Abstract
The refrigerated storage of raw milk throughout the dairy chain prior to heat treatment creates selective conditions for growth of psychrotolerant bacteria. These bacteria, mainly belonging to the genus Pseudomonas, are capable of producing thermoresistant extracellular proteases and lipases, which can cause spoilage and structural defects in pasteurized and ultra-high-temperature-treated milk (products). To map the influence of refrigerated storage on the growth of these pseudomonads, milk samples were taken after the first milking turn and incubated laboratory scale at temperatures simulating optimal and suboptimal preprocessing storage conditions. The outgrowth of Pseudomonas members was monitored over time by means of cultivation-independent denaturing gradient gel electrophoresis (DGGE). Isolates were identified by a polyphasic approach. These incubations revealed that outgrowth of Pseudomonas members occurred from the beginning of the dairy chain (farm tank) under both optimal and suboptimal storage conditions. An even greater risk for outgrowth, as indicated by a vast increase of about 2 log CFU per ml raw milk, existed downstream in the chain, especially when raw milk was stored under suboptimal conditions. This difference in Pseudomonas outgrowth between optimal and suboptimal storage was already statistically significant within the farm tank. The predominant taxa were identified as Pseudomonas gessardii, Pseudomonas gessardii-like, Pseudomonas fluorescens-like, Pseudomonas lundensis, Pseudomonas fragi, and Pseudomonas fragi-like. Those taxa show an important spoilage potential as determined on elective media for proteolysis and lipolysis.
Psychrotolerant bacteria are mainly ubiquitous organisms able to grow at refrigeration temperatures regardless of their optimal growth temperature (10, 22). Extracellular enzymes (mainly lipases and proteases) that are secreted by these organisms are known to cause spoilage of milk and dairy products, leading to important economic losses (21, 32). Lipases degrade the milk fat, causing rancid, soapy, and occasional bitter off-flavors through the formation of medium-chain fatty acids. Proteases that degrade casein cause a gray color, bitter off-flavors, and gelation of ultra high-temperature (UHT) products (7, 19, 21).
Psychrotolerant bacteria have become more important for the shelf life of heat-treated dairy products because of the development of these bacteria during prolonged refrigerated storage of raw milk on the farm and at the dairy plant. In an effort to reduce the total aerobic plate count of raw milk, a lower storage temperature (1 to 4°C) is upheld, leading to the perception that raw milk could be stored for a longer period before further processing. However, the combination of a longer storage time and a lower temperature creates a selective advantage for psychrotolerant bacteria, especially Pseudomonas members, that enter raw milk via biofilms in the milk tanks, contaminated water, and soil (6, 28). These pseudomonads are able to outgrow other bacteria, such as members of the Aeromonas, Listeria, Staphylococcus, and Enterococcus genera and the family Enterobacteriaceae, thus becoming the predominant microbiota in raw milk (29), constituting up to 70 to 90% of the psychrotrophic raw milk microbiota (1). Even though they are easily inactivated through pasteurization or UHT treatment, their heat-resistant enzymes persist upon processing of the milk (4).
A persisting problem in unraveling the exact nature of the spoilage microbiota is the unresolved taxonomic situation of the genus Pseudomonas. Pseudomonas members are still often identified based on phenotypic characteristics, a methodology that became outdated because of the general introduction of molecular DNA methodologies. However, a clear-cut phylogenetically based identification approach for Pseudomonas members is not available yet. Even recent studies therefore still rely on phenotypical methods for routine identification of isolates (9, 12, 23, 34). This study aims at better understanding the outgrowth of Pseudomonas species throughout the dairy chain (farm tank to transport to the dairy plant) under optimal and suboptimal cooling conditions, as well as assessing the qualitative species composition in the stored raw milk through a polyphasic identification approach. Furthermore, it combines the use of cultivation, spoilage potential characterization, and noncultivation monitoring of the psychrotolerant bacteria in raw milk to better assess the shelf life risks in the end product.
MATERIALS AND METHODS
Simulation of the cold dairy chain and sampling.
Three independent simulations (s1, s2, and s3) that imitate preprocessing conditions at two temperature extremes that represent optimally and suboptimally cooled storage conditions were set up.
Milk samples used for the simulations were constituted by mixing equal volumes of raw milk samples from a number (n) of different farms (for s1, n = 1; for s2, n = 8; and for s3, n = 7), collected from the farm bulk tank after the first milking turn. The 600-ml samples were incubated in a water bath, the temperature of which was regulated with a cryostat. A smaller bottle with 300 ml of the same mixed milk sample was used for temperature registration with the Ellab Tracksense PRO Basic logger system (Ellab Inc., Centennial, CO).
An overview of the different experimental conditions during the various simulations is given in Table 1. To simulate the storage at the farm—where in theory an optimal resident temperature of 3.5°C is envisaged—the milk mixtures were heated twice a day (morning and afternoon, with 8 h in between) to imitate the increased temperatures due to the milking peaks (= warming up of the tank milk when fresh milk enters the tank), 6°C and 10°C for optimal and suboptimal conditions, respectively (see the figure in the supplemental material). The conditions for these milking-linked temperatures were determined from data obtained in 205 Belgian dairy farms. The stored farm milk is collected after 2 to 3 days (4 days were simulated to include extreme conditions) and is stored again within 8 h after collection for a maximum of 24 h in an industrial tank before processing at the dairy plant. The temperature regime during the simulation of transport and storage at the dairy factory was a consensus decided upon by an expert panel with people from the dairy industry.
TABLE 1.
Simulation of the dairy chain from the farm milk tank, transport, and storage at the industrial milk tank on a 0.6-liter scalea
| Parameter | Simulated storage mode |
||
|---|---|---|---|
| Farm milk tank | Transport | Industrial milk tank | |
| Temp (°C) | |||
| Optimal | 3.5 | 6 | 6 |
| Suboptimal | 6 | 10 | 10 |
| Milking peak (°C) | |||
| Optimal | 6 | ||
| Suboptimal | 10 | ||
| Duration | 4 days | 8 h | 24 h |
| Use of stirring | Yes | No | Yes |
| Sampling | 2×b | 3× | 3× |
| Sampling time | |||
| RM1 | t0 | ||
| RM2 | t0 + 8 h | ||
| RM3 | t0 + 24 h | ||
| RM4 | t0 + 32 h | ||
| RM5 | t0 + 48 h | ||
| RM6 | t0 + 56 h | ||
| RM7 | t0 + 72 h | ||
| RM8 | t0 + 80 h | ||
| RM90 | t0 + 96 h | ||
| RM10 | t0 + 100 h | ||
| RM11 | t0 + 104 h | ||
| RM12 | t0 + 120 h | ||
| RM13 | t0 + 124 h | ||
| RM14 | t0 + 128 h | ||
Conditions are based on data from 205 Belgian dairy farms and advice from an expert panel from the dairy industry. t0, time zero.
