluate whether the mutation in clone 16 rhLCV was associated with any detectable change in viral replication or B-cell immortalization. Multiple preparations of LCL8664, rWT, and clone 16 rhLCV were produced from large batches of LCL induced for lytic replication with phorbol ester- and butyrate-containing medium. Multiple virus-producing cell lines were used to collect LCL8664, rWT, and clone 16 virus preparations (three, two, and three different LCL, respectively). Virus was concentrated from cell-free supernatants by high-speed centrifugation and resuspended in 1 ml of medium. Real-time PCR was used to determine the relative amount of viral DNA in each virus preparation. Multiple replicates of serial dilutions from all virus preparations were tested in the same real-time PCR assay for experiments 1 and 2, and the results were expressed as arbitrary DNA units (Fig. 4A, columns 5 and 10). Overall, there was no significant difference in the yield of viral DNA recovered from an equivalent number of LCL8664-, rWT-, or clone 16 rhLCV-infected LCL (data not shown), suggesting no major differences in viral replication.
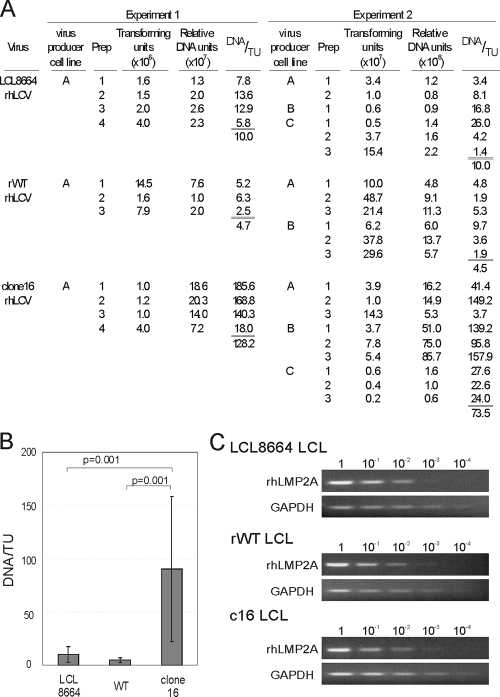
FIG. 4.
Comparison of B-cell-transforming activity by LCL8664, rWT, and clone 16 rhLCV. (A) Transforming activity in multiple virus preparations of LCL8664, rWT, and clone 16 rhLCV. Different virus-producing LCL infected with LCL8664 (lines A, B, and C), rWTrhLCV (lines A and B), and clone 16 rhLCV (lines A, B, and C) were used to make multiple virus preparations. Transforming activity (DNA/TU) was evaluated by calculating the number of relative viral DNA units required per transforming unit (TU). Virus preparations were PCR amplified together in two separate experiments, and the relative DNA units were normalized so that the transforming activity of the naturally occurring LCL8664 rhLCV was 10 DNA units/TU. (B) The efficiency of transforming activity (DNA/TU) for all virus preparations in experiments 1 and 2 is graphed. The overall average DNA/TU for LCL8664 rhLCV was 10.0 (n = 10), 4.6 for rWT rhLCV (n = 9), and 90.3 for clone 16 rhLCV (n = 13). P values (t test) less than 0.05 are shown. (C) rhLMP2A mRNA expression in LCL8664-, rWT-, and clone 16 rhLCV-infected LCL. Total RNA from LCL was reverse transcribed with gene-specific primers, and 10-fold serial dilutions (1 to 10−4) of cDNA were PCR amplified for rhLMP2A or GAPDH.
