Abstract
Bobtail squid from the genera Sepiola and Rondeletiola (Cephalopoda: Sepiolidae) form mutualistic associations with luminous Gram-negative bacteria (Gammaproteobacteria: Vibrionaceae) from the genera Vibrio and Photobacterium. Symbiotic bacteria proliferate inside a bilobed light organ until they are actively expelled by the host into the surrounding environment on a diel basis. This event results in a dynamic symbiont population with the potential to establish the symbiosis with newly hatched sterile (axenic) juvenile sepiolids. In this study, we examined the genetic diversity found in populations of sympatric sepiolid squid species and their symbionts by the use of nested clade analysis with multiple gene analyses. Variation found in the distribution of different species of symbiotic bacteria suggests a strong influence of abiotic factors in the local environment, affecting bacterial distribution among sympatric populations of hosts. These abiotic factors include temperature differences incurred by a shallow thermocline, as well as a lack of strong coastal water movement accompanied by seasonal temperature changes in overlapping niches. Host populations are stable and do not appear to have a significant role in the formation of symbiont populations relative to their distribution across the Mediterranean Sea. Additionally, all squid species examined (Sepiola affinis, S. robusta, S. ligulata, S. intermedia, and Rondeletiola minor) are genetically distinct from one another regardless of location and demonstrate very little intraspecific variation within species. These findings suggest that physical boundaries and distance in relation to population size, and not host specificity, are important factors in limiting or defining gene flow within sympatric marine squids and their associated bacterial symbionts in the Mediterranean Sea.
Population structure can be determined in nature by a number of intrinsic elements in the environment, including selective pressures resulting from abiotic variables. Many of these factors are the result of physical change in the environment over long periods of time (change in landscape, habitat fragmentation), while some, such as temperature, currents, and salinity, are constantly in flux (3, 7, 21). Small-scale changes in salinity and temperature can have significant effects and are exacerbated in marine environments, where osmotic balance and physical gradients are naturally established. Specifically, temperature may affect the distribution, viability, and fitness of organisms that have not readily adapted to such changing conditions (38, 57). This is pertinent to organisms living in symbiosis with each other, where one partner is dependent upon the other for a number of capabilities, such as physiological functions, nutritional requirements, and predator/prey avoidance (16, 20, 35). Interestingly, very few marine symbioses span across broad geographic ranges, since an enormous spectrum of abiotic and biotic conditions may prevent the ability of the symbiont to move between host populations. Studies of population structure within marine symbioses have increasingly pointed toward the underlying importance of understanding the nonliving elements of the environment as they apply to the distribution of symbiotic partners (either host or symbiont) across broadly distributed landscapes (42).
Population structure between host and symbionts in many symbioses, particularly mutualistic associations, often produce evidence of cospeciation between established partner lineages in both terrestrial and marine environments (1, 34, 37). The evidence for cospeciation or parallel cladogenesis is often attained through comparative phylogenetic analysis, which may neglect to unveil or account for various environmental factors. Abiotic characteristics of individual habitats may drive ecological or physiological processes, which in turn influence population distribution and interspecies interactions for both host and symbiont, particularly those that are environmentally transmitted. This may also influence either benefits or costs that arise due to the adaptability of each partner to accommodate not only environmental changes but also changes occurring with respect to each other. Considering the potential influencing power of the environment, it is prudent to explore the relative impact of nonliving habitat characteristics on symbiont-host distributions as it applies to the phylogeography and population history of both host and symbiont. This is particularly true for the mutualistic symbiosis between sepiolid squids (Mollusca: Cephalopoda) and bioluminescent bacteria in the genus Vibrio (Gammaproteobacteria: Vibrionaceae).
Symbiosis between sepiolid squids (Cephalopoda: Sepiolidae) and Vibrio bacteria has become an ideal model to study environmentally transmitted symbioses between cooperative microbes and their eukaryotic hosts (19, 44, 67). The best-known association within this family of squids is between members of the genus Euprymna and its symbiont Vibrio fischeri (21, 29). The symbiosis is established when environmentally transmitted, symbiosis-competent vibrios infect the light organs of newly hatched juvenile squid (30, 53). Upon entrance into the light organ, the bacteria flourish in a relatively nutrient-rich, protected environment and in turn achieve high densities within the organ (59). When a sufficiently high number of symbiont cells is achieved in the light organ, the bacteria become luminescent based on a communication mechanism known as quorum sensing (9, 12, 28, 31). Vibrio bacteria then induce a program of drastic morphological changes in the squid host, which helps to establish the symbiont population and completes development of the naïve light organ (33). Development and timing of this Vibrio population coincides with the nocturnal hunting behavior of the squid, in which the bacteriogenic luminescence is used in a predator/prey avoidance behavior known as counterillumination (20).
At dawn, the squid bury themselves in the sand and expel 90 to 95% of the symbiotic bacteria from the light organ (25, 54). This expulsion results in a “seeded” environment in which newly hatched, axenic juvenile squids can acquire symbiotically competent Vibrio bacteria. The result of this local acquisition of symbionts previously inhabiting hosts from the same population may lead to high degrees of specificity between symbiotic partners (37, 39). In the association between bobtail squids of the genus Euprymna (E. scolopes and E. tasmanica in the Indo-West Pacific Ocean) and Vibrio fischeri, this high degree of mutual exclusivity has been observed with V. fischeri (43, 67). Although V. fischeri is the only symbiont present in most Euprymna species, strain variation has been observed within and between populations, indicating that vibrios can migrate among host species complexes, changing the geographic mosaic of these symbionts within their host squids (21, 68). This distribution is affected mainly by other factors, such as temperature, which has been observed to influence the location of other marine populations in similar habitats (21, 22).