Samples taken prior to simulation of the milking peaks.
During these approximately 6-day simulation experiments, 14 milk samples for microbial analysis (RM1 to RM14) (Table 1) were collected at regular time intervals for the following: (i) total aerobic plate counting by pour plating of serial dilutions on plate count agar (PCA) (Oxoid Ltd., Basingstoke, United Kingdom) with incubation at 30°C for 3 days and (ii) presumptive psychrotolerant Pseudomonas counting by streaking of serial dilutions on a selective medium for Pseudomonas that contains cetrimide (10 mg liter−1), fucidin (10 mg liter−1), and cephalosporin (50 mg liter−1) (CFC agar) (Oxoid Ltd., Basingstoke, United Kingdom), with incubation at 22°C for 4 days.
Isolation.
Pseudomonas isolates were picked from CFC agar at 4 sampling occasions: (i) at the beginning (RM 1) and (ii) at the end of the simulation of a farm bulk tank (RM 8), (iii) at the end of simulation of the transport (RM 11), and (iv) at the end of simulation of storage at the dairy plant prior to processing (RM 14). Where possible, 30 isolates (constituting 10 to 20% of the total amount of colonies) were randomly picked from the same dilution plate at each isolation point. Isolates were subsequently stored in the research collection (R-collection) of the Laboratory of Microbiology (Ghent University) at −80°C under cryoprotection.
For simulation 3, not only was CFC agar used as an isolation medium for pseudomonads, but milk plate count agar (MPCA) (Oxoid Ltd., Basingstoke, United Kingdom) also was used. Only Gram-negative strains (tested with the KOH string test) were retained from MPCA (n = 114) and stored in the R-collection.
Polyphasic identification.
DNA from all isolates (n = 779) was obtained through simple alkaline lysis, and repetitive sequence-based PCR analysis with BOX primers was performed (17). To obtain a first grouping of the isolates, a Pearson correlation-based distance matrix was calculated from all BOX patterns, and the distance matrix was applied in a cluster analysis using the unweighted-pair group method using average linkages (UPGMA). Groups were visually delineated, and for each group, representatives were chosen and further analyzed with fatty acid methyl ester (FAME) analysis and sequencing of the 16S rRNA and rpoB genes to obtain an identification. FAME extraction and analysis were performed as described by Vancanneyt et al. (33). A preliminary identification of the bacteria, based on their FAME profiles, was obtained using the TSBA database (version 5.0) of the MIDI software program (MIDI microbial ID system). Only those representatives belonging to the genus Pseudomonas—according to FAME analysis—were further considered for sequencing of the 16S rRNA and rpoB genes. 16S rRNA gene sequencing was performed as described by Heyrman and Swings (16). Sequencing products were purified with the BigDye XTerminator purification kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions using sequential pipetting and a MixMate (Eppendorf) shaking device. Sequence analysis was performed as described by Coorevits et al. (5). Sequencing of the rpoB gene was executed as described by Tayeb et al. (31); however, for several isolates, no rpoB amplicon could be obtained following the author′s instructions. For those isolates, new primers targeting the rpoB gene were designed: rpoBF′ (5′-CAGTTCATGGACCAGAACAACCCG-3′) and rpoBR′ (5′-ACGCTGGTTGATGCAGGTGTTC-3′), aligning on positions 1552 and 2298 of the rpoB gene sequence of Pseudomonas aeruginosa UCBPP-PA14 (CP000438). A species allocation based on rpoB sequences was obtained by comparing (UPGMA, neighbor-joining algorithm) the representative sequences with publicly available sequences of Pseudomonas type strains, using the BioNumerics software program (version 5.2) (Applied Maths Inc., St. Martens Latem, Belgium).
Extraction of total bacterial DNA from raw milk.
Total bacterial DNA extraction from raw milk was performed using the Adiapure Paratb milk extraction kit (Adiagene, Paris, France) according to the manufacturer's instructions. This kit was designed to extract DNA of Mycobacterium avium subsp. paratuberculosis from milk using magnetic beads and mechanical lysis but proved to be suitable for total bacterial DNA extraction (data not shown). PCR-grade DNA was obtained by extracting the crude DNA with chloroform (1:1).
Denaturating gradient gel electrophoresis (DGGE).
The primers UN357f and UN518r (24) were used to amplify 194 bp of the V3 region of the 16S rRNA gene. A 40-nucleotide (nt) GC-rich sequence (GC-clamp) was attached to the 5′ end of the forward primer (24), resulting in a total amplicon size of 234 bp. One microliter DNA was used as a template in a total reaction volume of 50 μl containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3.5 mM MgCl2, 0.1 mM (each) deoxynucleoside triphosphate (dNTP), 10 pmol of each primer (Eurogentec, Seraing, Belgium), and 1 U Platinum Taq DNA polymerase (Invitrogen Ltd., Paisley, United Kingdom). Amplifications were performed in a DNA thermocycler, GeneAmp 9700 (Applied Biosystems, Foster City, CA). The PCR was set up as a touchdown PCR with annealing temperatures ranging from 65°C to 55°C as previously described by Muyzer et al. (24) to increase the specificity of the amplification and to reduce the formation of by-products. The PCR program started with a denaturation step at 95°C for 5 min, 32 cycles of denaturation at 95°C for 1 min, annealing for 1 min, and elongation at 72°C for 1 min (2 cycles for each annealing temperature and 12 cycles for the final annealing temperature), followed by a final elongation at 72°C for 7 min. Amplicon length was verified on a 1.5% (wt/vol) Seakem LE agarose gel (Cambrex Bio Science Rockland Inc., Rockland, ME) in 1× TAE buffer (40 mM Tris-acetate and 1 mM EDTA at pH 8.3) in comparison with a standard containing DNA fragments of defined lengths (1-kb ladder [0.4 μg μl−1]) (Invitrogen Ltd., Paisley, United Kingdom).