The efficiency of B-cell immortalization was quantified by calculating the number of DNA units required for each transforming unit. Tenfold serial dilutions of each virus preparation were tested for B-cell immortalization with multiple replicates at each virus dilution, and the number of transforming units (TU) per ml of virus preparation was established as the inverse dilution required for B-cell immortalization in 50% of the replicates (Fig. 4A, columns 4 and 9). The relative DNA units were normalized so that 10 DNA units of LCL8664 rhLCV was required for each transforming unit. In comparison, an average of 4.7 and 4.5 DNA units was required for each transforming unit of rWT rhLCV in experiments 1 and 2, respectively. The average result from these two experiments (4.6 DNA units/TU) was not significantly different from that for LCL8664 rhLCV (Fig. 4B), suggesting that the transforming efficiency of the cloned virus derived from rWT rhLCV BAC was comparable to that of the naturally derived strain LCL8664. In contrast, 128.2 and 73.5 DNA units were required for one TU of clone 16 rhLCV in experiments 1 and 2. The average for all clone 16 rhLCV virus preparations (90.3 DNA units/TU) was 9 times higher than for LCL8664 rhLCV and 19.6 times higher than for rWT rhLCV (Fig. 4B), suggesting that a higher MOI was required to immortalize PBMC with clone 16 than with LCL8664 or rWT rhLCV. The differences between clone 16 rhLCV and LCL8664 and between clone 16 rhLCV and rWT rhLCV were statistically significant (P = 0.001 [Fig. 4B]). These results suggested that rhBARF1 may enhance B-cell immortalization, since the mutated rhBARF1 in clone 16 rhLCV was associated with a slight reduction in transformation efficiency when assayed at a limiting MOI. The scar sequence in clone 16 and rWT rhLCV did not affect LMP2A expression, since LMP2A RNA levels were comparable in LCL immortalized with LCL8664, rWT, and clone 16 rhLCV, as measured by semiquantitative reverse transcriptase-mediated PCR assays (Fig. 4C). rhBARF1 could contribute to B-cell immortalization due to a direct effect in the infected B cell or an indirect effect through noninfected cells, such as T cells or monocytes, since unfractionated lymphocytes were used for these assays and BARF1 is rapidly secreted.
Blockade of CSF-1 signaling is functionally conserved in rhBARF1.
The clone 16 and rWT rhLCVs are isogenic except for rhBARF1, and different outcomes after experimental rhesus macaque infections would be linked to the functional differences in their rhBARF1 genes. EBV BARF1 has been implicated as an immune evasion gene during EBV infection of humans because it can bind colony-stimulating factor 1 (CSF-1) and competitively inhibit cytokine signaling through the CSF-1 receptor. In order to test whether rhBARF1 can block CSF-1 signaling and to determine the effect of the rhBARF1 truncation in clone 16 rhLCV, recombinant expression vectors for EBV BARF1, rhBARF1, and truncated rhBARF1 present in clone 16 rhLCV were constructed. All ORFs were tagged with a Flag epitope at the carboxy terminus. Recombinant expression vectors were transfected into 293 cells.
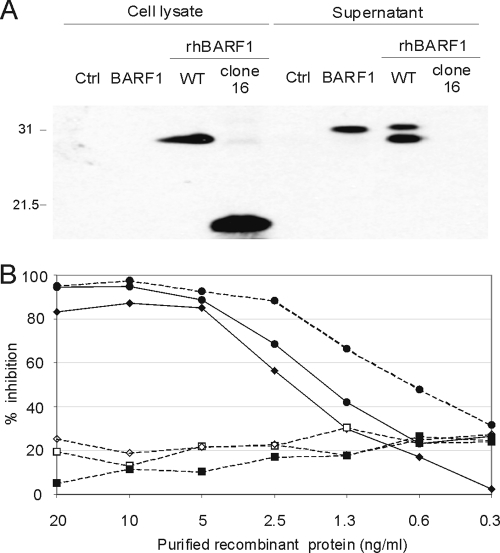
Transfected cell lysates and supernatants were examined by anti-Flag immunoblotting for expression of the recombinant proteins. As shown in Fig. 5A and previously reported (45), EBV BARF1 was readily secreted from cells; i.e., it was easily detected in cell supernatants but was not detected in cell lysates. Similarly, rhBARF1 was easily detected in the cell supernatant, but a significant fraction was also detected in the cell lysate, suggesting that rhBARF1 was not as rapidly secreted as the EBV protein (Fig. 5A). Secreted rhBARF1 also migrated in some blots as a doublet which may represent phosphorylation on serine and threonine or N and O glycosylation, as previously reported for EBV BARF1 (13). In contrast, truncated rhBARF1 from clone 16 rhLCV was detected only in the cell lysate and was not detected in the cell supernatant. Thus, the inability of clone 16 rhBARF1 to reach the extracellular space may contribute to a loss of function in vivo.