In comparison, mutualistic symbioses between Mediterranean squid from the genera Sepiola and Rondeletiola and Gram-negative bacteria from the genera Vibrio and Photobacterium (11, 18) demonstrate that species specificity is not always common among all sepiolid genera (17, 38). Furthermore, temperature has been observed to be a major factor determining symbiont distribution both within hosts as well as in their free-living state (22, 38, 57). Therefore, abiotic factors have a much larger influence in the establishment of environmentally transmitted symbiosis than just recognition and specificity to a particular squid host. The Mediterranean Sea harbors 10 species of squid from the genus Sepiola and one species from the genus Rondeletiola (R. minor), all of which possess a bacteriogenic light organ (6, 38, 41, 55). Sepiolid squid species in the Mediterranean exist in sympatry, with overlapping population boundaries along the northern coast of the Mediterranean (55). Also of interest pertaining to the Mediterranean sepiolid fauna is the fact that each squid can harbor two species of symbionts in their light organs (17, 38). Therefore, the aim of this study was to understand whether the population structure of different sympatric host species had a direct effect on symbiont distribution or if the combined influence of abiotic factors (temperature, water movement, and salinity) and multiple symbiont species influences the association among sympatric populations. This is in contrast to previous studies of allopatric populations, in which Vibrio bacteria were specialists with their specific host species (21). We hypothesized that a lack of specificity occurs between host squid and the Vibrio species found in Mediterranean sepiolids (V. fischeri, V. logei, and Photobacterium leiognathi) and that temperature was a major force in determining these host-symbiont assemblages. Using a phylogeographical approach, we tested whether any type of genetic structure existed among species of sepiolids in the Mediterranean Sea and if their Vibrio symbionts' distribution was influenced by host specificity or environment.
MATERIALS AND METHODS
Collection of specimens.
Specimens used in this study were collected from sites in southern France (Banyuls-sur-Mer), eastern Italy (Bari, Italy), and the North Atlantic (Bay of Biscay) (Table 1). Banyuls-sur-Mer is located in the south of France near the Spanish border at the foot of the Pyrenees Mountains. Specimens were collected by trawling (R. minor, S. robusta, and S. intermedia) or by hand during SCUBA dives at night (S. affinis). Italian sepiolid squid were collected by trawl net off the coast of Bari, Italy, while samples of R. minor were collected via mid-water trawl on several cruises in the Bay of Biscay. Other sequence data were incorporated for individual squid (S. robusta and S. affinis) from previously published sources as well (27, 40, 41). To increase the sample population size, other hosts and associated symbiotic strains from previous work were incorporated into the overall analysis (27, 40, 41). These samples were included in order to equalize the number of strains and host species for each collection site.
TABLE 1.
Collection sites of sepiolid squid species and their bacterial symbionts
| Location and date specimen collected | Host species | Symbiont species | Depth/temp (m/°C) |
|---|---|---|---|
| Banyuls-sur-Mer, France (42°35′29′′N, 3°2′22′′E), 2003-2008 | Sepiola affinis | Vibrio fischeri, V. logei | 3-10/20-22 |
| S. robusta | V. fischeri, V. logei | 20/13 | |
| S. intermedia | V. fischeri, V. logei | 20-30/13 | |
| Rondeletiola minor | Photobacterium leiognathi | 70/13 | |
| Bari, Italy (42°35′29′′N, 3°2′22′′E), 2005 | S. affinis | V. fischeri, V. logei | 3-10/21-24 |
| S. robusta | V. fischeri, V. logei | 10-20/14-18 | |
| S. intermedia | V. fischeri, V. logei | 10-20/14-18 | |
| Bay of Biscay, France (43°35′43′′N, 01°48′08′′E), 2007 | R. minor | P. leiognathi | 80-120/12-16 |
DNA isolation and sequencing.
All specimens of squid were anesthetized by placing the animal on ice and subsequently removing the light organ through ventral dissection. The light organ from each individual was then homogenized in a 2-ml tube to isolate the symbiotic bacteria inside. Each homogenate was placed through a series of dilutions in sterile filtered seawater before plating the bacteria. These homogenates were spread onto seawater tryptone agar plates (SWT; 0.5% tryptone, 0.3% yeast extract, 70% seawater, 0.3% glycerol, and 15% agar). After growth on SWT (12 to 24 h, 20 to 28°C), 10 to 15 individual colonies from each plate (representing one squid light organ) were selected, and each colony was then stab inoculated into an SWT agar vial for transport back to New Mexico State University. This sampling regime is applicable to any strain variation that may be observed in an individual squid light organ (68).
DNA was extracted from each individual isolate using the Qiagen DNeasy blood and tissue kit (Qiagen, Valencia, CA). Approximately 5 to 10 ng of the extracted genomic DNA was subsequently used in 4 separate PCRs to amplify the 16S rRNA locus of the symbiotic bacteria using Vibrio-specific primer sets (see Table S1 in the supplemental material). A total of 5 to 10 ng of the template DNA was also used to amplify a 650-base-pair region of the glyceraldehyde phosphate dehydrogenase locus (gapA) using both previously published Vibrio-specific primers (40) and primers newly designed specifically for V. logei (see Table S1). The gapA locus was used to analyze the bacterial symbiont populations in this study, as it has been shown to be amenable to phylogeographic and phylogenetic analyses for Vibrio populations (21, 40). This is due to the fact that the gapA locus is less conserved than other metabolic, constitutively expressed genes in the Vibrio genus and is capable of differentiating between strains of the same species of bacteria. The V. logei primers were designed and utilized due to broad problems associated with amplifying the gapA locus in a number of light organ isolates determined to be vibrios. DNA from host squid tissue was extracted by homogenizing a 20- to 25-mg piece of tissue dissected from the anterior mantle. The homogenate was processed using the same DNeasy blood and tissue kit (Qiagen, Valencia, CA). After DNA extraction, approximately 5 ng of host DNA was used in a PCR to amplify the cytochrome c oxidase subunit I (COI) and 12S and 16S rRNA loci (21, 27, 41) (see Table S1 in the supplemental material).