The PCR products were analyzed with the Dcode universal mutation detection system (Bio-Rad, Hercules, CA) on 6% (wt/vol) polyacrylamide gels containing a denaturating gradient from 40 to 50% or 40 to 60% urea and formamide (with 100% corresponding to 7 mol liter−1 urea and 40% formamide [wt/vol]). On each gel, three markers containing 8 reference species were loaded for normalization of the banding pattern using the BioNumerics software program. Migration was performed at 45 V for 16.5 h, and the 1× TAE running buffer temperature was kept constant at 60°C. Patterns were visualized after staining with 1× TAE (pH 8) containing the SybrGold nucleic acid gel stain (Invitrogen Ltd., Paisley, United Kingdom) under UV light and digitally captured using the G:BOX camera (Syngene, Cambridge, United Kingdom). The resulting patterns were analyzed with the BioNumerics software package (version 6.0; Applied Maths, Inc.). Similarities were calculated using DICE correlation, and an average linkage dendrogram was obtained (UPGMA).
Identification of DGGE fragments.
Identification of fragments in the DGGE pattern was performed using different approaches. (i) Marked fragments were excised from the gel using a sterile scalpel, and DNA was subsequently diffused overnight at 5°C in TE buffer (0.05 M Tris [pH 8], 0.02 M EDTA). The DNA was cloned using the pMOSBlue blunt-ended cloning kit (Amersham, Buckinghamshire, United Kingdom) according to the manufacturer's instructions. Plasmid DNA was prepared using the Qiagen (Crawley, United Kingdom) plasmid minikit. Clones were verified with DGGE after PCR amplification for correct positioning of the fragment on the gel, and the cloned gene sequence was determined using the primer T7 from the pMOSBlue blunt-ended cloning kit. Sequencing was performed with a 3130 XL genetic analyzer (Applied Biosystems, Foster City, CA) using the ABI Prism BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems). An identification for the assembled sequence was obtained using the online FASTA tool of EMBL (http://www.ebi.ac.uk/fasta33/), resulting in the 50 most closely related gene sequences retrieved from the EMBL sequence database.
(ii) Representative strains from the predominant taxa isolated from raw milk in this study and a previous study performed by Marchand et al. (20) and type strains representing these species (groups) (i.e., Pseudomonas fluorescens LMG 1794T, Pseudomonas gessardii LMG 21604T, Pseudomonas fragi LMG 2191T, and Pseudomonas lundensis LMG 13517T) were used as marker strains to obtain an identification for some prominent fragments in the DGGE pattern. Therefore, DNA was prepared from each of the type strains as described by Flamm et al. (11), and equal amounts (25 ng) of DNA were pooled and subjected to the DGGE assay as described above.
Data analysis.
Statistical analysis was performed on log total aerobic plate count (tapc) and log Pseudomonas count (pc) response data by means of generalized estimating equations (GEE) using the SPSS Statistics software package (version 17.0). This method allows analysis of correlated data that arise from longitudinal studies: in our study, the subsequent samples taken from the same batch in each simulation experiment. The correlation structure used was autoregressive AR(1). The explanatory variables were sampling time (RM) (Table 1) and condition (optimal/suboptimal).
Furthermore, principal component analysis (PCA) was performed using the BioNumerics software program, version 6.0, on the semiquantitative DGGE profiles and the numbers of isolates per cluster that were picked at the different sampling points (RM8, RM11, and RM14; RM1 was omitted from the analysis since no DGGE pattern could be obtained from these samples due to undetectable levels of bacteria).
Screening for spoilage potential.
Elective media were used as a screening method for proteolytic and lipolytic spoilage potential for all isolates (n = 779), as described by De Jonghe et al. (8). The inoculated media were incubated at 22°C for 72 h, and the display of enzymatic activity (a clear halo around the colony) was checked daily. Per batch of inoculated plates, the diameter (d) of the halo was determined to assess the degree of activity: strong (d > average halo in the same batch), intermediate (d < average halo in the same batch), or no activity (an absence of halo).
Nucleotide sequence accession numbers.
All sequences determined in this work were deposited in EMBL under the accession numbers FN650710 to FN650746 (16S rRNA gene sequences) and FN650748 to FN650791 (rpoB sequences).
RESULTS
Sampling and bacterial counts.
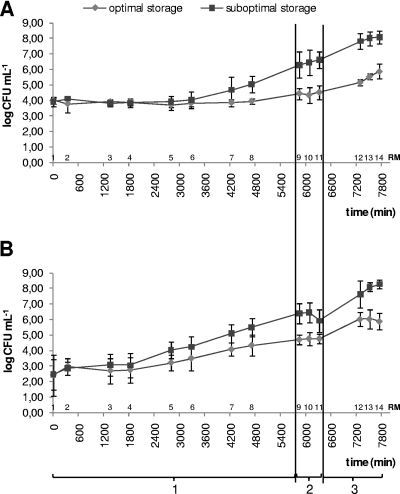
The results for both total aerobic plate count and Pseudomonas count are depicted in Fig. 1. The total aerobic plate count remained fairly stable in the farm tank even under suboptimal storage conditions (see Fig. 1A) (average tapc under optimal storage conditions per simulation [RM1 to RM8], 3.96 ± 0.58 log CFU ml−1 [s1], 2.55 ± 1.00 log CFU ml−1 [s2], and 3.17 ± 0.68 log CFU ml−1 [s3], and under suboptimal storage conditions, 4.37 ± 1.08 log CFU ml−1 [s1], 3.36 ± 1.36 log CFU ml−1 [s2], and 3.74 ± 1.19 log CFU ml−1 [s3]). An outgrowth to approximately 105 CFU ml−1 was visible only after 4 days of storage at the farm. The outgrowth of bacteria was observed during transport and storage at the dairy plant to approximately 106 and 108 CFU ml−1 for optimally and suboptimally cooled raw milk, respectively. However, as shown in Fig. 1B, Pseudomonas members had already started growing within the farm tank and showed an enhanced outgrowth under suboptimal storage conditions of approximately 1 log CFU ml−1 at the end of storage in the farm tank compared to growth under optimal conditions of storage. Further downstream in the simulation of the dairy chain, the difference between optimal and suboptimal storage conditions became even greater (2 log CFU ml−1) in the case of suboptimal storage for both the total aerobic plate count and the Pseudomonas count, reaching levels of 106 and 108 CFU ml−1 in optimally and suboptimally stored raw milk, respectively.
FIG. 1.
Total aerobic plate count (A) and total Pseudomonas count (B) as determined at 14 different times (RM1 to RM14) during all three simulations (simulation of farm tank [4 days, 8 samples] [1], simulation of transport [8 h, 3 samples] [2], and simulation of the dairy plant [24 h, 3 samples] (3).
Polyphasic identification.
