Abstract
In order to analyse the frequency of point mutations in whole plants, several constructs containing single nonsense mutations in the β-glucuronidase (uidA) gene were used to generate transgenic Arabidopsis thaliana plants. Upon histochemical staining of transgenic plants, sectors indicative of transgene reactivation appeared. Reversion frequencies were in the range of 10–7–10–8 events per base pair, exceeding the previous estimates for other eukaryotes at least 100-fold. The frequency was dependent on the position of the mutation substrate within the transgene and the position of the transgene within the Arabidopsis genome. An inverse relationship between the level of transgene transcription and mutation frequency was observed in single-copy lines. DNA-damaging factors induced the mutation frequency by a factor of up to 56 for UV-C, a factor of 3 for X-rays and a factor of 2 for methyl methanesulfonate. This novel plant mutation-monitoring system allowed us to measure the frequencies of point mutation in whole plants and may be used as an alternative or complement to study the mutagenicity of different environmental factors on the higher eukaryote’s genome.
Keywords: Arabidopsis/frequency/mutation/somatic
Introduction
All organisms suffer from a certain number of mutations as a result of normal cellular operations or interactions with the environment. A point mutation can be caused by either of two types of events: chemical modification of DNA directly changes the base, or a malfunction during DNA replication causes insertion of an incorrect base.
Previous studies have shown that mutation frequencies vary between organisms, between different genomic loci of a particular organism (Wolfe et al., 1989; Lichtenauer-Kaligis et al., 1996) and between different nucleotides of the same gene (Coulondre et al., 1978). Calculations of the mutation rate were usually based on either gain or loss of function of a particular gene. As summarized by Lewin (1997), spontaneous mutations in bacteria that inactivate gene function occur at a rate of ∼10–5–10–6 events per gene per generation. The rate of back mutation is correspondingly lower than that of forward mutation, typically by a factor of 10; for instance, the rate of loss of the ability to ferment lactose in Escherichia coli is 2 × 10–6, whereas the gain of this function is 2 × 10–7 per cell division or 10–10 per nucleotide (Klug and Cummings, 1986). Studies performed on mice and zebra fish using the lacI and rspL gene as a transgene, respectively, yielded mutation frequencies in the range of 0.6 × 10–5–1.7 × 10–5 per transgene, depending on the tissue analysed (Kohler et al., 1991; Amanuma et al., 2000).
Not only do organisms differ from each other in their spontaneous mutation frequency, but various agents also increase mutation frequencies differently (Sander et al., 1978; Katoh et al., 1994). Although many systems based on unicellular organisms are available for mutagenicity tests, assays using higher eukaryotes are of special relevance. The spontaneous mutation frequency in the tested transgenes was induced significantly by different mutagens in mice and fish (Kohler et al., 1991; Amanuma et al., 2000).
Somatic mutation events are of particular importance in plants, since they do not have a pre-determined germ line but form their reproductive structures from somatic meristems late in development. Thus, any somatic mutation potentially can be passed on to subsequent generations. In contrast to animals, plants cannot avoid the influence of environmental factors due to their settled life and therefore probably require a system dedicated to maintaining genome stability. Unfortunately, up to now, no system was available to measure somatic mutation events in plants.
In the present study, we undertook the development of such a system. Amber, opal and ochre stop codons were introduced at five different positions into the β-glucuronidase (uidA) gene. These termination codons completely prevented translation of active protein. Transgenic Arabidopsis thaliana plants carrying these inactivated uidA genes were generated. We observed spontaneous restoration of uidA activity due to reversion of the stop codons to the original codons. Since generation of transgenic plants is not limiting, we could analyse reversion frequencies in the non-essential test gene integrated into different chromosomal regions. A large variation of the mutation frequency of individual single-copy transformants could be determined, whereby the frequencies exhibited an inverse correlation to steady-state transcriptional levels of the transgenes. The frequency of reversions was increased in response to different DNA-damaging agents to different extents. Thus, with this system, the endogenous level of mutations within the transgene in A.thaliana plants can be easily and reliably detected. In addition, the influence of known mutagens on the frequency of point mutations was determined.
Results
Generation of transgenic plants harbouring a uidA gene inactivated by nonsense mutations
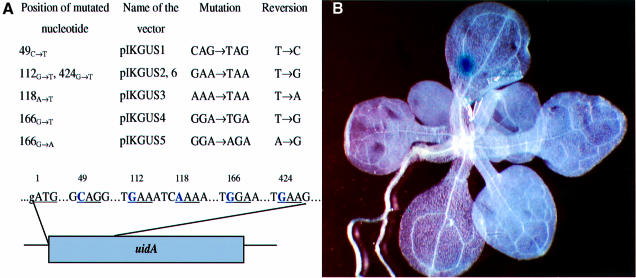
Using site-directed mutagenesis, three different stop codons were introduced into five sites of the 5′-proximal part of the uidA gene and the resulting constructs were inserted into a binary vector plasmid and introduced into A.thaliana ecotype Columbia plants (see Materials and methods) (Figure 1A). Sequence context has been taken into consideration to prevent leakage of the stop codon; in all cases, a stop codon was generated in a triplet followed by adenine or guanine, which had been shown to be essential for efficient termination of translation (Atkinson and Martin, 1994; Betzner et al., 1997). Reversion to the original nucleotide or mutation to another amino acid that allows protein function was expected to restore the activity of the gene. In addition, one construct causing an amino acid substitution (GGA to AGA in position 166, coding for glycine in place of arginine) was chosen for plant transformation (Figure 1A). This allowed us to study the frequency of reversions of A to G, in addition to the mutations from T to C, A or G.