FIG. 5.
Wild-type rhBARF1, but not the truncated rhBARF1, is readily secreted and capable of blocking CSF-1-dependent cell growth. (A) Expression of recombinant BARF1 homologues from EBV (BARF1), rWT rhLCV (rWT rhBARF1), and clone 16 rhLCV (clone 16 rhBARF1). ORFs were tagged with a carboxy-terminal Flag epitope, and expression vectors, or a pSG5-Flag vector control (Ctrl), were transiently transfected into 293 cells. Protein expression in cell lysates or cell-free supernatants was detected by immunoblotting with an anti-Flag monoclonal antibody. Relative migration of 31- and 21.5-kDa molecular mass markers is shown. (B) Inhibition of CSF-1-dependent cell growth by wild-type BARF1 and rhBARF1. BARF1 and rhLCV proteins were immunoprecipitated with an anti-Flag antibody from cell supernatants or cell lysates of transfected 293 cells or recombinant vaccinia virus-infected BSC40 cells. Decreasing amounts of recombinant protein were added to duplicate microwells of CSF-1-dependent BAC1 2F5 cells supplemented with 9 ng/ml of recombinant CSF1, and cell growth was measured by a colorimetric assay after 3 days. The background value from BAC1 2F5 cells without CSF-1 was subtracted from all values, and maximal growth was derived from BAC1 2F5 cells supplemented with CSF-1 only. The growth inhibition after addition of BARF1 (closed diamonds), rWT rhBARF1 (closed circles; from supernatant and cell lysate), clone 16 rhBARF1 (closed squares), rhBZLF1 (open squares), and rhBVDF1 (open diamonds) is plotted for each dose of recombinant protein. Proteins affinity purified from cell supernatants are shown with a solid line, and proteins affinity purified from cell lysates are shown with a dotted line.
Recombinant proteins were also tested for the ability to block growth of the CSF-1-dependent cell line BAC1 2F5. Recombinant BARF1 and rhBARF1 were affinity purified from transfected cell supernatants by anti-Flag immunoprecipitation. Recombinant clone 16 rhBARF1 was affinity purified by anti-Flag immunoprecipitation from transfected cell lysates. As controls, rhBARF1 was also immunoprecipitated from transfected cell lysates, and recombinant Flag tagged rhBZLF1 and rhBVRF2 were affinity purified by anti-Flag immunoprecipitation from BSC40 cells infected with the respective recombinant vaccinia viruses. Decreasing amounts of affinity-purified recombinant protein were added to replicate cultures of BAC1 2F5 cells supplemented with recombinant human CSF-1, and cell proliferation after 3 days was measured by a colorimetric assay for cell growth.
Recombinant rhBZLF1 and rhBVRF2 had a minimal effect on CSF-1-dependent growth of BAC1 2F5 cells, approximately 20% inhibition of BAC1 2F5 growth in the presence of CSF-1 with no added recombinant viral protein, and this effect was not significantly dose dependent (Fig. 5B, open diamonds and squares). In contrast, 20, 10, and 5 ng/ml of either BARF1 or rhBARF1 inhibited more than 90% of CSF-1-induced BAC1 2F5 cell growth (Fig. 5B, closed diamonds or circles and solid lines). The effect was dose dependent with a 50% mean inhibitory concentration of slightly less than 2.5 ng/ml for both BARF1 and rhBARF1. The truncated rhBARF1 from clone 16 rhLCV had no significant inhibitory effect (Fig. 5B, closed squares) and was indistinguishable from control rhBZLF1 and rhBVRF2 proteins. rhBARF1 purified from cell lysates also inhibited CSF-1-dependent growth (Fig. 5B, closed circles and dotted line), indicating that the purification procedure from transfected cell lysates was not responsible for the loss of rhBARF1 function. Thus, functional inhibition of CSF-1 signaling has been conserved in rhBARF1, and truncated rhBARF1 from clone 16 rhLCV was functionally defective in both its ability to inhibit CSF-1 induced signaling and its ability to be readily secreted from the cell.