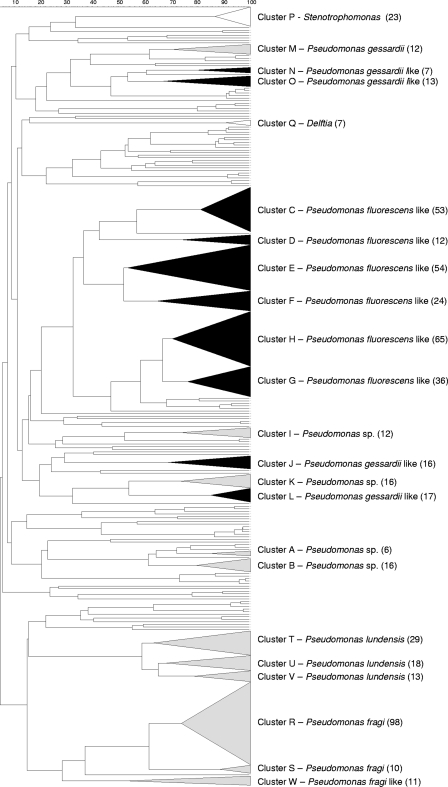
A BOX pattern could be generated for 684 out of 779 presumptive Pseudomonas isolates (87.8%) from the three dairy chain simulations. A grouping of these fingerprints is represented in Fig. 2. This dendrogram shows that 568 isolates (72.9%) were grouped into 23 BOX clusters and 116 isolates appeared separately. The strains were polyphasically identified as shown in Table SA in the supplemental material. Identification was based mainly on the rpoB gene sequence, which has recently been described as a useful taxonomic marker in the genus Pseudomonas (31) (Fig. 3; see also Table SA in the supplemental material). The majority of the isolates in the BOX clusters belonged to the Pseudomonas gessardii-like taxa, which were isolated only in simulation 2 (cluster J [n = 16], cluster L [n = 17], cluster N [n = 7], and cluster O [n = 13]), and Pseudomonas fluorescens-like taxa, which were isolated throughout all three simulations of the dairy chain (cluster C [n = 53], cluster D [n = 12], cluster E [n = 54], cluster F [n = 24], cluster G [n = 36], and cluster H [n = 65]). The annotation “-like” stands for probably novel species of which the rpoB gene sequences showed great similarity (approximately 98%) with those of the P. fluorescens or P. gessardii rpoB gene, as demonstrated in Fig. 3. Two other major groups (Fig. 2) were preliminarily identified as Pseudomonas lundensis (spread over clusters T [n = 13], U [n = 18], and V [n = 29]) and Pseudomonas fragi (spread over clusters R [n = 98] and S [n = 10]). Members of one (smaller) cluster (cluster W [n = 11]) were identified as P. fragi-like, and cluster M (n = 12) was identified as P. gessardii (Fig. 3). Four other clusters probably represent as yet undescribed species within Pseudomonas (cluster A [n = 6], cluster B [n = 16], cluster I [n = 12], and cluster K [n = 16]), since they could not be identified to the species level.
FIG. 2.
BOX-PCR dendrogram of the isolates obtained from the three simulations of the dairy chain. Different clusters are marked with a letter. The number of isolates in each cluster is mentioned between parentheses. Clusters belonging to the P. fluorescens group are visualized in black, and other clusters are shown in gray. P, Stenotrophomonas sp.; M, Pseudomonas gessardii; N and O, P. gessardii-like 2; Q, Delftia sp.; C and D, P. fluorescens-like 1; E and F, P. fluorescens-like 2; H and G, P. fluorescens-like 3; I, Pseudomonas sp. 2; J to L, P. gessardii-like 1; K, Pseudomonas sp. 3; A and B, Pseudomonas sp. 1; T, U, and V, P. lundensis; R and S, P. fragi; W, P. fragi-like.
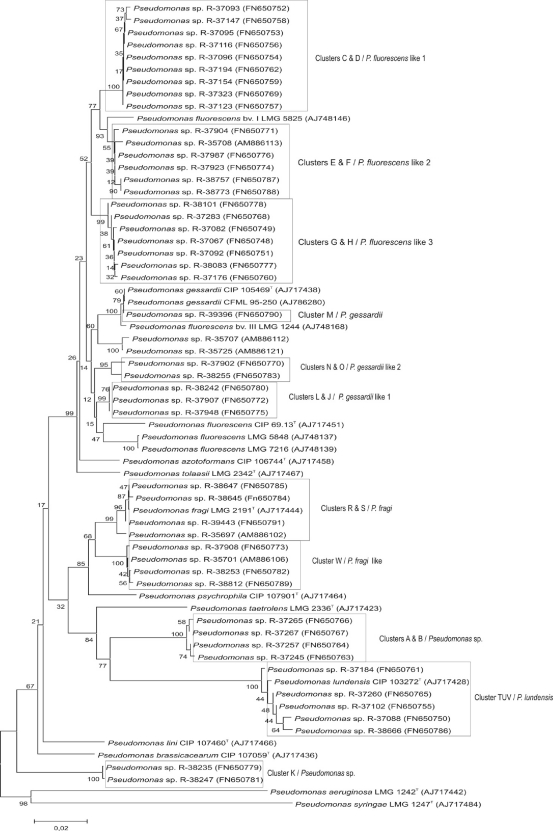
FIG. 3.
Neighbor-joining tree based on rpoB sequences of the milk isolates and closest relatives. The unrooted tree was constructed using the MEGA software program, version 4.0 (30); bootstraps (%) are based on 1,000 replications. Scale bar, 0.02 substitutions/site. The BOX clusters from which the representative strains originate (see Fig. 2) are also given.
In Table 2, the absolute numbers of isolates, picked from the different isolation points throughout the simulation experiment under both storage conditions, are given for each taxon (Fig. 2). P. fragi (clusters R and S) and P. fragi-like (cluster W) were markedly isolated more frequently at the end of the simulation of the dairy chain, as were P. gessardii-like 2 (clusters N and O) and Pseudomonas sp. 3 (cluster K) (56%, 55%, 50%, and 50% of all isolates in that taxon, respectively). Stenotrophomonas sp. and Delftia sp., however, were isolated only at the very beginning of the dairy chain simulation (96% and 100% of all isolates in that taxon, respectively).
TABLE 2.