Fig. 1. (A) Stop codons generated in the open reading frame of the uidA gene. Nucleotides shown in bold were mutated by site-directed mutagenesis. Numbers represent the position of the nucleotides in the open reading frame (ORF) starting from A of the ATG start codon. In-frame triplets are underlined. (B) Stop codon reversion visualized in a 4-week-old A.thaliana plant (subline 166G→T-1).
In order to evaluate whether mutations to new amino acids could restore the activity of the gene, triplets with changes of single nucleotides coding for possible amino acid substitutions were generated in place of the three stop codons described above (Table I). Nicotiana plumbaginifolia protoplasts were transfected with equal amounts of DNA from different constructs. The activity of the uidA gene was tested using a fluorometric assay (Rossi et al., 1993). None of the amino acid substitutions gave rise to an activity >2.2% of the respective non-mutated version. These background levels may be due to a low activity of the protein containing a mutated amino acid. The protein apparently can be activated only via reversion to the original amino acid, and thus only the original nucleotide was expected to be found in revertants, at least at the three analysed positions.
Table I. Enzymatic activity of mutant uidA proteins.
| 112 (pIKGUS2) | 118 (pIKGUS3) | 166 (pIKGUS4) | |
|---|---|---|---|
| Endogenous codon | GAA (100.0%) (Glu) | AAA (100.0%) (Lys) | GGA (100.0%) (Gly) |
| Mutant codons | TAA (<1.0%) stop | TAA (<1.0%) stop | TGA (<1.0%) stop |
| CAA (<1.0%) (Gln) | GAA (<1.0%) (Glu) | CGA (1.2%) (Arg) | |
| AAA (<1.0%) (Lys) | CAA (1.4%) (Gln) | AGA (1.3%) (Arg) | |
| TAT (<1.0%) (Tyr) | TAT (<1.0%) (Tyr) | TGC (1.7%) (Cys) | |
| TCA (1.5%) (Ser) | TAC (1.9%) (Tyr) | TGG (<1.0%) (Trp) | |
| TTA (2.2%) (Leu) | TCA (<1.0%) (Ser) | TCA (1.6%) (Ser) | |
| TTA (<1.0%) (Leu) | TTA (<1.0%) (Leu) |
The headings of the columns are the numbers of the first nucleotide in the mutated triplet. Nucleotides in bold are point mutations introduced into the uidA ORF. The relative β-glucuronidase activity of each mutant measured in a transient protoplast assay is presented in parentheses.
For analysis of mutation frequency, several homozygous lines, called sublines, were generated for every uidA mutant.
Frequencies of reversions in A.thaliana
The activity of the uidA gene can be monitored easily by histochemical staining of plant tissue (Jefferson, 1987). Staining of whole uidA stop codon-containing plants revealed the presence of blue sectors on a white background (Figure 1B). To test whether the blue sectors indeed represent reversion events, plant material from blue sectors obtained after staining was excised from ∼1000 plants of the line 166G→T. DNA prepared from this tissue was used to amplify the 5′ part of the uidA gene by PCR. PCR products were cloned and sequenced. Out of 250 independent clones, 98 contained reversions to the original sequence of the gene while 152 had retained the stop codon, thus reflecting the heterozygous genotype for reversion and nonsense codon, respectively. Interestingly, two of the clones contained a second mutation in a codon adjacent to the reverted stop codon. Sixteen clones derived from white tissue all contained the stop codon.
Plants from selected transgenic lines were grown either on sterile medium or on soil under standard conditions (Materials and methods). Frequencies of mutations of selected transgenic plant lines were determined in 300–1000 histochemically stained plants per line, by relating the number of blue spots to the total number of stained plants and to the number of transgene copies (fp/c; frequency per plant per one uidA gene copy) (see definitions in Materials and methods).
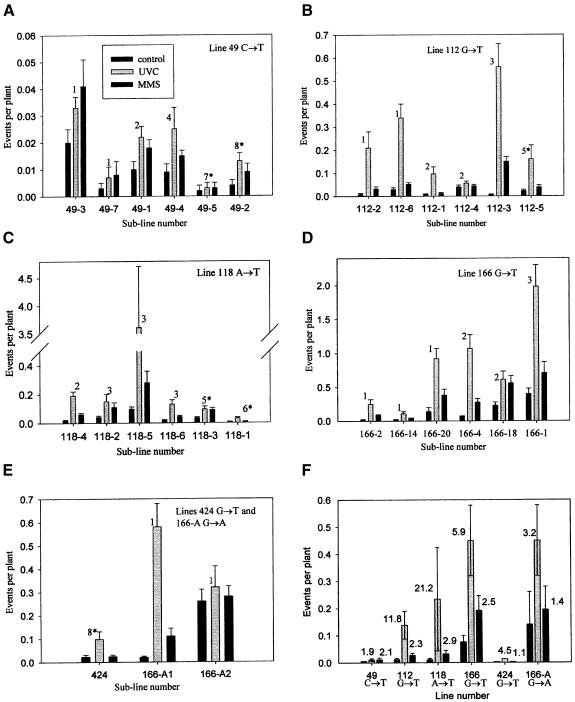
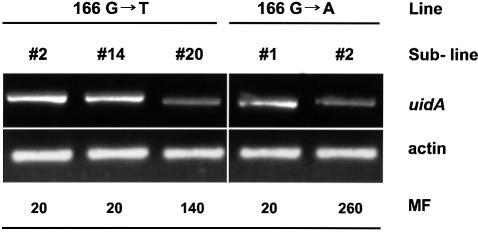
The fp/c varied between plants harbouring different transgenic constructs (Table II; Figure 2A–E). Different sublines harbouring the same transgene at presumably different chromosomal positions also exhibited a wide range of mutation frequencies. The fp/c varied from 0.0003 to 0.14 in sublines 49C→T-5 and 166G→T-20, respectively. The most important observation is that the frequency of reversion between sublines, in which identical single- copy transgenes are located in different positions in the genome, varied significantly. For example, two sublines of the 166G→A line with one copy of the transgene each differed in reversion frequency by a factor of 13; subline 166G→T-20 of line 166G→T revealed 0.14 reversion events per plant, whereas subline 166G→T-2, also with one copy of the transgene, reverted with an fp/c of 0.02. Interestingly, we observed a negative correlation between the mutation frequency and the level of transgene expression (Figure 3). This was obvious in single-copy sublines of line 166G→T where the transcription level was higher for sublines 166G→T-2 and 166G→T-14, but lower for subline 166G→T-20. The inverse relationship was also evident in sublines of the 166G→A line: the transcription level of the transgene in subline 166G→A-1 exhibiting lower mutation frequency was lower than in subline 166G→A-2 with the higher mutation frequency (Figure 3).