DISCUSSION
We describe the first molecular clone for the EBV-related rhLCV and a BAC-based system to make recombinant rhLCV for reverse genetic studies in the rhesus animal model. Our BAC clone was derived from the same rhLCV isolate, LCL8664, that results in persistent infection after oral inoculation of immunocompetent macaques and is tumorigenic after experimental infection of immunodeficient macaques (30, 38). The LCL8664 rhLCV strain has been fully sequenced, and restriction enzyme analysis of the rhLCV BAC was consistent with a complete viral genome. The rhLCV BAC could be readily manipulated in E. coli, and the rhBARF1 ORF in the clone 16 rhLCV BAC was fully repaired to generate a wild-type BAC clone for rhLCV. The BAC-derived rWT rhLCV replicated and immortalized B cells with an efficiency comparable to that of the naturally derived LCL8664 rhLCV strain, suggesting that this rhLCV molecular clone will be a suitable prototype for reverse genetic studies in the rhesus macaque animal model for EBV infection.
Interestingly, we found that the rhLCV BAC was more difficult to replicate in epithelial cells than the EBV BAC, but this could be overcome by overexpressing BRLF1. We did not directly test the effect of BRLF1 expression in B cells infected with the recombinant rhLCV, since transforming virus could be obtained by scaling up LCL culture volumes without additional BRLF1. The requirement for additional BRLF1 expression to induce replication in epithelial cells may be due to LF2, which is present in the rhLCV BAC but has been deleted from the B95-8 EBV strain. Thus, the full repertoire of viral genes in the LCL8664 rhLCV strain may have presented a technical hurdle unanticipated from work with the B95-8 EBV BAC. Having a full viral repertoire of viral genes represented in the rhLCV BAC will be important for in vivo experiments.
In these studies, we also characterized a recombinant rhLCV with a loss-of-function rhBARF1 mutation. BARF1 may be a particularly interesting LCV gene to study in vivo. BARF1 was initially described as a lytic infection mRNA expressed with early gene characteristics (52). Wei et al. subsequently showed that BARF1 transfection conferred increased tumorigenicity to rodent fibroblasts (51) and later showed that BARF1 transfection could also transform growth of EBV-negative B lymphoma cells (50). The same group has shown that the amino-terminal 54 amino acids of BARF1 were sufficient to both alter cell growth properties and induce bcl2 expression (43).
It is not obvious how growth-transforming properties of a lytic infection protein might contribute to B-cell immortalization and malignancy associated with latent EBV infection. However, more recent studies suggest that BARF1 expression may not be limited to lytic infection. BARF1 mRNA can be detected in the absence of lytic replication from tissue culture LCL and peripheral blood lymphocytes of EBV-seropositive humans, as well as EBV-associated nasopharyngeal carcinoma and gastric carcinoma tissues (11, 15, 26, 42). This raises the tantalizing hypothesis that BARF1 effects on cell growth may have an important role in latently infected cells and contribute to EBV-induced B-cell immortalization, EBV persistence in the peripheral blood, or EBV-induced malignancy.
Other investigators determined that BARF1 had sequence homology to a conserved domain in the CSF-1 receptor (45). They showed that a recombinant immunoglobulin protein fused to EBV BARF1 could block CSF-1-dependent proliferation of murine bone marrow cells. Thus, it was hypothesized that BARF1 was a viral immune evasion protein important for blocking CSF-1-dependent signaling. Expression during lytic replication, when a large number of immunogenic viral proteins are produced, and the rapid secretion of soluble BARF1 that could act on immune cells can be easily incorporated into an immune evasion model for BARF1 function during EBV infection of humans.