Percentages of isolates from each taxon that were picked under different storage conditions and at every isolation point throughout the three simulations of the cold dairy chain
| Identification (na) | Cluster(s) | % of isolates |
|||||
|---|---|---|---|---|---|---|---|
| Storage condition |
Sample |
||||||
| Optimal | Suboptimal | RM1 | RM8 | RM11 | RM14 | ||
| Pseudomonas sp. 1 (22) | A, B | 100 | 73 | 27 | |||
| P. fluorescens-like 1 (65) | C, D | 69 | 31 | 3 | 37 | 28 | 32 |
| P. fluorescens-like 2 (78) | E, F | 60 | 40 | 3 | 49 | 32 | 17 |
| P. fluorescens-like 3 (104) | G, H | 62 | 38 | 25 | 23 | 31 | 21 |
| Pseudomonas sp. 2 (12) | I | 83 | 17 | 17 | 25 | 25 | 33 |
| P. gessardii-like 1 (33) | J, L | 52 | 48 | 39 | 27 | 33 | |
| P. gessardii-like 2 (20) | N, O | 35 | 65 | 25 | 25 | 50 | |
| Pseudomonas sp. 3 (16) | K | 12.5 | 87.5 | 31 | 19 | 50 | |
| Pseudomonas gessardii (12) | M | 75 | 25 | 8 | 17 | 58 | 17 |
| Stenotrophomonas sp. (23) | P | 48 | 52 | 96 | 4 | ||
| Delftia sp. (7) | Q | 100 | 100 | ||||
| P. fragi (108) | R, S | 36 | 64 | 10 | 17 | 17 | 56 |
| P. lundensis (60) | TUV | 52 | 48 | 3 | 22 | 40 | 35 |
| P. fragi-like (11) | W | 55 | 45 | 27 | 18 | 55 | |
n, no. of isolates.
Isolates identified as P. gessardii-like 2 (clusters N and O), Pseudomonas sp. 1 (cluster A and B), and Pseudomonas sp. 3 (cluster K) were isolated markedly more frequently under suboptimal storage conditions (65%, 100%, and 87.5% of all isolates in that taxon, respectively) than P. fluorescens-like 1 (clusters C and D), Pseudomonas sp. 2 (cluster I), and P. gessardii (cluster M), which were picked up in much higher numbers in raw milk stored under optimal conditions (69%, 83%, and 75% of all isolates in that taxon, respectively) (Table 2). In general, a different microbial diversity was isolated under suboptimal storage conditions.
Cultivation-independent DGGE.
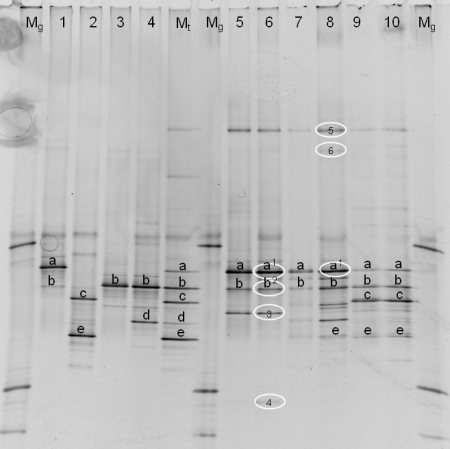
In Fig. 4, the DGGE fragments that were successfully cloned and sequenced are represented, as are patterns of type strains of the individual species that compose the type strain marker (P. gessardii LMG 21604T, P. fragi LMG 2191T, P. lundensis LMG 13517T, and P. fluorescens LMG 1794T). These strains were chosen because they represent four species or highly related taxa which were predominantly isolated in the culture-dependent approach. The buildup and interpretation of the type strain marker is explained in Table SB in the supplemental material.
FIG. 4.
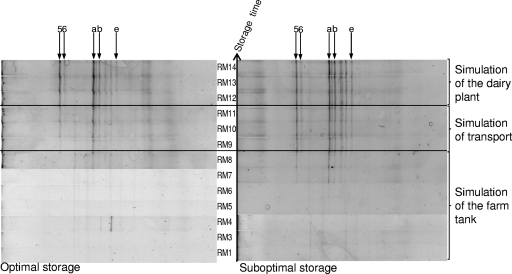
Identification of the DGGE patterns (40 to 50% denaturating gel). The patterns were obtained from milk samples taken at the end of the three simulations. Lanes: Mg, marker for analysis of the gel; 1, P. gessardii (LMG 21604T); 2, P. fragi (LMG 2191T); 3: P. lundensis (LMG 13517T); 4, P. fluorescens (LMG 1794T); Mt, type strain marker (combines lanes 1 to 4); 5, s1 (optimal storage); 6, s1 (suboptimal storage); 7, s2 (optimal storage); 8, s2 (suboptimal storage); 9, s3 (optimal storage); 10, s3 (suboptimal storage). Fragments at the same positions on the gel are marked with letters (a to e). Identifications based on sequencing of cloned fragments are encircled, as follows: 1, Pseudomonas argentinensis/Pseudomonas fulgida/Pseudomonas teesidea/P. gessardii/Pseudomonas cedrella/Pseudomonas libanensis (99.5 and 100% similarity for s1 and s2, respectively); 2, Pseudomonas vranovensis/P. lundensis (98.4% similarity); 3, Pseudomonas sp.; 4, Acinetobacter sp.; 5, P. fluorescens group (100% similarity); 6, Acinetobacter haemolyticus (99.0% similarity).
The right side of the gel in Fig. 4 showed a parallel overview of DGGE patterns obtained at the end of each simulation (RM14) during both optimal and suboptimal storage. It can be concluded from this figure that pseudomonads are the dominant microbiota at the end of the cold chain of raw milk, since they are represented by the most intense bands in the DGGE assay. Members of the P. fluorescens group/P. lundensis (fragments a, b, and 5) could be detected in each of the 3 simulations in both optimally and suboptimally stored milk. P. fragi (fragments c and e) was detected in optimally stored milk from simulation 3 and in suboptimally cooled milk from simulations 2 and 3; however, no P. fragi isolates were obtained from simulation 2. An unknown Pseudomonas sp. was detected in optimally and suboptimally stored milk from simulation 1, although it was isolated only from suboptimally stored raw milk. Two fragments were identified as Acinetobacter species (Fig. 4). Those fragments were more typical of suboptimally cooled raw milk at the end of the simulation of the dairy chain (Fig. 4).
Finally, an example of the outgrowth of Pseudomonas species is represented for simulation 2 in Fig. 5. The outgrowth of Pseudomonas species reached detectable numbers at the start of simulation of transport for optimally stored raw milk (RM9) and at the end of the simulation of storage at the farm tank for suboptimally cooled raw milk (RM8), i.e., when the Pseudomonas count reached approximately 105 CFU ml−1, as can be derived from Fig. 1. This figure demonstrates the applicability of the type strain marker that was developed in this study to monitor the outgrowth of important Pseudomonas species groups throughout storage in the dairy chain.
