Abstract
The objective of this study was to analyze multifunctional autoprocessing repeats-in-toxin (MARTX) toxin domain organization within the aquatic species Vibrio vulnificus as well as to study the evolution of the rtxA1 gene. The species is subdivided into three biotypes that differ in host range and geographical distribution. We have found three different types (I, II, and III) of V. vulnificus MARTX (MARTXVv) toxins with common domains (an autocatalytic cysteine protease domain [CPD], an α/β-hydrolase domain, and a domain resembling that of the LifA protein of Escherichia coli O127:H6 E2348/69 [Efa/LifA]) and specific domains (a Rho-GTPase inactivation domain [RID], a domain of unknown function [DUF],a domain resembling that of the rtxA protein of Photorhabdus asymbiotica [rtxAPA], and an actin cross-linking domain [ACD]). Biotype 1 isolates harbor MARTXVv toxin types I and II, biotype 2 isolates carry MARTXVv toxin type III, and biotype 3 isolates have MARTXVv toxin type II. The analyzed biotype 2 isolates harbor two identical copies of rtxA1, one chromosomal and the other plasmidic. The evolutionary history of the gene demonstrates that MARTXVv toxins are mosaics, comprising pieces with different evolutionary histories, some of which have been acquired by intra- or interspecific horizontal gene transfer. Finally, we have found evidence that the evolutionary history of the rtxA1 gene for biotype 2 differs totally from the gene history of biotypes 1 and 3.
Vibrio vulnificus is an “accidental” pathogen, inhabiting brackish water ecosystems in temperate and tropical regions (22). The species has been subdivided into three biotypes on the basis of genetic, phenotypic, and host range differences (4, 35). Biotypes 1 and 2 are distributed worldwide while biotype 3 is geographically restricted to Israel (1, 4, 11, 35). Three O serovars have been described in biotype 2 (serovar A, serovar E, and serovar I), and one has been identified in biotype 3 (serovar O) while biotype 1 has not been fully serotyped (2, 5, 11).
The three biotypes of V. vulnificus can cause disease in humans after ingestion of contaminated seafood or immersion of an open wound in brackish waters (22). In healthy people, the ingestion of V. vulnificus leads to vomiting and diarrhea while external exposure produces wound infection or severe ulcerations; however, in immunocompromised people, particularly those with chronic liver disease, the pathogen can invade the bloodstream and cause septicemia, leading to death in almost 50% of the cases (22). Interestingly, biotype 2 can also infect fish (mainly eels) and shrimps and cause death by hemorrhagic septicemia, a disease called warm-water vibriosis (35). Human isolates of biotype 2 belong to the most virulent fish serovar, serovar E (1).
Several studies have been performed with biotype 1 isolates to elucidate the genetic basis of V. vulnificus virulence for humans. These isolates produce a wide range of putative virulence factors, among which is a repeats-in-toxin (RTX) toxin, which belongs to the subfamily of multifunctional autoprocessing RTX (V. vulnificus MARTX [MARTXVv]toxin, or RtxA1) toxins. rtxA1 mutants are less virulent for mice and have lower cytotoxicity for different cell lines (17). The rtxA1 gene is practically identical in the two sequenced isolates of biotype 1 (7, 15) (98.9% and 99.5% nucleotide and amino acid identity, respectively) and codes for a protein of 5,206 amino acids (aa), which shows extensive regions of similarity with the MARTX toxin of Vibrio cholerae (MARTXVc toxin), the most extensively studied MARTX toxin to date (28). Both toxins share the N- and C-terminal repetitive regions, which seem to be common to all MARTX toxins, a Rho-GTPase inactivation domain (RID), an autocatalytic cysteine protease domain (CPD), and an α/β-hydrolase domain. Meanwhile, they differ in that the biotype 1 MARTXVv (MARTXVvbt1) toxin lacks an actin cross-linking domain (ACD) and presents three additional putative domains, DUF3 (domain of unknown function), Mcf (a domain similar to an Mcf toxin of Photorhabdus luminescens), and PMT C1/C2 (a domain similar to a portion of the Pasteurella mitogenic toxin PMT) (27).