Table II. Frequency of spontaneous and induced reversions in different A.thaliana lines carrying mutated uidA genes.
| Line and subline | Copynumber | fp/c | Fold induction | |
|---|---|---|---|---|
| |
|
|
UV-C |
MMS |
| 49C→T-3 | 1 | 0.020 | 1.5 | 2.0 |
| 7 | 1 | 0.003 | 3.3 | 3.3 |
| 1 | 2 | 0.005 | 2.0 | 2.0 |
| 4 | 4 | 0.003 | 3.0 | 2.0 |
| 5 | 7a | 0.0003 | 1.5 | 1.5 |
| 2 | 8a | 0.0005 | 2.5 | 2.5 |
| 112G→T-2 | 1 | 0.010 | 21.0 | 3.0 |
| 6 | 1 | 0.030 | 11.3 | 1.7 |
| 1 | 2 | 0.005 | 10.0 | 1.0 |
| 4 | 2 | 0.020 | 1.5 | 1.0 |
| 3 | 3 | 0.003 | 56.0 | 15.0 |
| 5 | 5a | 0.004 | 8.0 | 2.0 |
| 118A→T-4 | 2 | 0.005 | 19.0 | 6.0 |
| 2 | 3 | 0.013 | 3.8 | 2.8 |
| 5 | 3 | 0.033 | 36.1 | 2.8 |
| 6 | 3 | 0.007 | 6.5 | 2.0 |
| 3 | 5a | 0.006 | 3.3 | 3.0 |
| 1 | 6a | 0.002 | 3.0 | 1.0 |
| 166G→T-2 | 1 | 0.020 | 12.5 | 4.0 |
| 14 | 1 | 0.020 | 5.5 | 2.0 |
| 20 | 1 | 0.140 | 6.6 | 2.7 |
| 4 | 2 | 0.030 | 17.8 | 4.7 |
| 18 | 2 | 0.120 | 2.7 | 2.4 |
| 1 | 3 | 0.130 | 5.0 | 1.8 |
| 424G→T-1 | 8a | 0.003 | 4.5 | 1.1 |
| 166G→A-1 | 1 | 0.020 | 29.0 | 5.5 |
| 2 | 1 | 0.260 | 1.2 | 1.1 |
Single transgene locus lines are in bold.
aComplex integration pattern with possibly multiple loci.
Fig. 2. (A–E) Frequencies of spontaneous and induced reversions in sublines of lines 49C→T, 112G→T, 118A→T, 166G→T, 424G→T and 166G→A. The numbers on the top of the bars represent copy numbers of the transgenes; the transgenes in the sublines marked with an asterisk are integrated in complex patterns of single or multiple loci. (F) Average frequencies of reversions as calculated per plant per single transgene copy. Bars represent average frequencies of reversions with the standard error. Numbers at the top of the bars represent the fold induction.
Fig. 3. Steady-state level of the uidA transcript in single-copy sublines of lines 166G→T and 166G→A. MF, mutation frequency, presented as number of events per 1000 plants.
The higher the transgene copy number, the more possible targets exist for reversion to the dominant phenotype. Therefore, the frequency of spontaneous point mutations was expected to depend on the copy number of the analysed gene in the genome. However, only for the 166G→T line were the mutation frequencies higher in sublines with two and three copies of the transgene than in most of the single-copy lines (Table II; Figure 2). For none of the other lines could we notice any correlation between the copy number of the mutation target and the reversion frequency; for example, subline 112G→T-5 with five copies of the transgene had a frequency of reversion (0.02 spots per plant) comparable with that of the single-copy subline 112G→T-6 (0.03 spots per plant). This indicates that other factors, such as location of the integrated gene, the type and position of the mutation within the gene or silencing effects, also influence the mutation frequency.
Indeed, the position within the uidA gene of the mutating bases also influences the average fp/c. For instance, most sublines of line 49C→T exhibited a low mutation frequency, averaging 0.005 ± 0.003; sublines of 166G→T showed an average fp/c of 0.076 ± 0.024 (Figure 2F).
UVC, MMS and X-rays increased reversion frequencies to different extents
To investigate whether induced DNA damage would increase the mutation frequency (as required for a mutagen-monitoring system), we applied UV-C, X-rays and methyl methanesulfonate (MMS) to the plants carrying the nonsense mutations. We used mutagen doses just below those that caused visible damage. For optimal sensitivity, mutagen application had to be late during development so that many genomes and transgene copies per plant were already present to detect possible reversions (see Materials and methods).
Irradiation of plants with UV-C (1000 J/m2 for 30 s) caused a significant increase in mutation frequency in all lines (up to 56-fold), with the exception of most sublines of line 49C→T, which showed a very low level of induction of mutations (Figure 2). Such differences may be explained by the specificity of the UV-C irradiation targets. The creation of CT and TT dimers at the target nucleotide may influence the mutation frequencies at that site. Indeed, reversion frequencies in lines 112G→T, 118A→T and 166G→T, in which pyrimidine dimer formation (CT or TT) between the first nucleotide in the nonsense codon (T) and the 5′ adjacent nucleotide (C or T) is a possibility, are increased by exposure to UV-C to higher extents than in lines 49C→T and 166G→A (Figures 1A and 2).