FIG. 5.
DGGE: comparison between optimal and suboptimal storage (40 to 60% denaturating gel) (s2). Identifications based on marker fragments (letters) and sequence information (numbers) are represented as in Fig. 4 and Table SB in the supplemental material. 5, P. fluorescens group; 6, Acinetobacter haemolyticus; a, P. fluorescens group; b, P. fluorescens group/P. lundensis; e, P. fragi.
Data analysis.
GEE analysis of the colony count data revealed that the increase in total aerobic microbiota (established by determining the tapc) became statistically significant from RM9 onwards (the start of the simulation of transport to the dairy factory) under both optimal and suboptimal storage conditions. However, the outgrowth of Pseudomonas (established by determining the pc) was already statistically significant from RM7 onwards (the beginning of day 4 in the farm tank) for optimal storage conditions and from RM4 onwards (at the end of day 2 in the farm tank) for suboptimal storage conditions. The difference in the tapc between optimal and suboptimal storage became statistically significant from RM8 onwards (the end of day 4 at the farm tank), whereas for pc this was the case from RM 5 onwards the (beginning of day 3 at the farm tank).
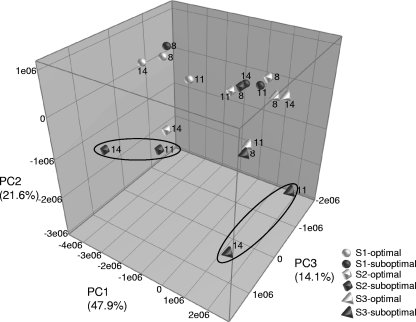
PCA on the composite data set encompasses the (semi) quantitative bacterial diversity data of the different milk samples (RM8, RM11, and RM14) (Table 1) obtained by both cultivation-dependent and DGGE approaches. The three-dimensional (3D) plot (Fig. 6) showed that in two of the three simulations, the Pseudomonas population structure between optimal and suboptimal storage conditions is affected from the simulation of transport to the dairy factory onwards (RM11 to RM14). This was not observed in simulation 1, which showed an overall lesser diversity in Pseudomonas microbiota, probably because this simulation was performed on an individual tank milk sample as opposed to simulations 2 and 3. However, the latter two simulations are in better correspondence with real practice during transport to and storage at the dairy silo since they represent mixed milk samples.
FIG. 6.
Tridimensional PCA plot of milk samples from all three simulations (both optimal and suboptimal storage conditions). The numbers in the figure refer to the milk samples that were analyzed (RM) (see Table 1).
Screening for spoilage potential.
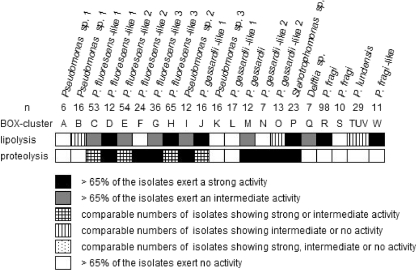
The results for screening of individual strains for spoilage potential on elective media are visualized in Fig. 7. A number of taxa demonstrated lipolytic and proteolytic spoilage potential: Pseudomonas sp. 2, P. gessardii, the different P. fluorescens-like taxa, and a subgroup of P. gessardii-like 1 (BOX cluster J) showed both proteolytic and lipolytic activity, as opposed to P. lundensis and P. gessardii-like 2, which demonstrated mainly proteolytic activity, and P. fragi and P. fragi-like, which showed only an important lipolytic activity. Stenotrophomonas sp. also showed an important spoilage potential, but this can be largely ignored since these strains were not growing out under the simulated storage conditions.
FIG. 7.
Screening for spoilage potential of all isolates on elective media for proteolysis and lipolysis.
DISCUSSION
This study aimed at a better understanding of the outgrowth of Pseudomonas members in raw milk. To achieve this, we determined which storage conditions favor or minimize this outgrowth and thoroughly identified and characterized the isolated strains to assess their enzymatic spoilage potential in heat-treated milk.
From a simulation of different raw milk storage conditions, it was observed that the farmer's efforts to lower the total colony count of raw milk by cooling the raw milk to approximately 4°C or lower seem to pay off, since the tapc remained stable under optimal storage conditions in the farm tank (approximately 4 log CFU ml−1). Still, such low temperatures are not always achieved. Surprisingly, suboptimal storage of raw milk at the farm (approximately 6°C) did not appear to have a great effect on the total aerobic plate count as long as the raw milk was not stored longer than 3 days at the farm. In Belgium, it is mandatory for the dairy companies to collect the raw milk every 2 to 3 days, thereby controlling total aerobic microbiota, as shown in our simulation experiments. However, psychrotolerant bacteria are not as hampered by low storage temperatures (18), with Pseudomonas members making up 90% of the total psychrotolerant microbiota of raw milk (3, 9). In this study, Stenotrophomonas sp. and Delftia acidovorans were isolated only at the beginning of the simulation of the dairy chain, meaning that these species either cannot grow out under refrigerated storage conditions or are overgrown by the better-adapted Pseudomonas species. Further downstream in the simulation of the dairy chain—transport and storage at the dairy plant—the outgrowth of both pseudomonads and total microbiota continues, resulting in a striking difference of 2 log CFU ml−1 between optimal and suboptimal storage. The total aerobic plate count and Pseudomonas count reached the same level at the end of the simulation of the dairy chain (106 and 108 CFU ml−1 for optimally and suboptimally cooled milk, respectively), indicating that cold storage of raw milk selects for the outgrowth of the Pseudomonas microbiota.
A culture-independent molecular approach, using a universal 16S rRNA gene targeted DGGE, allowed confirmation that the pseudomonads, and specifically the P. fluorescens group (comprising members of P. fluorescens, P. gessardii, and highly related taxa, among others [25]), P. lundensis, and P. fragi (and highly related taxa), are the only psychrotolerant bacteria able to grow out in the cooled raw milk. DGGE and cultivation monitoring proved to be complementary approaches, since some Pseudomonas species were detected with the DGGE assay that could not be isolated in a particular simulation or storage condition. Furthermore, Acinetobacter appeared in DGGE patterns at the end of some simulations. DGGE monitoring allowed visualization that the final predominant Pseudomonas species composition is already formed during transport under suboptimal cooling conditions, whereas under optimal cooling conditions, this is formed in the dairy plant silo. This means that under suboptimal storage conditions, the bacterial population can be more active in producing spoilage enzymes, which may have a more pronounced effect on the spoilage potential of the end product. DGGE has proven to be a powerful tool for giving an overall picture of spoilage-related changes in microbial communities, such as in pasteurized milk, as demonstrated by He et al. (15). However, the difficulties in obtaining an accurate species assignment, as encountered in our study, were also confirmed in that study.