PCRs to amplify the above-specified loci from both host squid and Vibrio bacteria were performed as 25-μl reaction mixtures containing 5 to 10 ng of extract DNA, 0.2 μM both forward and reverse primers, 0.2 U of Taq polymerase (GoTaq Flexi DNA polymerase; Promega, San Luis Obispo, CA), 2.5 mM magnesium chloride (MgCl2), a final concentration of 200 μM dGTP, dATP, dCTP, and dCTP (50 μM each), and 1× buffer (Tris-HCl, KCl, and 0.1% Triton X-100). PCRs were completed in an MJ Research Dyad Disciple thermal cycler (Waltham, MA). Following PCRs, amplified fragments were purified using either a QIAquick PCR purification kit (Qiagen, Valencia, CA) or the QIAEX II gel extraction kit (Qiagen, Valencia, CA). The purified PCR products were used for presequencing reactions in an MJ Dyad Disciple thermal cycler (Waltham, MA) using BigDye Terminator 3.1 (Applied Biosystems, Foster City, CA). Reactions were cleaned of excess dideoxynucleotides and fluorescent dyes by centrifuging the samples through 400 μl of dehydrated Sephadex (Sigma, St. Louis, MO) and subsequently collected in a 96-well sequencing plate. All samples were sequenced using the Applied Biosystems 3100 automated capillary sequencer (Applied Biosystems, Foster City, CA) at New Mexico State University. Sequences were aligned and edited using Sequencher, version 4.6 (Gene Codes Corporation, Ann Arbor, MI).
Analysis of variance.
The baseline variance of initial sample populations of both host squid and symbiotic bacteria was measured using an analysis of molecular variance (AMOVA) using the program ARLEQUIN, version 3.0 (15). Variance calculated in this program was indicative of variation within populations at the different collection sites (North Atlantic, France, and Italy) and between each of the populations sampled. Variation observed within a population at each site was calculated with a base-pair-by-base-pair polymorphic measure, theta.
Nested clade analysis.
Analysis of the sequence data for building haplotype networks was completed in TCS version 1.12 (10). TCS v. 1.12 utilizes a statistical parsimony-based procedure based on Templeton's rules (61). The resulting symbiont and host networks were nested, and potential ambiguities and complementary problems were addressed and resolved using the nesting rules according to Templeton et al. (60, 63). The final nested clade information was then analyzed by the program GEODIS (50). GEODIS analyzes the haplotype network and interprets the nesting to describe intrinsic variation between clades, assuming the specific levels of nesting according to the associated geographical locations.
Phylogenetic analysis.
Corroboration of the hierarchical nesting scheme that was revealed through the formation of the haplotype network and the nesting of unique clades was generated using the parsimony-based phylogenetics program POY, version 4.1.2 (66). Equal weights for transitions-transversions were assigned for all trees, for both host squid populations and bacterial symbionts, by the use of the respective loci (combined analyses using all molecular loci).
RESULTS
Genetic variation and haplotype prevalence.
Variation in the sequence loci from both host squid and Vibrio/Photobacterium symbionts was measured from a subsampling of 178 symbiont strains. V. fischeri and V. logei strains were isolated from all Sepiola species, and P. leiognathi only was isolated from R. minor host populations (for a total of 56 individual squids sampled). Vibrio/Photobacterium identification was determined using the 1,600-base-pair 16S rRNA locus. After each symbiont strain was identified as either V. fischeri, V. logei, or P. leiognathi, the 650-base-pair gapA locus was used in the nested clade analysis for the bacterial symbionts. Numerical distribution of species for each geographical location reflects the relative prevalence of each symbiont species in the populations sampled in this study (Table 2). A high occurrence of V. fischeri symbionts was found in both French and Italian populations (Fig. 1).
TABLE 2.
Symbiont species present at each collection sitea
| Location | Symbiont species (no. of isolates)b | % of total symbiont population |
|---|---|---|
| Banyuls-sur-Mer, France | V. fischeri (74) | 78 |
| V. logei (16) | 16 | |
| P. leiognathi (12) | 6 | |
| Bari, Italy | V. fischeri (58) | 85 |
| V. logei (10) | 15 | |
| Bay of Biscay, Atlantic | P. leiognathi (8) | 100* |
Species identification is based on the 16S rRNA sequence for all symbiotic bacteria isolated from squid light organs. Percentages are based on the total population within one particular site. *, all symbionts isolated from the R. minor host were Photobacterium leiognathi. No Vibrio species were isolated from these hosts.
n = 178.
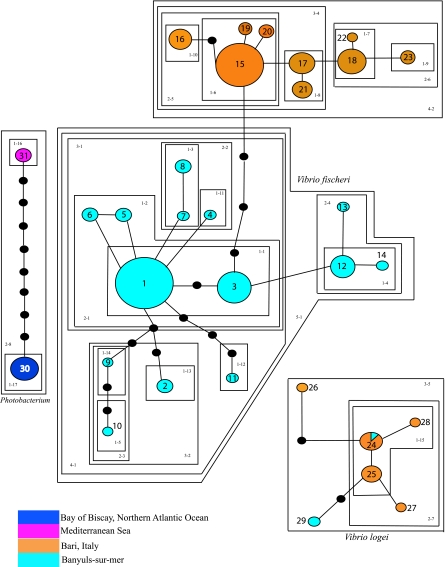
FIG. 1.
TCS-generated haplotype network and nested clades for Mediterranean vibrios and Photobacterium leiognathi symbionts. Each line represents one base pair difference, with black circles representing an unsampled intermediate haplotype. Each numbered circle represents a distinct haplotype, with color denoting collection site. The size of each circle is proportional to the number of individuals representing each given haplotype. Nested clades are marked by two-number identifiers within each box, signifying the representative nested clade.
Both Vibrio and Photobacterium populations used in this study represented a total of 31 haplotypes. There were, respectively, 23, 6, and 2 species-specific haplotypes found in the analysis belonging to V. fischeri, V. logei, and P. leiognathi. One haplotype (haplotype 24) was shared among V. logei symbionts from both French and Italian localities (Fig. 1). In total, 24 strains identified in the initial 16S rRNA species identification were not used in the formation of haplotype networks due to incomplete gapA sequence information.