Plants grown on liquid media with MMS (50 µM) also exhibited increased mutation frequencies in comparison with control plants (Figure 2). Treatment with MMS stimulated the reversion frequencies by factors of 1.1–5.5 in different lines. The average level of induction of reversions varied between 1.4- and 2.9-fold and thus was most probably independent of the type of stop codon and the type of mutated nucleotide (T or A). Irradiation with X-rays (absorbed dose of 25 Gy) induced mutations to a similar level of 1.4- to 4.8-fold (data not shown).
Discussion
Transgenic A.thaliana plants containing a chimeric uidA gene inactivated by nonsense mutations were used to assay the frequency of reversion events. The restoration of a functional uidA gene by reversion led to a gene product that could be detected by histochemical staining. This allowed us to visualize and quantify reversion events in whole plants. The results obtained with this set-up led to the following conclusions: (i) all mutations analysed were precise reversions to the endogenous nucleotide; (ii) the spontaneous reversion frequency was in the range 10–7–10–8 events per base pair; (iii) the reversion frequency was most probably dependent on the position of the transgene in the genome and the position and type of the stop codon introduced into the uidA gene; (iv) the levels of transgene transcription were generally lower in the single-copy lines that exhibited higher levels of reversion frequency; and (v) the reversion frequency increased upon treatment with UV-C, MMS and X-rays to up to 10–6 events per base pair; the extent of this increase was dependent on the mutagen applied.
The reversion assay
For the reversion assay to function, two main criteria had to be fulfilled. The leakage of the stop codon mutations had to be low in comparison with the expression of reverted sequences, and the reversion frequency had to be high enough to yield scorable events in reasonably sized plant populations. Indeed, both requirements were met. Upon histochemical staining, all ‘mutation plants’ were white, indicating that there was very little read-through translation of the stop codons. This was confirmed by measuring enzyme activity in protoplasts transfected with the mutated genes (Table I). Since the activity of the transgenes with stop codons did not differ markedly from those with amino acid substitutions, we consider mutation to a suppressor tRNA recognizing the stop codon as less frequent than mutation of the stop codon. This is consistent with the finding that deliberate introduction of suppressor tRNA genes, even on a strong promoter, yielded low activity of a stop codon-mutated uidA gene (Betzner et al., 1997). Moreover, all analysed sequences that did not retain the stop codon showed reversion to the original nucleotide. These results thus indicate that each of the mutated triplets codes for amino acids critical for the proper function of the enzyme.
Mutation frequencies in plants and other organisms
The spontaneous somatic reversion frequency in the uidA transgene in Arabidopsis was measured to be in the range of 10–7–10–8 events per base pair (Materials and methods). This can be translated into a frequency of forward mutations of 10–6–10–7 events per base pair. Of special interest is the comparison of our results with data on somatic mutation in animal systems as well as with mutation frequencies in bacteria and yeast (Table III). Comparison of the experimental series shows that the mutation frequency in plants is two to three orders of magnitude higher than in the other tested organisms. This may point to higher sensitivity or precision of the system we have used and/or indeed to a much higher frequency of somatic mutations in plants.
Table III. Comparison of somatic mutation frequencies in different organisms.
| Species | fb | Reference |
|---|---|---|
| Bacteria | ∼6 × 10–10 | Hall (1991) |
| Yeast | ∼7 × 10–10 | Galli and Schiestl (1999) |
| Mouse | ∼5 × 10–10 | Kohler et al. (1991) |
| Gondo et al. (1996) | ||
| Rat | ∼4 × 10–9 | Dycaico et al. (1994) |
| Parsons and Heflich (1997) | ||
| Fish | ∼3 × 10–10 | Amanuma et al. (2000) |
| Arabidopsis | 10–6–10–7 | this study |
fb = mutation frequency, number of mutation events per base pair of analysed gene, normalized to one copy of the (trans)gene; if mutation frequency was given per gene analysed, the number was divided by 1000, the average in base pairs of a (trans)gene.
It should be considered that the measurements in the rodent and fish systems were indirect; they required packaging of the transgene into λ phage or electroporation of rescued plasmids with subsequent propagation in E.coli, where extra mutations within the lacI and rpsL test genes may have accumulated (Kohler et al., 1991; Gondo et al., 1996; Amanuma et al., 2000). An additional problem may be the presence of two to 30 to 350 copies of the transgene as targets for mutations in the quoted animal systems (Kohler et al., 1991; Gondo et al., 1996; Amanuma et al., 2000). In contrast, for the calculation of the mutation frequency in Arabidopsis, only plants with a single or few transgene copies were included. On the other hand, the comparatively high somatic mutation frequencies, as measured by us, may indeed be plant specific. Plants may have a higher genomic flexibility since, due to their settled life, they cannot avoid being exposed to various environmental mutagenic factors. A higher mutation frequency and higher repair capacity in plants may be advantageous in evolution.
In contrast to somatic mutation, the germline mutation frequency in plants resembles that of animals. Mutations in single genes causing visible phenotypes have been measured to be in the range of 10–5–10–6 per gamete per generation in Zea mays (Stadler, 1930). These rates amount to 10–8–10–9 changes per base pair in a gene of 1000 nucleotides. The rate of deleterious mutations in Arabidopsis was calculated to be ∼10–9 per base pair per year (Wolfe et al., 1987; Schultz et al., 1999). The germline mutation frequency in transgenic rodents, as calculated for a single transgene copy, was ∼10–9 per base pair (Kohler et al., 1991; Dycaico et al., 1994). Mutations causing inherited human genetic diseases were in the range of 10–8–10–9 per base pair (Atkinson and Martin, 1994). It may not be surprising that the germline mutation frequency in higher eukaryotes is similar, as these similarities may reflect the necessity for precise transmission of the genetic information to subsequent generations.