Our study showed that, at least for research purposes, a polyphasic approach is still indispensable to achieving an accurate identification of Pseudomonas isolates. The rpoB gene sequence (27, 31) was shown to give the highest taxonomic resolution at the species level. An important remark remains that the rpoB gene still needs to be validated as a taxonomic marker for species delineation (by extensive DNA-DNA hybridizations) because the boundaries of the various species of Pseudomonas still remain obscure. The dominant Pseudomonas microbiota was identified as members of the P. fluorescens group (P. fluorescens-like and P. gessardii-like), P. lundensis, and P. fragi(-like). Within some taxa delineated by rpoB sequencing, several subgroups that may represent novel species (three in the P. fluorescens-like taxon, two in the P. gessardii-like taxon, and one in the P. fragi-like taxon) could be defined. Moreover, three taxa, identified as Pseudomonas sp., may also represent novel species within the genus Pseudomonas.
In general, the retrieved dominant microbiota from this study largely extends earlier work by Marchand et al. (20), who identified some of these species (or species groups) as the predominant proteolytic spoilers isolated from raw milk. Further novel species allocations must be verified by more in-depth taxonomic studies also comprising DNA-DNA hybridization experiments. It is clear though that the taxonomic situation of the genus Pseudomonas, and especially that of the P. fluorescens group, is very confusing and needs revision (2, 20, 26). In this context, the recently published multilocus sequence analysis (MLSA) data are very promising (27).
The dominant Pseudomonas microbiota that was established in this study also shows an important proteolytic and lipolytic spoilage potential as determined using elective media. Not only does the general outgrowth of members of known Pseudomonas species represent a potential danger for milk spoilage, but also the isolation of some presumptive novel species under suboptimal storage conditions indicates that these conditions favor the development of a larger diversity of Pseudomonas microbiota with possible spoilage potential. These new insights make it more difficult to assess the spoilage potential of processed dairy products made from such milk. It is not clear from our simulation experiments whether members of Acinetobacter species, among which is A. haemolyticus, as demonstrated by sequence analysis of a DGGE band (fragment 6 in Fig. 4), indicate a possible risk of a still more complex microbiota with proteolytic and lipolytic traits (14). The presence of Acinetobacter in particular also poses a safety issue since Acinetobacter haemolyticus is able to produce Shiga toxin, a toxin that can cause bloody diarrhea upon ingestion (13).
Our second objective, achieving a thorough identification of the isolates, had implications for the interpretation of the DGGE banding patterns. The type strain marker approach presented here, together with excision and sequencing of some fragments, for the interpretation of complex DGGE banding patterns showed that this method has the potential to monitor the evolution of the diversity of the Pseudomonas psychrotolerants in dairy processing. However, prior detailed knowledge of the actual composition of the microbiota in the sample (in this case provided by a parallel culturing procedure and a thorough polyphasic identification of the strains) is an absolute prerequisite for selecting the marker species for correct interpretation of present and future DGGE banding patterns. Otherwise, the historical misconception of the predominant role for P. fluorescens would have been falsely confirmed by our DGGE results (see Table SB in the supplemental material). The use of the rpoB gene as a target gene for DGGE analysis might therefore improve the identification capacities of the DGGE approach. Furthermore, the rpoB gene appears as a single copy in the bacterial genome, thereby avoiding the interpretation problems caused by the allelic nature of the 16S rRNA gene.
In the third simulation experiment, not only was the Pseudomonas-specific medium CFC agar used for isolation, but also the more general MPCA medium (only the Gram-negative isolates were taken into consideration) was used to test whether members of other important taxa might have been missed on CFC agar. Surprisingly, the general diversity of Pseudomonas members retrieved from MPCA was less than that from CFC agar and comprised mainly P. fragi- (-like) and to a lesser extent Pseudomonas sp. 2, P. gessardii, and a few isolates belonging to P. lundensis and P. fluorescens-like groups (data not shown). This may indicate that these bacteria are better equipped to compete with other raw milk microbiota during cultivation. It also indicates the important bias which can be introduced by the use of general growth medium.
To our knowledge, this is the first study that attempted to monitor (on a laboratory scale) the outgrowth of Pseudomonas microbiota throughout the first part of the dairy chain. A striking result is that P. fragi(-like) and to a lesser extent P. lundensis were predominantly isolated at the end of the simulation of the dairy chain (visualized by DGGE). This might indicate that these organisms may be controlled by adequate cooling and/or rapid processing at the dairy plant. This study further shows that minimizing the outgrowth of spoilage microbiota must be the result of contributions in every step in the dairy chain, and control of raw milk quality should not be restricted to the farm tank level. It is highly recommended to reduce the storage time of raw milk prior to processing to a minimum and to keep the storage temperature as low as possible (preferentially 3.5°C or lower) throughout the dairy chain.
Conclusions.
The use of refrigerated conditions throughout milk processing in order to maintain a safe product has created a specific niche in which the psychrotolerant spoilage microbiota can thrive. This work studied the implications associated with the stringency of this cold storage by comparing the Pseudomonas microbiota that can grow out under optimal and suboptimal conditions. It appeared that prolonged storage under suboptimal conditions indeed significantly affected the growth rate of the Pseudomonas strains, resulting in a 2 log CFU ml−1 difference from results with optimal storage before processing.
This study demonstrated that the combined use of cultivation, spoilage potential characterization, and noncultivation monitoring of the psychrotolerant bacteria in raw milk helps to better assess the shelf life risks in the end product.
Supplementary Material
Acknowledgments
This work was supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT), Program for Agricultural Investigation.
We thank Ann Vanhee, Els Verween, and Emly Samyn for their excellent technical assistance. We are grateful to the staff of the dairies for their help organizing the sampling campaign and to the sampled dairy farms for their hospitality.