The distribution of haplotypes of symbiotic V. fischeri demonstrates grouping in geographical “clusters,” which was also reflected in the final nesting. In the Mediterranean Sea, French and Italian clusters were clearly recognizable in the nesting scheme (Fig. 1). The genetic distance between populations of V. fischeri from the two Mediterranean sites was comparatively small; the closest association between a French V. fischeri haplotype and an Italian V. fischeri haplotype was a 3-base-pair difference or 3 unsampled haplotypes (Fig. 1). This sampling discrepancy occurs between the second most numerous French haplotype and the largest Italian haplotype (haplotype 15) (Fig. 1). Final nesting was the most parsimonious arrangement, which resulted in no confirmable false-positive inferences regarding colonization events or other population-level indices (64). Only one closed-loop structure arose in the construction of the symbiont haplotype network, which was resolved according to Templeton's rules (62) (Fig. 1).
The V. logei network was comparably small, reflecting the total proportion of V. logei bacteria within the total symbiont sample population. This network consists of 6 distinct haplotypes (24-29) (Fig. 1), with one haplotype containing both French and Italian strains (haplotype 24). The nesting of these haplotypes followed an interior-to-exterior scheme, with relatedness, not geography, being the most parsimonious nesting result.
Complete cladograms for symbiotic bacteria from the sampled populations infer a lack of evidence for restriction of gene flow between populations (Fig. 1). Clades 1-3, 1-4, and 2-3 show potential restriction in gene flow and possible habitat fragmentation, which may be a reflection of the terminal, exterior location of these clades within the total cladogram. Overall, the nesting diagrams for the symbiont populations demonstrate geographical structure. Nesting at the fourth level (clades 4-1 and 4-2) (Fig. 1) is indicative of an association between genetic relatedness and local environment.
All Vibrio symbionts formed one clade in the phylogenetic analysis based on location (see Fig. S1 in the supplemental material). This confirms the geographical bias of the haplotype network and subsequent nesting diagrams (Fig. 1). Interestingly, Vibrio isolates (both V. fischeri and V. logei) were contained with the Photobacterium clade, with Italian V. logei sister to Italian V. fischeri. All French V. fischeri isolates analyzed were sister to the Italian V. fischeri/V. logei clade.
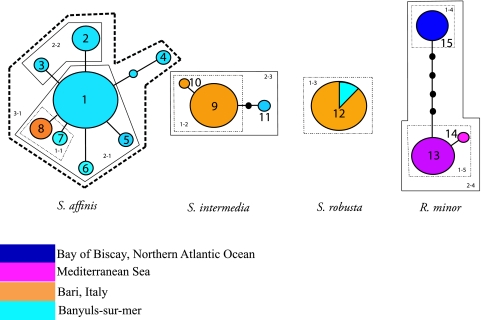
For host squids, a total of 15 haplotypes were found in all sample populations (Fig. 2). Initial networks that were calculated using a single-gene model (COI) revealed a relatively restricted network, supporting strong species delineation. To diversify the network, two additional genes (12S and 16S rRNA) were incorporated into the analysis to uncover unique variation inherent in other genes within species and among host populations. S. affinis had the largest number of COI haplotypes from all host species sampled, with a total of 9 distinct haplotypes, resulting in the most diverse and possibly least parsimonious set of haplotypes of all genes examined. Both 12S and 16S rRNA data had a more limited set of haplotypes for S. affinis (6 and 7 haplotypes, respectively), compared to 8 when using all 3 loci (Fig. 2). S. intermedia and R. minor each showed a relatively low number of representative haplotypes. These particular species demonstrate a smaller, more inclusive set of haplotypes, with both S. intermedia and R. minor having only 3 haplotypes. R. minor haplotypes are split between two distant geographic locales, Banyuls-sur-Mer and the Bay of Biscay (Fig. 2). Interestingly, all S. robusta hosts (8 total) sampled in this study possessed the same haplotype for all three loci investigated (haplotype 12). Haplotypes were all species specific, with no introgression of any individual haplotype between squid species sampled for all loci. Additionally, combined phylogenetic analysis of all loci confirmed the structure and orientation of the nested haplotypes as they occur within the populations sampled, with S. affinis and S. intermedia sister to S. robusta and with all R. minor individuals being sister to the Sepiola clade (see Fig. S2 in the supplemental material).
FIG. 2.
Haplotype network and corresponding nesting schematic of individual species haplotype networks for both Mediterranean and Northern Atlantic (Bay of Biscay) squids. All species were genetically distinct at the COI, 12S, and 16S loci by a 10 bp-difference, precluding any connection between separate species.
Host squid networks and nested diagrams demonstrated no introgression between sympatric species (Fig. 2). No connectable distances were found between species, regardless of distance or geographical location. The largest number of base pair differences among any one species was two in the population of S. affinis, nesting at the highest level of 3 (clade 3-1) (Fig. 2). Evidence for range expansion or fragmentation was unsubstantiated in this analysis due to a lack of direct evidence for the geographical separation of any of the host species (recall that all species are sympatric).
Variation within total populations of both hosts and symbionts was calculated using the measurement theta (θ). Symbiont theta values indicate higher per-base-pair variation, ranging from 0.00 to 0.0331. All symbiont populations, except the relatively small sample size of P. leiognathi, demonstrated some degree of genetic variation within each population (Table 3). The largest amount of within-population variation (θ = 0.0331) was observed with V. fischeri sampled from Banyuls-sur-Mer, France, from S. affinis (Table 3). Conversely, no genetic variation was found in symbiotic P. leiognathi isolated from the Bay of Biscay (Table 3). Additionally, no significant variation within populations of P. leiognathi was observed over the time period sampled, as only 3 derivative novel haplotypes arose chronologically in the course of this study (Fig. 1).
TABLE 3.