The reversion frequency is dependent on the chromosomal location of the transgene, possibly at the level of transgene transcription and the mutated base within the transgene
Surprisingly, mutation frequencies in individual sublines, all carrying identical stop codons, differed dramatically. The copy number of the transgenes may have affected these differences. In most cases, however, higher copy numbers apparently influenced the frequency of mutation negatively. This may be due to gene silencing of the transgenes and, therefore, may be an indirect effect, which was not analysed further. Reversion frequencies in single-copy transgenes, however, are directly comparable and were found to differ drastically between transformants (see Table II). Since the transgenes almost certainly integrated in different chromosomal sites, the mutation frequency in these particular transformants may be a function of the chromosomal position of the transgene. This finding is reminiscent of data on intrachromosomal homologous recombination in transgenes located in different chromosomal environments in tobacco (Puchta et al., 1995) and may reflect different structural or dynamic properties of the chromatin at the locus of the transgene. Influences of these properties on mutation behaviour may be expected at either of two levels: accessibility for damage (except for cases of base misincorporation upon DNA replication) and accessibility for repair.
At least part of this accessibility may be due to the DNA unwinding upon transcription. In order to address this question, we analysed steady-state levels of uidA mRNA and noted a negative correlation between the mutation frequency and the transcription level in single-copy lines. Thus, highly transcribed regions may be repaired more efficiently, leading to lower mutation levels in comparison with the regions with a low transcription level. Turker et al. (1993) found a 4-fold lower frequency of accumulation of mutations in the transcribed mouse adenine phosphoribosyltransferase gene than in a contiguous non- transcribed downstream region. In addition, the level of mutations in non-coding regions may generally be higher than in coding regions, because the latter are under some selective pressure (Wolfe et al., 1989). Substitution frequencies for several groups of physically linked genes were found to be nearly identical for genes in the same linkage group but different between different groups (Wolfe et al., 1989). The expression of the uidA gene, used in our study, is regulated by the strong and constitutive viral 35S promoter. Generalization of our uidA mutation frequencies may therefore actually yield an underestimated general mutation frequency. We believe that the data on the frequency of mutations obtained from the transgenic assay are representative of those of endogenous genes. Indeed, the frequencies of mutations within the uidA transgene were found to be in the same range as those found for several Arabidopsis genes, i.e. 10–7 per base pair (calculated from McCallum et al., 2000).
Another important finding was the difference in the reversion frequency in various locations of the transgene itself. The susceptibility of one or the other nucleotide to DNA-damaging agents most probably is dependent on the sequence context. Mutations were shown frequently to cluster around pyrimidine dimer sites creating hotspots for mutation (Hutchinson, 1987), as discussed below.
UV-C, MMS and X-rays increase the reversion frequency to different extents
Irradiation of the plants by UV-C led to an increase in the reversion frequencies by factors of 2.0–56.0, whereas MMS increased the frequency by only 1.1–5.5 times. This drastic difference could originate from the kind of damage created by different agents and depend on the repair system involved in the process. Under strongly mutagenic conditions, many systems of the cell may be recruited collectively to the site of damage.
UV-C has two major effects: at the DNA level it creates pyrimidine dimers; at the level of the cell it leads to production of free radical species. Even one single dimer was shown to be able to abolish expression of a transfected gene (Protic-Sabljic and Kraemer, 1985). Pyrimidine dimers can be repaired via different mechanisms such as photoreactivation, base excision repair (BER), nucleotide excision repair (NER) or dimer bypass repair (Britt, 1995). Repair by photolyase involves interception of the bridge between two pyrimidines, thus restoring the original sequence, whereas repair by the other mechanisms may lead to mutation. Excision repair, in contrast to photoreactivation, does not directly reverse DNA damage, but instead substitutes the damaged nucleotides with new, undamaged ones and thereby causes transitional strand breaks (Britt, 1995). Free radicals, when converted to toxic forms such as hydroxyl radicals, can directly damage DNA, creating strand breaks. In addition, repair via illegitimate recombination repair of strand breaks introduced close to the mutated nucleotide can lead to misincorporation of nucleotides and thereby to the restoration of the original sequence of the uidA gene. Indeed, 10- to 100-fold higher frequencies of reversions were obtained in irradiated lines in which a pyrimidine was located immediately 5′ of the stop codon (Figure 1A). Such clustering of mutations around pyrimidine dimer sites has also been found to create hotspots in E.coli (Hutchinson, 1987).
The influence of MMS on the frequency of reversions was notably different from that of UV-C. MMS, a monoalkylating agent with affinity for nucleophilic centres in organic macromolecules, has been shown previously to induce point mutations (Singer, 1986). Although target sites exist in all nucleotides, MMS causes changes primarily in purines (Pegg, 1984; Singer, 1986). Never theless, we noted an increase in T:A→G:C transversions and A:T→G:C transitions in plants exposed to MMS. If alkylated purines close to the thymidine in the stop codon are repaired through NER, possible mistakes during the repair could cause reversion to the original sequence of the uidA gene. On the other hand, alkylation of the adenine located on the opposite strand could result in an A:T→C:G transversion, which restores the endogenous sequence. In sharp contrast to the UV-C-treated population, induction of point mutations by MMS was nearly equal in all tested lines, confirming predictions made above.