Footnotes
Published ahead of print on 29 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, D. M., J. T. Barach, and M. L. Speck. 1975. Heat resistant proteases produced in milk by psychrotrophic bacteria of dairy origin. J. Dairy Sci. 58:828-834. [DOI] [PubMed] [Google Scholar]
- 2.Bossis, E., P. Lemanceau, X. Latour, and L. Gardan. 2000. The taxonomy of Pseudomonas fluorescens and Pseudomonas putida: current status and need for revision. Agronomie 20:51-63. [Google Scholar]
- 3.Champagne, C. P., R. R. Laing, D. Roy, A. A. Mafu, and M. W. Griffiths. 1994. Psychrotrophs in dairy products: their effects and their control. Crit. Rev. Food Sci. Nutr. 34:1-30. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L., R. M. Daniel, and T. Coolbear. 2003. Detection and impact of protease and lipase activities in milk and milk powders. Int. Dairy J. 13:255-275. [Google Scholar]
- 5.Coorevits, A., et al. 2008. Comparative analysis of the diversity of aerobic spore-forming bacteria in raw milk from organic and conventional dairy farms. Syst. Appl. Microbiol. 31:126-140. [DOI] [PubMed] [Google Scholar]
- 6.Cousin, M. A. 1982. Presence and activity of psychrotrophic microorganisms in milk and dairy products: a review. J. Food Prot. 45:172-207. [DOI] [PubMed] [Google Scholar]
- 7.Datta, N., and H. C. Deeth. 2001. Age gelation of UHT-milk—a review. Food Bioprod. Process. 79:197-210. [Google Scholar]
- 8.De Jonghe, V., et al. 2010. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int. J. Food Microbiol. 136:318-325. [DOI] [PubMed] [Google Scholar]
- 9.Dogan, B., and K. J. Boor. 2003. Genetic diversity and spoilage potentials among Pseudomonas spp. isolated from fluid milk products and dairy processing plants. Appl. Environ. Microbiol. 69:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy, B. P. 1960. The use and meaning of the term ‘psychrophilic.’ J. Appl. Bacteriol. 23:189-190. [Google Scholar]
- 11.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flint, S., and N. Hartley. 1996. A modified selective medium for the detection of Pseudomonas species that cause spoilage of milk and dairy plants. Int. Dairy J. 6:223-230. [Google Scholar]
- 13.Grotiuz, G., A. Sirok, P. Gadea, G. Varela, and F. Schelotto. 2006. Shiga toxin 2-producing Acinetobacter haemolyticus associated with a case of bloody diarrhea. J. Clin. Microbiol. 44:3838-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hantsis-Zacharov, E., and M. Halpern. 2007. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl. Environ. Microbiol. 73:7162-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, H., J. Dong, C. N. Lee, and Y. Li. 2009. Molecular analysis of spoilage-related bacteria in pasteurized milk during refrigeration by PCR and denaturing gradient gel electrophoresis. J. Food Prot. 72:572-577. [DOI] [PubMed] [Google Scholar]
- 16.Heyrman, J., and J. Swings. 2001. 16S rDNA sequence analysis of bacterial isolates from biodeteriorated mural paintings in the Servilia tomb (Necropolis of Carmona, Seville, Spain). Syst. Appl. Microbiol. 24:417-422. [DOI] [PubMed] [Google Scholar]
- 17.Heyrman, J., J. Verbeeren, P. Schumann, J. Swings, and P. De Vos. 2005. Six novel Arthrobacter species isolated from deteriorated mural paintings. Int. J. Syst. Evol. Microbiol. 55:1457-1464. [DOI] [PubMed] [Google Scholar]
- 18.Lafarge, V., et al. 2004. Raw cow milk bacterial population shifts attributable to refrigeration. Appl. Environ. Microbiol. 70:5644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law, B. A., A. T. Andrews, and M. E. Sharpe. 1977. Gelation of UHT-sterilized milk by proteases from a strain of Pseudomonas fluorescens, isolated from raw milk. J. Dairy Res. 44:145-148. [Google Scholar]
- 20.Marchand, S., et al. 2009. Seasonal influence on heat resistant proteolytic capacity of Pseudomonas lundensis and Pseudomonas fragi, predominant milk spoilers isolated from Belgian raw milk samples. Environ. Microbiol. 11:467-482. [DOI] [PubMed] [Google Scholar]
- 21.Meer, R. R., J. Bakker, F. W. Bodyfelt, and M. W. Griffiths. 1991. Psychotrophic Bacillus spp. in fluid milk products: a review. J. Food Prot. 54:969-979. [DOI] [PubMed] [Google Scholar]
- 22.Morita, R. Y. 1975. Psychrophilic bacteria. Bacteriol. Rev. 39:146-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munsch-Alatossava, P., and T. Alatossava. 2006. Phenotypic characterization of raw milk-associated psychrotrophic bacteria. Microbiol. Res. 161:334-346. [DOI] [PubMed] [Google Scholar]
- 24.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palleroni, N. J. 2005. Genus I. Pseudomonas, p. 323-379. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part B. Springer-Verlag, New York, NY. [Google Scholar]
- 26.Palleroni, N. J., R. Kunisawa, R. Contopoulou, and M. Doudoroff. 1973. Nucleic acid homologies in the genus Pseudomonas. Int. J. Syst. Bacteriol. 23:333-339. [Google Scholar]
- 27.Peix, A., M. H. Ramirez-Bahena, and E. Velazquez. 2009. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 9:1132-1147. [DOI] [PubMed] [Google Scholar]
- 28.Simões, M., L. C. Simões, and M. J. Vieira. 2009. Species association increases biofilm resistance to chemical and mechanical treatments. Water Res. 43:229-237. [DOI] [PubMed] [Google Scholar]
- 29.Sørhaug, T., and L. Stepaniak. 1997. Psychrotrophs and their enzymes in milk and dairy products: quality aspects. Trends Food Sci. Technol. 8:35-41. [Google Scholar]
- 30.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 31.Tayeb, L. A., E. Ageron, F. Grimont, and P. A. Grimont. 2005. Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res. Microbiol. 156:763-773. [DOI] [PubMed] [Google Scholar]
- 32.Ternström, A., A. M. Lindberg, and G. Molin. 1993. Classification of the spoilage flora of raw and pasteurized bovine milk, with special reference to Pseudomonas and Bacillus. J. Appl. Bacteriol. 75:25-34. [DOI] [PubMed] [Google Scholar]
- 33.Vancanneyt, M., S. Wit, W. R. Abraham, K. Kersters, and H. L. Fredrickson. 1996. Fatty acid content in whole-cell hydrolysates and phospholipid fractions of pseudomonads: a taxonomic evaluation. Syst. Appl. Microbiol. 19:528-540. [Google Scholar]
- 34.Wiedmann, M., D. Weilmeier, S. S. Dineen, R. Ralyea, and K. J. Boor. 2000. Molecular and phenotypic characterization of Pseudomonas spp. isolated from milk. Appl. Environ. Microbiol. 66:2085-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.