Within-population variance (theta, θ) for each host species at each respective collection sitea
| Host species | Collection site | No. of hosts | Host θ | Symbiont θ |
|---|---|---|---|---|
| S. affinis | Banyuls-sur-Mer | 18 | 0.0292 | 0.0331 |
| Bari | 9 | 0.0220 | 0.0297 | |
| S. intermedia | Banyuls-sur-Mer | 4 | 0.0026 | 0.0055 |
| Bari | 7 | 0.0000 | 0.0082 | |
| S. robusta | Banyuls-sur-Mer | 4 | 0.0000 | 0.0071 |
| Bari | 6 | 0.0000 | 0.0025 | |
| R. minor | Banyuls-sur-Mer | 4 | 0.0184 | 0.0000* |
| Northern Atlantic | 4 | 0.0000 | 0.0000* |
Values were calculated only for hosts from which at least two Vibrio or Photobacterium strains were isolated. *, all symbionts are P. leiognathi. Variation among this species was observed only between populations in the Mediterranean versus the North Atlantic (Bay of Biscay).
Theta values for host squids ranged from 0.00 to 0.0292 (range indicative of all 3 loci sampled for the entire host population) (Table 3). S. robusta hosts from both French and Italian sample sites demonstrated no genetic variation among individuals and therefore have a θ value of 0. Relatively low theta values for host variation across all loci indicate little or no divergence within populations of species, regardless of the location of populations.
DISCUSSION
Symbiont population structure.
Symbiotic bacteria that were sampled from collected host squids exhibited much more dynamic patterns across geographical locations than their respective host squids. Only one closed-loop structure was observed with the initial haplotype network construction within the V. fischeri population from Banyuls-sur-Mer (France). This ambiguity occurred between the most highly represented haplotype (Fig. 1, haplotype 1) and less-represented satellite haplotypes (haplotypes 5 and 6). These structures in future haplotype networks need to be carefully considered due to the potential for false-positive results (23, 48).
The most obvious trend apparent in the nesting of individual haplotypes is a strong correlation between relatedness and geography, with different amounts of variation depending on the species sampled. In V. fischeri populations, geographical grouping is evident (Fig. 1; see also Fig. S1 in the supplemental material). Within this species, no haplotypes were shared between Banyuls-sur-Mer and Bari. This suggests that genetic differences are due to geographical boundaries for symbiotic bacteria. In V. logei populations, a shared haplotype between sample sites was found, indicating that a small amount of introgression has occurred between geographical locations and that not all populations are in complete isolation from each other. V. logei is a known psychrophile and has been shown to prefer colder habitats (17, 38, 65), which may reduce the amount of movement during warmer seasons. Differences also exist between V. fischeri and V. logei in their concentration throughout different times of the year (22). Thus, introgression of certain haplotypes may occur when abiotic factors may be conducive for bacteria to have higher residence times in the water column when habitats share similar features. Since seasonal temperature fluctuations seem to be driving population dynamics within other symbiotic Vibrio communities (21, 58), it is likely that introgression may readily occur in Mediterranean vibrios when habitats are similar.
Niche-specific differences driven by abiotic factors like temperature may also play a major role in defining specific genetic differences which have ultimately led to a clear distinction between the two Vibrio species in the northern Mediterranean (57, 58). This has direct implications for the population structure of the squid-Vibrio symbiosis. Deeper-dwelling, colder-water sepiolid species (i.e., S. robusta and S. intermedia) may be discriminately exposed to a higher proportion of V. logei, thereby favoring colonization by one Vibrio species over another (38, 42, 58). Although this in situ evidence is only suggestive, additional studies of such populations could add evidence as to how symbiont structure is formed in these particular sepiolids over both time and habitat preference.
P. leiognathi isolates from Mediterranean R. minor hosts all shared the same haplotype (Fig. 1, haplotype 29) compared to those from the North Atlantic (Fig. 1, haplotypes 29 and 30). This genetic distinction may be due to differences in abiotic factors that shape their ecology, as well as a large geographical distance between sample sites (Atlantic versus Mediterranean seas). The locational disparity may also be exacerbated by the very narrow Strait of Gibraltar, which symbiotic bacteria would need to pass through to access either host site while adjusting to differential conditions between the Atlantic Ocean and the Mediterranean Sea (Table 1) (45, 46). Additionally, water movement into the Mediterranean Sea from the Atlantic Ocean would most likely prevent significant physical movement and gene flow of most organisms out of the Mediterranean (2, 5, 26, 32, 57).
Host population structure.
The population structure of host squid species used in this study suggests strong species delineations within sympatric populations (Fig. 2; see also Fig. S2). Individual squids within each species shared similar haplotypes regardless of collection site (Fig. 2). There was also no significant temporal or geographical change in host populations based on the genetic information, considering squid were sampled within a period of 6 years (Table 1). Lack of significant change over time (between 3 to 7 years) suggests a slow mutation rate in the COI and rRNA-coding loci or active gene flow between populations such that all species populations are homogeneous (8).
Genetically similar populations occurring in geographically distinct areas suggest a number of possible evolutionary scenarios. One is a recent dispersal or radiation event which separated genetically similar populations into various geographical locations. Since the Mediterranean Sea has been shown to have surface water currents driven by strong seasonal changes, this may provide a mechanism for juvenile squids to be moved to different locations at specific times of the year (56). Another possible consideration is that certain loci do not provide sufficient phylogenetic resolution to expose fine differences between geographically different populations of host squid (4). This second scenario is less likely, given that previous studies of Indo-West Pacific sepiolids and other Mediterranean fauna using mitochondrial loci provide strong support for determining population structure (13, 14, 21, 47). Additional mitochondrial genes (12S and 16S rRNA) reinforce this species exclusivity through parallel networks resulting in only two additional haplotypes when combined with the COI sequence data (haplotypes 9 and 13) (Fig. 2).
Influence of abiotic factors.