Interestingly, upon irradiation of plants with X-rays (25 Gy), a powerful DNA-damaging agent, we could detect only a small increase in frequency of mutations as compared with the influence of UV-C (data not shown). X-rays are known to create mostly strand breaks, which are repaired by illegitimate or homologous recombination. Our point mutation system, which ‘measures’ direct damage to nucleotides, most probably could not detect strand breaks created by X-rays, at least under the conditions used. The small increase in reversion frequencies that we could observe may be due to direct damage of nucleotides or to non-precise repair of strand breaks. Indeed, recombinational repair of double-strand breaks (DSBs), traditionally believed to be an error-free DNA repair pathway, was recently shown to increase the frequency of mutations in a nearby gene; the reversion frequency of Saccharomyces cerevisiae trp1 alleles (either nonsense or frameshift) near an HO-endonuclease cleavage site was increased at least 100-fold in cells that have experienced an HO-mediated DSB (Holbeck and Strathern, 1997).
The influence of different mutagens thus far was analysed qualitatively. For the introduction of our system as a mutagenicity test, a dose–response relationship will of course have to be established.
Outlook
The genetic variation created in part through point mutation is a prerequisite of both natural and artificial selection. Therefore, understanding the mechanisms of genetic change is relevant to both theoretical evolution and genetic engineering. In addition, the transgenic plants constitute the first system to study point mutations in situ in the whole organism. Our ‘mutation plants’ will serve as a useful tool in investigating the general mechanisms of DNA damage and repair in plants.
Materials and methods
Generation of the stop codon constructs
Three stop codons were introduced by site-directed mutagenesis into five different positions of a 35S promoter-driven β-glucuronidase gene. In order to increase the functional efficiency of the newly generated stop codons, the context of nucleotides in the neighbourhood of the new stop codons was taken into consideration (Angenon et al., 1990; Stansfield et al., 1995). In all cases, the base directly following the newly synthesized terminator codon was adenine or guanine. Mutations were introduced by PCR into the uidA gene contained in the pGUS23 vector (Puchta and Hohn, 1991) (Figure 1A). PCR fragments were cut with AccI endonuclease at the promoter region and with BsmI, BclI or SnaBI at the coding region, and ligated into the uidA gene replacing the endogenous sequence. Mutations were confirmed by sequencing. The mutated genes were excised with EcoRI endonuclease and inserted into the binary vector pJL 513, which was partially digested with EcoRI. This vector, a gift from J.Lucht, contains as a transformation marker the bar gene, the gene coding for resistance to the herbicide ‘BASTA’, under the control of the 1′ promoter. The resulting plasmids, called pIKGUS1–6, contained six different mutations (Figure 1A).
In addition, 17 mutations were generated in three different positions of the uidA gene (pIKGUS2, 3 and 4) in order to assess whether missense mutations would restore β-glucuronidase activity (Table I).
Plant transformation and growth
Mutated uidA gene constructs cloned in E.coli–Agrobacterium shuttle vectors were mobilized into Agrobacterium tumefaciens, as described by Mattanovich et al. (1989). The resulting strains were used to transform A.thaliana ecotype Columbia, as described by Bent and Clough (1998), with some modifications. Vacuum-infiltrated plants were grown in growth chambers at 22°C with a 16 h light:8 h dark regime until seed set. Seeds were germinated on soil and treated at the age of 7, 12 and 20 days with the herbicide BASTA (0.075%) to rescue the plants in which T-DNA integration had occurred. Pure lines homozygous for the transgenes (only single-copy ones) were generated upon repeated selfing. For the estimation of spontaneous mutation frequency, plants of different transgenic lines were grown on sterile MM+S medium or on soil in isolated growth chambers at 22°C with a 16 h light:8 h dark regime.
Histochemical staining
Histochemical staining, as described by Jefferson (1987), was carried out with plants at the full rosette stage (4–5 weeks after germination). Plants were vacuum infiltrated for 2 × 10 min in sterile staining buffer containing 100 mg of 5-bromo-4-chloro-3-indolyl glucuronide (X-glu, Jersey Labs Inc., USA) in 300 ml of 100 mM phosphate buffer pH 7.0, 0.05% NaN3, 0.1% Triton X-100. Afterwards plants were incubated at 37°C for 48 h and bleached with ethanol (Figure 1B).
Transient protoplast transfection
Protoplasts were prepared from leaves of N.plumbaginifolia. Equal amounts of DNA from 17 different constructs with amino acid substitutions were used for evaluation of the activity of the uidA gene. To 15 ml Falcon tubes were added in the following order: 5 µg of DNA, 0.3 ml of a suspension containing 2 × 106 protoplasts/ml and 0.3 ml of 40% PEG 6000. After careful mixing, the tubes were allowed to stand at room temperature for 5 min. Then 4 ml of K3 medium (Bilang et al., 1994) were added and the tubes were incubated overnight in a horizontal position at 27°C in the dark. Before harvesting the transfected protoplasts, 10 ml of W5 buffer (150 mM NaCl, 125 mM CaCl2, 5 mM KCl, 6 mM glucose) were added to each tube to dilute the high osmolarity of the K3 medium and thereby to facilitate sedimentation of the protoplasts. Protein extracts were prepared by resuspending protoplasts in 200 µl of GUS extraction buffer (50 mM NaH2PO4 pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.1% sarcosyl). Aliquots of 20 µl were taken immediately for fluorometric reporter gene assay (Rossi et al., 1993).
Southern blot analysis
Total DNA was prepared from whole A.thaliana plants using a Nucleon PhytoPure total DNA isolation kit (Amersham Life Science) in accordance with the manufacturer’s protocol. A 5 µg aliquot of sample DNA was digested with either HindIII (single cutter) or XhoI (non-cutter) restriction nucleases, separated on a 0.7% agarose gel and transferred onto Hybond N membranes (Amersham, Little Chalfont, UK) using the manufacturer’s protocol. Hybridization and washing of blots was performed according to Sambrook et al. (1989). The full-length GUS gene was used as a probe after 32P-labelling with the random primer DNA labelling kit (Gibco-BRL, Gaithersburg, MD), according to the manufacturer’s instructions. For the lines used in the experiment, the copy numbers of the transgenes were determined (data not shown).