BARF1 also shows striking sequence homology with CD80, a cell surface protein important for delivering T-cell-costimulatory signals. BARF1 structural studies confirmed a three-dimensional homology between the viral protein and the T-cell-costimulatory receptor, CD80, suggesting that secreted BARF1 may be capable of inhibiting T-cell responses through blockade of CD80 costimulation (46). Interestingly, a recent clinical trial showed that treatment with belatacept, a blocking monoclonal antibody for CD80, was associated with a marked increase of EBV-associated lymphoproliferative disease in renal transplant patients, providing clinical evidence that blocking CD80 costimulation can functionally inhibit EBV immunity (14, 47). Despite the apparent clinical importance of CD80-mediated costimulation for effective EBV immunity and the striking structural homology between BARF1 and CD80, no published studies have provided laboratory evidence that BARF1 can functionally inhibit CD80-mediated costimulation of T cells. It is not clear whether the homology between BARF1 and CD80 is coincidental, whether experimental designs with BARF1 and CD80 have been inadequate to demonstrate a functional effect, or whether BARF1 targets a closely related family member instead of CD80.
Thus, in vitro studies implicate three potential functions for BARF1: (i) altering cell growth, (ii) inhibiting innate immunity by blockade of CSF-1, and (iii) blocking CD80 costimulation of T cells. We believe that evolution supports the hypothesis that BARF1 was acquired by LCV primarily for immune evasion purposes. Every EBV gene has a homologue in rhLCV, but we found 11 EBV/rhLCV genes that had no homologue in the marmoset LCV, i.e., 11 viral genes were acquired during LCV evolution from New World nonhuman primates to Old World nonhuman primates and humans (39). Viral genes important for fundamental properties common to all LCV, e.g., B-cell tropism and B-cell immortalization, should already be present in New World LCV. In contrast, the evolution of a more complex immune system from marmosets to humans may have forced LCV to acquire more sophisticated immune evasion genes to successfully infect more highly evolved primates. Two other LCV genes acquired during the evolution from New to Old World primates support this hypothesis, BCRF1 (vIL-10) and BNLF2a. The function of BNLF2a was unknown at the time the marmoset LCV was sequenced, but since then BNLF2a has been identified as an inhibitor of HLA class I surface expression and antigen presentation (9, 25).
The present study demonstrates that the ability to block CSF-1 signaling has been functionally conserved in rhBARF1, and rhBARF1 appears to be as potent as EBV BARF1. This is consistent with BARF1 structural studies which predicted that the CSF-1R and CD80 homology should be conserved in the rhBARF1 (46). We have been unable to determine whether growth-transforming properties are conserved in rhBARF1 because we have as yet been unable to demonstrate similar effects from EBV BARF1. These types of studies may be highly dependent on the specific cells and conditions used for the cell growth assays. It is interesting that we find a decrease in B-cell-immortalizing efficiency by recombinant clone 16 rhLCV, which carries the truncated rhBARF1. Our results are not inconsistent with the previous report that EBV deleted for BARF1 was immortalization competent (8). Our rhBARF1 mutant is also immortalization competent and indistinguishable from natural or rWT rhLCV in routine B-cell immortalizations performed at a high multiplicity of infection, but a subtle phenotypic difference was detected when more quantitative assays were performed at a limiting MOI. It remains to be determined whether the slightly reduced transforming activity is due to loss of an indirect effect of rhBARF1 on other cell types in the immortalization assay or due to a more direct effect of rhBARF1 on the immortalized B cells. We developed these quantitative assays to determine titers of rhLCV because they did not require a foreign marker such as a fluorescent protein that may complicate interpretations of animal infections, e.g., immune responses to GFP, and they offer the capability of quantitating more subtle effects on B-cell immortalization.
The clone 16 rhLCV, which has a truncated rhBARF1, will be an interesting mutant to study in experimental infection of rhesus macaques. We demonstrated that the mutated rhBARF1 has lost CSF-1 blocking function. We predict that the truncation would also destroy CD80 homology. In both cases, the lack of efficient secretion would also prevent functional blockade between cells. The truncated rhBARF1 is predicted to retain growth-transforming properties, since the amino-terminal 54 amino acids of BARF1 are reported to be sufficient for growth transformation and bcl2 induction (43).