The genetic basis underlying fish virulence is related to a recently described plasmid of 68 to 70 kb (18). This plasmid codes for resistance to the innate immune system of fishes and is currently under study. The plasmid also carries an rtxA1 gene, which codes for a toxin that seems to be a hybrid between the MARTXVvbt1 toxin and the MARTXVc toxin because it lacks the RID domain and presents an ACD domain (18). Interestingly, the chromosome of the only biotype 2 isolate studied in detail so far also harbors a copy of rtxA1 although it is as yet unknown whether this copy is similar to the plasmidic or the MARTXVvbt1 toxin one (18). The relevance of both copies for virulence in fishes and mice is currently under study.
Considering the aforementioned results, we hypothesized that the rtxA1 gene of V. vulnificus is a mosaic gene, which varies within and among biotypes because it has acquired, probably through horizontal gene transfer (HGT) and subsequent integration into the original allele, some DNA sequences from different bacterial species. To test this idea, we studied the domain structure of MARTXVv toxin as well as the evolution of the rtxA1 gene in V. vulnificus using different approaches. First, we sequenced the complete rtxA1 gene in seven isolates and a 828-bp fragment from the 5′ region of the gene in a collection of 115 strains of different biotypes and serovars (Table 1). Second, we analyzed the mosaic domain organization of the predicted protein by comparing it with the sequences previously published in V. vulnificus (strains YJ016, CMCP6, CECT 4999, and CECT 4602), V. cholerae (strain N16961), V. splendidus (12B01), and Aeromonas hydrophila (strain ATCC 7966) (27). The sequences of the MARTX toxin of V. cholerae M66-2, AM19226, N16961, MO10, and O395 as well as of Listonella anguillarum M93 (20) were also included although the domain organization of these proteins has not been published previously. Finally, we analyzed the phylogeny of the rtxA1 gene from whole gene sequences, from the sequences of the selected common fragment, and from each separate domain using the previously published homologous sequences from V. cholerae, V. splendidus, A. hydrophila, and L. anguillarum as outgroups.
TABLE 1.
V. vulnificus strains used in the study: source, biotype, and country of isolation
| Strain(s) | Biotype/serovara | Origin or reference | Country of isolation |
|---|---|---|---|
| ATCC 33816, CECT 5164, CECT 5168, CECT 5169, CECT 529Tb,c,d | t1/NT | Human blood | USA |
| CECT 5166 | Bt1/NT | Wound infection | USA |
| CECT 5165 | Bt1/NT | Seawater | USA |
| E4, JE | Bt1/NT | Seafood | USA |
| MLT364, MLT404, MLT406, MLT362, VV1003, VV352, VV425 | Bt1/NT | Environment | USA |
| V4 | Bt1/NT | Human blood | Australia |
| CS9133 | Bt1/NT | Human blood | South Korea |
| CECT 5167, N87, KH03, YN03 | Bt1/NT | Human blood | Japan |
| L49 | Bt1/NT | Brackish water | Japan |
| YJ106d | Bt1/NT | Human blood | Taiwan |
| CG118 | Bt1/NT | Seawater | Taiwan |
| CG100d, CG106 | Bt1/NT | Oyster | Taiwan |