Semiquantitative RT–PCR analysis of transcription
Total RNA was extracted from 100 mg of 3-week-old plants by using standard protocols. Reverse transcription was performed according to the manufacturer’s protocol (Pharmacia) for reverse transcriptase beads using 1 µg of total RNA. The cDNA was prepared in a total amount of 50 µl. For the estimation of PCR sensitivity, different dilutions of cDNA were used. A 1 µl aliquot of cDNA was diluted 1:10, 1:50 and 1:100. Specific primers for the amplification of uidA and actin1 were used and produced products of the predicted size: 530 bp for uidA and 259 bp for AtActin-1. The latter was used as constitutive control. Different numbers of cycles (from 20 to 40) were tested for an evaluation of the transcript levels, and 30 cycles proved to be optimal for this purpose. The cDNAs for all tested genes were amplified using hot-start PCR under the following conditions: (i) 95°C for 5 min for one cycle; (ii) 94°C for 30 s, 57°C for 30 s, 72°C for 45 s for 30 cycles; and (iii) 72°C for 10 min for one cycle. Equal loading of each amplified gene sequence was determined by the control AtActin-1 PCR product.
DNA amplification and sequencing
PCR analyses were performed in a 50 µl volume containing 20 mM Tris pH 8.3, 50 mM KCl, 2 mM MgCl2, 0.2 mM of each dNTP, 50 ng of template DNA, 0.25 µM of each primer and 5 U of Taq DNA polymerase. DNA amplifications were performed in a Perkin-Elmer model 9600 thermal cycler with the following profile: (i) 95°C for 5 min for one cycle; (ii) 94°C for 1 min, 60°C for 1 min, 72°C for 1 min for 40 cycles; and (iii) 72°C for 10 min for one cycle. PCR primers for site-directed mutagenesis were designed based on the uidA gene sequence (not shown).
Blue sectors were excised from ∼1000 plants of subline 166G→T-1 and pooled. Following DNA isolation, the uidA gene was amplified with the specific primers. The PCR fragments were cloned into vector pCR 2.1 (Invitrogen Corporation, CA). About 250 independent clones were sequenced.
DNA sequencing reactions were performed with dRhodamine terminators from PE Applied Biosystems using a Perkin-Elmer GeneAmp PCR system 9600 thermocycler and analysed using an ABI PRISM 377 DNA sequencer.
Treatment of Arabidopsis seedlings with UV, X-rays and MMS
Two-week-old Arabidopsis seedlings grown on soil were irradiated with 1000 J/m2 for 30 s of UV-C light (254 nm) using a Minerallight-Lamp R-52 (UV-Products Inc., San Gabriel, CA) or with 25 Gy of X-rays.
Plants of the same age but grown on sterile medium were transferred into liquid medium containing or lacking 50 µM MMS. Two weeks later, the plants were stained histochemically and blue spots were counted.
Calculation of the number of genomes per plant
Total DNA of the respective transgenic lines was isolated from whole plants at the full rosette stage, using the Nucleon PhytoPure plant DNA extraction kit from Amersham Life Science. The yield of total DNA (in micrograms per plant) was compared with the mean DNA content (0.28 pg) of a diploid A.thaliana cell, to give an estimate of 2 × 106–8 × 106 (dependent on DNA preparation) genomes per plant (Swoboda et al., 1993).
Calculation of the reversion frequency
To evaluate mutation frequency per plant per copy number, the following formula was used: fp/c = rp/nc, where fp/c is the mutation frequency per plant per copy of the transgene, rp is the number of reversion events per plant, as an average of 300–1000 plants, and nc is the number of transgene copies in the genome.
For evaluation of the mutation frequency per base pair, the following formula was used: fb = rp/nc × 2ng, where fb is the mutation frequency per base pair, rp and nc are as described above and ng is the number of haploid genomes present in A.thaliana plants at the stage of analysis.
Acknowledgments
Acknowledgements
We thank O.Mittelsten Scheid, P.Pelczar, J.M.Lucht and J.Paszkowski for critical reading of the manuscript. We are grateful to J.Hays and J.Drake for stimulating discussions, C.Ramos and M.Müller for excellent technical assistance, and H.Angliker for DNA sequencing. The authors acknowledge an EMBO long-term fellowship to I.K. and an ESF ‘Plant Adaptation’ fellowship to O.K., as well as a grant from the SNSF, under the terms of the programme of Cooperation in Science and Research with CEEC and NIS, to I.K. and O.K.