The Mediterranean Sea is a relatively shallow, contained body of ocean, which is accessed by other bodies of water at only a small number of places: the Strait of Gibraltar, the Dardanelles, and the Bosporus. This results in the lack of water movement between the Mediterranean and any other significantly large body of water. The northern Mediterranean is also absent of ocean-like currents, which could facilitate movement around the coastal margins of the Mediterranean (49, 51). Previous work evaluating the population genetic structure of sepiolid squid and their symbionts in the Indo-West Pacific demonstrated some specificity between allopatric Euprymna species and their Vibrio symbionts, with observed movement of V. fischeri strains between host populations that had similar environmental regimes (21). This supports the idea that although Euprymna hosts can share V. fischeri from other Euprymna hosts (host specificity) throughout the Indo-West Pacific, Vibrio bacteria have the ability to move around and infect new populations if environmental factors facilitate their transport, survival, and fitness (57, 58).
Thus, one major difference between the Indo-West Pacific and the Mediterranean that may lead to a lack of specificity between host squids and their vibrio symbionts is temperature. The Mediterranean Sea is subject to shallow thermoclines, especially during summer months, which can drastically change temperatures within the water column (Table 1) (52). These seasonal temperature differences may create population differences due to temperature adaptation that separates the habitat ranges of prospective symbionts (22, 38, 57). Thus, particular temperature conditions of host habitats may directly influence which species/strain of symbiont predominates in their light organs (38, 39, 57, 58). Newly hatched juveniles of species which are found at relatively deeper depths, such as S. ligulata, S. intermedia, and S. robusta, are more likely to be inoculated by V. logei, which has a selective advantage at colder temperatures (38). Since sepiolid squids are found continuously year-round, various generations of Sepiola may have been colonized by different Vibrio bacteria, depending on when and where they were infected (6, 36, 38). In our study, the ratio of V. fischeri to V. logei found in S. affinis (found in shallower, warmer waters) (Table 1) was significantly higher, suggesting that the warm-water-preferring V. fischeri has an advantage in terms of its potential to form symbioses with sepiolid squids in those areas. This has also been observed in laboratory experiments, in which temperature rather than host specificity is a strong force in forming these associations (38).
The influence of abiotic factors on the establishment and persistence of this symbiosis in sepiolid squid has only recently emerged as a major force in determining the natural structure of the symbiosis (57, 58). This study has demonstrated a lack of specificity between sympatric species of light organ-bearing sepiolid squids and their Vibrio symbionts in the Mediterranean. Adaptation to multiple squid populations in the Mediterranean seems to be driven mainly by environmental determinants, but host specificity is still important in the establishment of the population genetic structure, since Vibrio or Photobacterium species are the sole symbionts of sepiolid squids in both the Indo-West Pacific and Mediterranean seas (41). The combined influence of host specificity and abiotic factors contribute to the complexity of the population genetic structure of environmentally transmitted host-symbiont associations. Understanding the behavior of symbiotic bacteria among eukaryotic hosts and the phenomena that determines their phylogeographical matrix may help aid in our understanding of the population dynamics of other related Vibrio species in nature (including pathogens such as V. cholerae and V. parahaemolyticus) and what factors are important for their survival in both their free-living and symbiotic states.
Supplementary Material
Acknowledgments
We thank B. W. Jones and J. E. Lopez for their help with population genetic software and analysis. We also thank N. Moltschaniwskyj for her assistance with the molecular analysis of the sepiolid squid and S. V. Boletzky for helpful discussions concerning Mediterranean cephalopod fauna. A very special thanks to B. Hesse, C. LaBrune, and J.-L. Martinez (Laboratoire Arago, Banyuls-sur-Mer, France), G. Cricket (University of Perpignan, France), and G. Bello (Instituto Arion, Bari, Italy) for helping collect squid specimens used in this study.
Funding for this study was provided by NIH- NIAID 1SC1AI081659, NIH-NIAID 3SC1AI81659-02S1, and NSF-IOS 0744498 to M.K.N.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ashen, J. B., and L. J. Goff. 2000. Molecular and ecological evidence for species specificity and coevolution in a group of marine algal-bacterial symbioses. Appl. Environ. Microbiol. 66:3024-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astraldi, M., S. Balopoulos, J. Candela, J. Font, M. Gacic, G. P. Gasparini, B. Manca, A. Theocharis, and J. Tintore. 1999. The role of straits and channels in understanding the characteristics of Mediterranean circulation. Prog. Oceanogr. 44:65-108. [Google Scholar]
- 3.Aurelle, D., T. Cuillemaud, P. Alfonso, T. Morato, P. Wirtz, R. S. Santos, and M. L. Cancela. 2003. Genetic study of Coris julis (Osteichtyes, Perciformes, Labridae) evolutionary history and dispersal abilities. C. R. Biol. 326:771-785. [DOI] [PubMed] [Google Scholar]
- 4.Ballard, J. W. O. 2000. When one is not enough: introgression of mitochondrial DNA in Drosophila. Mol. Biol. Evol. 17:1126-1130. [DOI] [PubMed] [Google Scholar]