How might experimental infection of rhesus macaques be affected by the loss of rhBARF1 function in the clone 16 rhLCV? In experimental oral inoculations of naïve rhesus macaques, it takes approximately 21 days for LCL8664 rhLCV to penetrate the oral mucosa and become detectable by DNA PCR in the peripheral blood (38). This is in line with classical epidemiologic studies where sailors developed symptoms of infectious mononucleosis approximately 42 days after leaving port (24). This eclipse period, from viral inoculation to weeks 3 to 6, when viremia or symptoms of acute primary infection can be first identified, is virtually impossible to study in humans. However, this is a critical period in the virus life cycle, when the virus must overcome obstacles not present during persistent infection nor easily reproduced in tissue culture, e.g., mucosal penetration, viral amplification, and invasion of the peripheral blood compartment. The critical events taking place during the eclipse period required for successful EBV infection of humans remain to be defined. rhBARF1 expression and immune evasion could play an important role if lytic replication is an important component for penetration of the oral cavity and viral expansion into the peripheral blood. If the rhBARF1 immune evasion function is lost, then lytic replication may be slowed by a more effective immune response during the eclipse phase, i.e., aborted or delayed appearance of detectable virus in the peripheral blood.
Alternatively, rhBARF1 immune evasion may be important for virus transmission, when lytic replication is required for virus shedding into oral secretions. If rhBARF1 is mutated and lytically infected cells are more susceptible to immune control, then less virus may be shed and effective transmission may be at peril. In this case, evolution may select for viruses with BARF1 immune evasion, since viruses that transmit more readily will have a strong biological advantage.
Another possibility is that BARF1 may be expressed during latent infection, and the detection of BARF1 transcripts in the peripheral blood of healthy, EBV-seropositive donors may suggest that either the immune evasion function or cell growth effects are important for successful persistence in peripheral blood B cells. Experimental infection of rhesus macaques with the clone 16 rhLCV, which carries a truncated rhBARF1, will reveal whether the loss of immune evasion function might be important for successful virus entry during acute primary infection, for persistent B-cell infection, or for virus reactivation, shedding, and transmission. A genetic system for generating recombinant rhLCV ushers in a new era for studying the role of specific LCV genes during infection of a natural host.
Supplementary Material
Acknowledgments
This work was supported by grants from the U.S. Public Health Service (CA68051 and DE18926). Services from the New England Primate Research Center were supported by a base grant to the institution (USPHS P51RR00168).
We thank Greg Smith and Lynn Enquist for providing the original BAC vector that was subsequently modified for constructing the rhLCV BAC; Amir Moghaddam, Young-gyu Cho, Pierre Rivaillier, and Mark Fogg for providing technical and scientific input; Angela Carville for providing rhesus macaque lymphocytes; and E. Richard Stanley for providing the BAC1 2F5 cells.
Footnotes
Published ahead of print on 17 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Auersperg, N. 1964. Long-term cultivation of hypodiploid human tumor cells. J. Natl. Cancer Inst. 32:135-163. [PubMed] [Google Scholar]
- 2.Blake, N. W., et al. 1999. Inhibition of antigen presentation by the glycine/alanine repeat domain is not conserved in simian homologues of Epstein-Barr virus nuclear antigen 1. J. Virol. 73:7381-7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockman, W. W., and D. Nathans. 1974. The isolation of simian virus 40 variants with specifically altered genomes. Proc. Natl. Acad. Sci. U. S. A. 71:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brune, W., M. Messerle, and U. H. Koszinowski. 2000. Forward with BACs: new tools for herpesvirus genomics. Trends Genet. 16:254-259. [DOI] [PubMed] [Google Scholar]