References
- Amanuma K. et al. (2000) Transgenic zebrafish for detecting mutations caused by compounds in aquatic environments. Nature Biotechnol., 18, 62–65. [DOI] [PubMed] [Google Scholar]
- Angenon G. et al. (1990) Analysis of the stop codon context in plant nuclear genes. FEBS Lett., 271, 144–146. [DOI] [PubMed] [Google Scholar]
- Atkinson J. and Martin,R. (1994) Mutations to nonsense codons in human genetic disease: implications for gene therapy by nonsense suppressor tRNAs. Nucleic Acids Res., 22, 1327–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A. and Clough,S. (1998) Agrobacterium germ-line transformation: transformation of Arabidopsis without tissue culture. In Gelvin,S. and Schilperoort,R. (eds), Plant Mol. Biol. Manual, Vol. B7.Kluwer Academic, Dordrecht, The Netherlands, pp. 1–14. [Google Scholar]
- Betzner A. et al. (1997) Transfer RNA-mediated suppression of amber stop codons in transgenic Arabidopsis thaliana. Plant J., 11, 587–595. [DOI] [PubMed] [Google Scholar]
- Bilang R. et al. (1994) PEG-mediated direct gene transfer and electroporation. In Gelvin,S. and Schilperoort,R. (eds), Plant Mol. Biol. Manual, Vol. A1.Kluwer Academic, Dordrecht, The Netherlands, pp. 1–16. [Google Scholar]
- Britt A. (1995) Repair of DNA damage induced by ultraviolet radiation. Plant Physiol., 108, 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C. et al. (1978) Molecular basis of base substitution hotspots in E.coli. Nature, 274, 775–780. [DOI] [PubMed] [Google Scholar]
- Dycaico M., Provost,G., Kretz,P., Ransom,S., Moores,J. and Short,J. (1994) The use of shuttle vectors for mutation analysis in transgenic mice and rats. Mutat. Res., 307, 461–478. [DOI] [PubMed] [Google Scholar]
- Galli A. and Schiestl,R. (1999) Cell division transforms mutagenic lesions into deletion-recombinagenic lesions in yeast cells. Mutat. Res., 429, 13–26. [DOI] [PubMed] [Google Scholar]
- Gondo Y. et al. (1996) A novel positive detection system of in vivo mutations in rpsL (strA) transgenic mice. Mutat. Res., 360, 1–14. [DOI] [PubMed] [Google Scholar]
- Hall B. (1991) Spectrum of mutations that occur under selective and non-selective conditions in E.coli. Genetica, 84, 73–76. [DOI] [PubMed] [Google Scholar]
- Holbeck S. and Strathern,J. (1997) A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics, 147, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson F. (1987) A review of some topics concerning mutagenesis by ultraviolet light. Photochem. Photobiol., 45, 897–903. [DOI] [PubMed] [Google Scholar]
- Jefferson R. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep., 5, 387–405. [Google Scholar]
- Katoh Y. et al. (1994) Mutagenic effects of nitropyrenes in a soybean test system. Mutat. Res., 320, 59–68. [DOI] [PubMed] [Google Scholar]
- Klug W. and Cummings,M. (1986) Concepts of Genetics. 2nd edn. Merrill Publishing Co. A.Bell & Howell Co., Columbus, OH. [Google Scholar]
- Kohler S. et al. (1991) Spectra of spontaneous and mutagen-induced mutations in the lacI gene in transgenic mice. Proc. Natl Acad. Sci. USA, 88, 7958–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. (1997) Genes VI. Oxford University Press, Oxford, NY. [Google Scholar]
- Lichtenauer-Kaligis E. et al. (1996) Comparison of spontaneous hprt mutation spectra at the nucleotide sequence level in the endogenous hprt gene and five other genomic positions. Mutat. Res., 351, 147–155. [DOI] [PubMed] [Google Scholar]
- Mattanovich D. et al. (1989) Efficient transformation of Agrobacterium spp. by electroporation. Nucleic Acids Res., 17, 6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum C. et al. (2000) Targeted screening for induced mutations. Nature Biotechnol., 18, 455–457. [DOI] [PubMed] [Google Scholar]
- Parsons B. and Heflich,R. (1997) Genotypic selection methods for the direct analysis of point mutations. Mutat. Res., 387, 97–121. [DOI] [PubMed] [Google Scholar]
- Pegg A. (1984) Methylation of the O6 position of guanine in DNA is the most likely initiating event in carcinogenesis by methylating agents. Cancer Invest., 2, 223–231. [DOI] [PubMed] [Google Scholar]
- Protic-Sabljic M. and Kraemer,K. (1985) One pyrimidine dimer inactivates expression of transfected gene in xeroderma pigmentosum cells. Proc. Natl Acad. Sci. USA, 82, 6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. and Hohn,B. (1991) A transient assay in plant cells reveals a positive correlation between extrachromosomal recombination rates and length of homologous overlap. Nucleic Acids Res., 19, 2693–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchta H. et al. (1995) Somatic intrachromosomal homologous recombination events in populations of plant siblings. Plant Mol. Biol., 28, 281–292. [DOI] [PubMed] [Google Scholar]
- Rossi L. et al. (1993) Efficient and sensitive assay for T-DNA-dependent transient gene expression. Plant Mol. Biol. Rep., 11, 220–229. [Google Scholar]
- Sambrook J. et al. (1989) Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sander C. et al. (1978) Mutagenic and chromosome-breaking effects of azide in barley and human leukocytes. Mutat. Res., 50, 67–75. [DOI] [PubMed] [Google Scholar]
- Schultz S. et al. (1999) Spontaneous deleterious mutation in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 96, 11393–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. (1986) O-Alkyl pyrimidines in mutagenesis and carcino genesis: occurrence and significance. Cancer Res., 46, 4879–4885. [PubMed] [Google Scholar]
- Stadler L. (1930) The frequency of mutation of specific genes in maize. (Abstract) Anat. Rec., 47, 381. [Google Scholar]
- Stansfield I. et al. (1995) The end in sight: terminating translation in eukaryotes. Trends Biochem. Sci., 20, 489–491. [DOI] [PubMed] [Google Scholar]
- Swoboda P. et al. (1993) Somatic homologous recombination in planta: the recombination frequency is dependent on the allelic state of recombining sequences and may be influenced by genomic position effects. Mol. Gen. Genet., 237, 33–40. [DOI] [PubMed] [Google Scholar]
- Turker M. et al. (1993) Region-specific rates of molecular evolution: a fourfold reduction in the rate of accumulation of ‘silent’ mutations in transcribed versus nontranscribed regions of homologous DNA fragments derived from two closely related mouse species. J. Mol. Evol., 36, 31–40. [DOI] [PubMed] [Google Scholar]
- Wolfe K. et al. (1987) Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNAs. Proc. Natl Acad. Sci. USA, 84, 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. et al. (1989) Mutation rates differ among regions of the mammalian genome. Nature, 337, 283–285. [DOI] [PubMed] [Google Scholar]