- 5.Bargelloni, L., J. A. Alarcon, M. C. Alvarez, E. Penzo, A. Magoulas, C. Reis, and T. Patarnello. 2003. Discord in the family Sparidae (Teleostei): divergent phylogeographical patterns across the Atlantic-Mediterranean divide. J. Evol. Biol. 16:1149-1158. [DOI] [PubMed] [Google Scholar]
- 6.Bello, G. 1995. A key for the identification of the Mediterranean sepiolids (Mollusca: Cephalopoda). Bull. Inst. Oceanogr. (Monaco) 16:41-56. [Google Scholar]
- 7.Bollmann, A., and H. J. Laanbroek. 2002. Influence of oxygen partial pressure and salinity on the community composition of ammonia-oxidizing bacteria in the Schelde estuary. Aquat. Microb. Ecol. 28:239-247. [Google Scholar]
- 8.Brierley, A. S., P. G. Rodhouse, J. P. Thorpe, and M. R. Clarke. 1993. Genetic evidence of population heterogeneity and cryptic speciation in the ommastrephid squid Martialia hyadesi from the Patagonian Shelf and Antarctic Polar Frontal Zone. Mar. Biol. 116:593-602. [Google Scholar]
- 9.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clement, M., D. Posada, and K. Crandall. 2000. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9:1657-1660. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap, P. V., and M. J. McFall-Ngai. 1987. Initiation and control of the bioluminescent symbiosis between Photobacterium leiognathi and leiognathid fish. Ann. N. Y. Acad. of Sci. 503:269-283. [DOI] [PubMed] [Google Scholar]
- 12.Dunlap, P. V. 1999. Quorum regulation of luminescence in Vibrio fischeri. J. Mol. Microbiol. Biotech. 1:5-12. [PubMed] [Google Scholar]
- 13.Duran, S., G. Giribet, and X. Turon. 2004. Phylogeographical history of the sponge Crambe crambe (Porifera, Poecilosclerida): range expansion and recent invasion of the Macronesian islands from the Mediterranean Sea. Mol. Ecol. 13:109-122. [DOI] [PubMed] [Google Scholar]
- 14.Duran, S., C. Palacín, M. A. Becerro, X. Turon, and G. Giribet. 2004. Genetic diversity and population structure of the commercially harvested sea urchin Paracentrotus lividus (Echinodermata, Echinoidea). Mol. Ecol. 13:3317-3328. [DOI] [PubMed] [Google Scholar]
- 15.Excoffier, L. G., and S. Schneider. 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 16.Felbeck, H., J. J. Childress, and G. N. Somero. 1981. Calvin-Benson cycle and sulphide oxidation enzymes in animals from sulphide-rich habitats. Nature 293:291-293. [Google Scholar]
- 17.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerrero-Ferreira, R. C., and M. K. Nishiguchi. 2007. Biodiversity among luminescent symbionts from squid of the genera Uroteuthis, Loliolus, and Euprymna (Mollusca: Cephalopoda). Cladistics 23:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanlon, R. T., M. F. Claes, S. E. Ashcraft, and P. V. Dunlap. 1997. Laboratory culture of the sepiolid squid Euprymna scolopes: a model system for bacteria-animal symbiosis. Biol. Bull. 192:364-374. [DOI] [PubMed] [Google Scholar]
- 20.Jones, B. W., and M. K. Nishiguchi. 2004. Counterillumination in the bobtail squid, Euprymna scolopes (Molluca: Cephalopoda). Mar. Biol. 144:1151-1155. [Google Scholar]
- 21.Jones, B. W., J. E. Lopez, J. Huttenberg, and M. K. Nishiguchi. 2006. Population structure between environmentally transmitted Vibrios and bobtail squids using nested clade analysis. Mol. Ecol. 15:4317-4329. [DOI] [PubMed] [Google Scholar]
- 22.Jones, B. W., A. Maruyama, C. C. Ouverney, and M. K. Nishiguchi. 2007. Spatial and temporal distribution of the Vibrionaceae in coastal waters of Hawaii, Australia, and France. Microb. Ecol. 54:314-323. [DOI] [PubMed] [Google Scholar]
- 23.Knowles, L. L., and W. P. Maddison. 2002. Statistical phylogeography. Mol. Ecol. 11:2623-2635. [DOI] [PubMed] [Google Scholar]
- 24.Kotoulas, G., A. Magoulas, N. Tsimenides, and E. Zouros. 1995. Marked mitochondrial DNA differences between Mediterranean and Atlantic populations of the swordfish, Xiphias gladius. Mol. Ecol. 4:473-482. [Google Scholar]
- 25.Lee, K., and E. G. Ruby. 1994. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefkaditou, E., and P. Kaspiris. 2005. Distribution and abundance of sepiolids (Mollusca: Cephalopoda) off the Northeastern Greek coasts. Belg. J. Zool. 135:199-204. [Google Scholar]
- 27.Lopez, J. 2003. Symbiosis among sepiolid squid: origins of the symbiotic light organ and examination of the host population structure at the intraspecies level. M.S. thesis. New Mexico State University, Las Cruces, NM.
- 28.Lupp, C., and E. G. Ruby. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFall-Ngai, M. J. 1994. Animal-bacterial interactions in the early life history of marine invertebrates: the Euprymna scolopes/Vibrio fischeri symbiosis. Am. Zool. 34:554-561. [Google Scholar]
- 30.McFall-Ngai, M. J., and E. G. Ruby. 1998. Sepiolids and vibrios: when first they meet. Bioscience 48:257-265. [Google Scholar]
- 31.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 32.Millot, C. 1999. Circulation in the western Mediterranean Sea. J. Mar. Sys. 20:423-442. [Google Scholar]
- 33.Montgomery, M. K., and M. J. McFall-Ngai. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development in the squid Euprymna scolopes. Development 120:1719-1729. [DOI] [PubMed] [Google Scholar]
- 34.Munkasci, A. B., J. J. Pan, P. Villesen, U. G. Mueller, M. Blackwell, and D. J. McLaughlin. 2004. Convergent coevolution in the domestication of coral mushrooms by fungus-growing ants. Proc. R. Soc. Lond. B 21:1777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muscatine, L., and J. W. Porter. 1977. Mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454-460. [Google Scholar]
- 36.Nesis, K. N. 1982. Cephalopods of the world. TFH Publications, Neptune City, NJ.