- 5.Calderwood, M. A., A. M. Holthaus, and E. Johannsen. 2008. The Epstein-Barr virus LF2 protein inhibits viral replication. J. Virol. 82:8509-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, W. L., and P. A. Barry. 2003. Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J. Virol. 77:5073-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, A., M. Divisconte, X. Jiang, C. Quink, and F. Wang. 2005. Epstein-Barr virus with the latent infection nuclear antigen 3B completely deleted is still competent for B-cell growth transformation in vitro. J. Virol. 79:4506-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, J. I., and K. Lekstrom. 1999. Epstein-Barr virus BARF1 protein is dispensable for B-cell transformation and inhibits alpha interferon secretion from mononuclear cells. J. Virol. 73:7627-7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croft, N. P., et al. 2009. Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle. PLoS Pathog. 5:e1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decaussin, G., F. Sbih-Lammali, M. de Turenne-Tessier, A. Bouguermouh, and T. Ooka. 2000. Expression of BARF1 gene encoded by Epstein-Barr virus in nasopharyngeal carcinoma biopsies. Cancer Res. 60:5584-5588. [PubMed] [Google Scholar]
- 12.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Turenne-Tessier, M., and T. Ooka. 2007. Post-translational modifications of Epstein Barr virus BARF1 oncogene-encoded polypeptide. J. Gen. Virol. 88:2656-2661. [DOI] [PubMed] [Google Scholar]
- 14.Durrbach, A., et al. 2010. A phase III study of belatacept versus cyclosporine in kidney transplants from extended criteria donors (BENEFIT-EXT study). Am. J. Transplant. 10:547-557. [DOI] [PubMed] [Google Scholar]
- 15.Fiorini, S., and T. Ooka. 2008. Secretion of Epstein-Barr virus-encoded BARF1 oncoprotein from latently infected B cells. Virol. J. 5:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogg, M. H., A. Kaur, Y. G. Cho, and F. Wang. 2005. The CD8+ T-cell response to an Epstein-Barr virus-related gammaherpesvirus infecting rhesus macaques provides evidence for immune evasion by the EBNA-1 homologue. J. Virol. 79:12681-12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franken, M., et al. 1996. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J. Virol. 70:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardella, T., P. Medveczky, T. Sairenji, and C. Mulder. 1984. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 50:248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 20.Grillot-Courvalin, C., S. Goussard, F. Huetz, D. M. Ojcius, and P. Courvalin. 1998. Functional gene transfer from intracellular bacteria to mammalian cells. Nat. Biotechnol. 16:862-866. [DOI] [PubMed] [Google Scholar]
- 21.Habis, A., G. B. Baskin, M. Murphey-Corb, and L. S. Levy. 1999. Simian AIDS-associated lymphoma in rhesus and cynomolgus monkeys recapitulates the primary pathobiological features of AIDS-associated non-Hodgkin's lymphoma. AIDS Res. Hum. Retroviruses 15:1389-1398. [DOI] [PubMed] [Google Scholar]
- 22.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 23.Hitt, M. M., et al. 1989. EBV gene expression in an NPC-related tumour. EMBO J. 8:2639-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoagland, R. J. 1955. The transmission of infectious mononucleosis. Am. J. Med. Sci. 229:262-272. [DOI] [PubMed] [Google Scholar]
- 25.Horst, D., et al. 2009. Specific targeting of the EBV lytic phase protein BNLF2a to the transporter associated with antigen processing results in impairment of HLA class I-restricted antigen presentation. J. Immunol. 182:2313-2324. [DOI] [PubMed] [Google Scholar]
- 26.Houali, K., et al. 2007. A new diagnostic marker for secreted Epstein-Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clin. Cancer Res. 13:4993-5000. [DOI] [PubMed] [Google Scholar]
- 27.Kanda, T., M. Yajima, N. Ahsan, M. Tanaka, and K. Takada. 2004. Production of high-titer Epstein-Barr virus recombinants derived from Akata cells by using a bacterial artificial chromosome system. J. Virol. 78:7004-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazzara, G. P., A. Destree, and A. Mahr. 1993. Generation and analysis of vaccinia virus recombinants. Methods Enzymol. 217:557-581. [DOI] [PubMed] [Google Scholar]