- 37.Nishiguchi, M. K., E. G. Ruby, and M. J. McFall-Ngai. 1998. Competitive dominance during colonization is an indication of coevolution in an animal-bacterial symbiosis. Appl. Environ. Microbiol. 64:3209-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishiguchi, M. K. 2000. Temperature affects species distribution in symbiotic populations of Vibrio. Appl. Environ. Microbiol. 66:3550-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishiguchi, M. K. 2002. Host recognition is responsible for symbiont composition in environmentally transmitted symbiosis. Microb. Ecol. 44:10-18. [DOI] [PubMed] [Google Scholar]
- 40.Nishiguchi, M. K., and V. S. Nair. 2003. Evolution of symbiosis in Vibrionaceae: a combined approach using molecules and physiology. Int. J. Syst. Evol. Microbiol. 53:2019-2026. [DOI] [PubMed] [Google Scholar]
- 41.Nishiguchi, M. K., J. L. Lopez, and S. van Boletzky. 2004. Enlightenment of old ideas from new investigations: more questions regarding the evolution of bacteriogenic light organs in squids. Evol. Dev. 6:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nyholm, S. V., and M. K. Nishiguchi. 2008. The evolutionary ecology of a sepiolid squid-Vibrio association: from cell to environment. Vie Milieu Paris 58:175-184. [PMC free article] [PubMed] [Google Scholar]
- 43.Nyholm, S., and M. J. McFall-Ngai. 2003. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl. Environ. Microbiol. 69:3932-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nyholm, S., and M. J. McFall-Ngai. 2004. The winnowing: establishing the squid-Vibrio symbiosis. Nat. Rev. Microbiol. 2:632-642. [DOI] [PubMed] [Google Scholar]
- 45.Patarnello, T., F. A. M. J. Volckaert, and R. Castilho. 2007. Pillars of Hercules: is the Atlantic-Mediterranean transition a phylogeographical break? Mol. Ecol. 16:4426-4444. [DOI] [PubMed] [Google Scholar]
- 46.Pérez-Losada, M., A. Guerra, G. R. Carvalho, A. Sanjuan, and P. W. Shaw. 2002. Extensive population subdivision of the cuttlefish Sepia officinalis (Mollusca: Cephalopoda) around the Iberian Peninsula indicated by microsatellite DNA variation. Heredity 89:417-424. [DOI] [PubMed] [Google Scholar]
- 47.Pérez-Losada, M., M. J. Nolte, K. A. Crandall, and P. W. Shaw. 2007. Testing hypotheses of population structuring in the Northeast Atlantic Ocean and Mediterranean Sea using the common cuttlefish Sepia officinalis. Mol. Ecol. 16:2667-2679. [DOI] [PubMed] [Google Scholar]
- 48.Petit, R. J. 2008. The coup de grace for the nested clade phylogeographic analysis? Mol. Ecol. 17:516-518. [DOI] [PubMed] [Google Scholar]
- 49.Pinardi, N., and E. Masetti. 2000. Variability of the large scale general circulation of the Mediterranean from observations and modeling: a review. Palaeogeogr. Palaeoclimatol. Palaeoecol. 158:153-173. [Google Scholar]
- 50.Posada, D., K. A. Crandall, and A. R. Templeton. 2000. GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol. Ecol. 9:487-488. [DOI] [PubMed] [Google Scholar]
- 51.Robinson, A. R., W. G. Leslie, A. Theocharis, and A. Lascaratos. 2001. Mediterranean Sea circulation, p. 283-298. In J. H. Steele, K. K. Turekian, and S.A. Thorpe (ed.), Encyclopedia of ocean sciences. Academic Press, London, United Kingdom.
- 52.Roussanov, V., E. Stanev, V. Artale, and N. Pinardi. 1995. A seasonal model of the Mediterranean Sea general circulation. J. Geophys. Res. 100:13.515-13.538. [Google Scholar]
- 53.Ruby, E. G., and M. J. McFall-Ngai. 1992. A squid that glows in the night: development of an animal-bacterial mutualism. J. Bacteriol. 174:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruby, E. G., and K. Lee. 1998. The Vibrio fischeri-Euprymna scolopes light organ association: current ecological paradigms. Appl. Environ. Microbiol. 64:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salman, A., T. Katagan, and H. Avni Benli. 2002. Cephalopod fauna of the Eastern Mediterranean. Turk. J. Zool. 26:47-52. [Google Scholar]
- 56.Send, U., J. Font, G. Krahmann, C. Millot, M. Rhein, and J. Tintore. 1999. Recent advances in observing the physical oceanography of the western Mediterranean Sea. Prog. Oceanogr. 44:37-64. [Google Scholar]
- 57.Soto, W., J. Gutierrez, M. R. Remmenga, and M. K. Nishiguchi. 2009. Synergistic affects of temperature and salinity in competing strains of symbiotic Vibrio fischeri. Microb. Ecol. 57:140-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soto, W., C. P. Lostroh, and M. K. Nishiguchi. 2009. Physiological responses to stress in the Vibrionaceae: aquatic microorganisms frequently affiliated with hosts, p. 409-426. In J. Seckbach and M. Grube (ed.), Cooperation and stress in biology. Springer, New York, NY.
- 59.Stabb, E. V. 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis, p. 204-218. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 60.Templeton, A. R., E. Beorwinkle, and C. F. Sing. 1987. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. I. Basic theory and analysis of alcohol dehydrogenase activity in Drosophila. Genetics 117:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Templeton, A. R., K. A. Crandall, and C. F. Sing. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. III. Cladogram estimation. Genetics 132:619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Templeton, A. R., and C. F. Sing. 1993. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics 134:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Templeton, A. R., E. Routman, and C. A. Phillips. 1995. Separating population structure from population history: a cladistic analysis of the geographical distribution of mitochondrial DNA haplotypes in the tiger salamander, Abystoma tigrunum. Genetics 140:767-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Templeton, A. R. 1998. Nested clade analyses of phylogeographic data: testing hypotheses about gene flow and population history. Mol. Ecol. 7:381-397. [DOI] [PubMed] [Google Scholar]
- 65.Thompson, J. R., M. A. Randa, L. A. Marcelino, A. Tomita-Mitchell, E. Lim, and M. F. Polz. 2004. Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70:4103-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Varón, A., L. S. Vinh, and W. C. Wheeler. 2010. POY version 4: phylogenetic analysis using dynamic homologies. Cladistics 26:72-85. [DOI] [PubMed] [Google Scholar]
- 67.Visick, K. L., and M. J. McFall-Ngai. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J. Bacteriol. 182:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wollenberg, M. S., and E. G. Ruby. 2009. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes from two Oahu populations. Appl. Environ. Microbiol. 75:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.