- 30.Moghaddam, A., et al. 1997. An animal model for acute and persistent Epstein-Barr virus infection. Science 276:2030-2033. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, C., J. W. Pollard, and E. R. Stanley. 1987. Isolation and characterization of a cloned growth factor dependent macrophage cell line, BAC1.2F5. J. Cell. Physiol. 130:420-427. [DOI] [PubMed] [Google Scholar]
- 32.Parker, B. D., A. Bankier, S. Satchwell, B. Barrell, and P. J. Farrell. 1990. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology 179:339-346. [DOI] [PubMed] [Google Scholar]
- 33.Peng, R., et al. 2000. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J. Virol. 74:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raab-Traub, N., T. Dambaugh, and E. Kieff. 1980. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell 22:257-267. [DOI] [PubMed] [Google Scholar]
- 35.Rangan, S. R., L. N. Martin, B. E. Bozelka, N. Wang, and B. J. Gormus. 1986. Epstein-Barr virus-related herpesvirus from a rhesus monkey (Macaca mulatta) with malignant lymphoma. Int. J. Cancer 38:425-432. [DOI] [PubMed] [Google Scholar]
- 36.Rao, P., H. Jiang, and F. Wang. 2000. Cloning of the rhesus lymphocryptovirus viral capsid antigen and Epstein-Barr virus-encoded small RNA homologues and use in diagnosis of acute and persistent infections. J. Clin. Microbiol. 38:3219-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rickinson, A., and E. Kieff. 2001. Epstein-Barr virus, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 38.Rivailler, P., et al. 2004. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood 104:1482-1489. [DOI] [PubMed] [Google Scholar]
- 39.Rivailler, P., Y. G. Cho, and F. Wang. 2002. Complete genomic sequence of an Epstein-Barr virus-related herpesvirus naturally infecting a new world primate: a defining point in the evolution of oncogenic lymphocryptoviruses. J. Virol. 76:12055-12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rivailler, P., H. Jiang, Y. G. Cho, C. Quink, and F. Wang. 2002. Complete nucleotide sequence of the rhesus lymphocryptovirus: genetic validation for an Epstein-Barr virus animal model. J. Virol. 76:421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivailler, P., C. Quink, and F. Wang. 1999. Strong selective pressure for evolution of an Epstein-Barr virus LMP2B homologue in the rhesus lymphocryptovirus. J. Virol. 73:8867-8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seto, E., et al. 2005. Epstein-Barr virus (EBV)-encoded BARF1 gene is expressed in nasopharyngeal carcinoma and EBV-associated gastric carcinoma tissues in the absence of lytic gene expression. J. Med. Virol. 76:82-88. [DOI] [PubMed] [Google Scholar]
- 43.Sheng, W., G. Decaussin, S. Sumner, and T. Ooka. 2001. N-terminal domain of BARF1 gene encoded by Epstein-Barr virus is essential for malignant transformation of rodent fibroblasts and activation of BCL-2. Oncogene 20:1176-1185. [DOI] [PubMed] [Google Scholar]
- 44.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strockbine, L. D., et al. 1998. The Epstein-Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J. Virol. 72:4015-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tarbouriech, N., F. Ruggiero, M. de Turenne-Tessier, T. Ooka, and W. P. Burmeister. 2006. Structure of the Epstein-Barr virus oncogene BARF1. J. Mol. Biol. 359:667-678. [DOI] [PubMed] [Google Scholar]
- 47.Vincenti, F., et al. 2010. A phase III study of belatacept-based immunosuppression regimens versus cyclosporine in renal transplant recipients (BENEFIT study). Am. J. Transplant. 10:535-546. [DOI] [PubMed] [Google Scholar]
- 48.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 49.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei, M. X., J. C. Moulin, G. Decaussin, F. Berger, and T. Ooka. 1994. Expression and tumorigenicity of the Epstein-Barr virus BARF1 gene in human Louckes B-lymphocyte cell line. Cancer Res. 54:1843-1848. [PubMed] [Google Scholar]
- 51.Wei, M. X., and T. Ooka. 1989. A transforming function of the BARF1 gene encoded by Epstein-Barr virus. EMBO J. 8:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, C. X., G. Decaussin, J. Daillie, and T. Ooka. 1988. Altered expression of two Epstein-Barr virus early genes localized in BamHI-A in nonproducer Raji cells. J. Virol. 62:1862-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.