| CG110, CG111 | Bt1/NT | Seawater | Taiwan |
| CECT 4867 | Bt1/NT | Diseased eel | Sweden |
| 94-9-118 | Bt1/NT | Human expectoration | Denmark |
| 94-9-130 | Bt1/NT | Water | Denmark |
| 94-9-119c | Bt1/NT | Human wound | Denmark |
| A2, A4, A5, A6, A7, PD-1, PD-3, PD-5, PD-12, PD-2-66, V1 | Bt1/NT | Eel tank water | Spain |
| CECT 4606d | Bt1/NT | Healthy eel | Spain |
| Riu-1c, Riu-3 | Bt1/NT | Seawater | Spain |
| 94385 | Bt1/NT | Leg wound | Spain |
| CECT 4608 | Bt1/NT | Eel farm water | Spain |
| CECT 5768, A13, A21, A22, CECT 5689d, CECT 5769, A15, A16, A17, A18, CECT 5198, CECT 5343, A10, A11, A14c,d | Bt2/SerA | Diseased eel | Spain |
| 4/7/17, 4/7/19, 21B, 22, 26, 27, 21A | Bt2/SerA | Diseased eel | Denmark |
| CECT 4864, CECT 4999d, CECT 4605, CECT 4607, CECT 4604d, CECT 4602, CECT 4603, CECT 4601 | Bt2/SerE | Diseased eel | Spain |
| PD-2-47, PD-2-50, PD-2-51, PD-2-55, PD-2-56, CECT 5763 | Bt2/SerE | Eel tank water | Spain |
| C1, CECT 5762 | Bt2/SerE | Healthy eel | Spain |
| Riu-2d | Bt2/SerE | Seawater | Spain |
| CIP8190 | Bt2/SerE | Human blood | France |
| CECT 4868 | Bt2/SerE | Diseased eel | Norway |
| 90-2-11 | Bt2/SerE | Diseased eel | Denmark |
| 4-8-112 | Bt2/SerE | Human wound | Denmark |
| 94-9-123 | Bt2/SerE | Seawater | Denmark |
| CCUG38521 | Bt2/SerE | Human blood | Sweden |
| Ö122 | Bt2/SerE | Diseased eel | Sweden |
| CECT 4866 | Bt2/SerE | Human blood | Australia |
| CECT 4865 | Bt2/SerE | Diseased shrimp | Taiwan |
| UE516 | Bt2/SerE | Diseased eel | Taiwan |
| CECT 898, CECT 897, CECT 4174d, CECT 4862 | Bt2/SerE | Diseased eel | Japan |
| 95-8-161c,d, 95-8-162 | Bt2/SerI | Diseased eel | Denmark |
| CECT 4869 | Bt2/SerI | Diseased eel | Belgium |
| 95-8-7 | Bt2/SerI | Diseased eel | Denmark |
| 95-8-6d | Bt2/SerI | Diseased eel | Denmark |
| 97, 162, 11028c,d, vv12, vv32 | Bt3/SerO | Diseased human | Israel |
| CT218c,d,e | Bt2/SerE | 18 |
Bt, biotype; NT, nontypeable; SerE, serovar E; SerA, serovar A; SerI, serovar I; SerO, serovar O.
Type strain.
Strain used to sequence the whole rtxA gene.
Strain used to locate the rtxA gene.
Cured derivative biotype 2 isolate.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The V. vulnificus strains used in this study are listed in Table 1. Strains were routinely grown in Trypticase soy broth or agar plus 5 g liter−1 NaCl (TSB-1 or TSA-1, respectively; Pronadisa, Spain) at 28°C for 24 h. For genomic DNA extraction, bacteria were grown overnight with shaking at 28°C. The strains were maintained both as lyophilized and as frozen stocks at −80°C in marine broth (Difco) plus 20% (vol/vol) glycerol.
DNA extractions.
Genomic DNA was extracted according to the mini-prep protocol for genomic DNA extraction (3). Plasmid DNA was extracted following the TENS protocol (36) with the modifications of Roig and Amaro (24).
Location of the rtxA1 gene.
The rtxA1 gene was located in the chromosome or in the plasmids of the strains CECT 529T, YJ106, CG100, CECT 4606, CECT 5689, A14, CECT 4999, CECT 4604, Riu-2, CECT 4174, 95-8-161, 95-8-6, 11028, and CT218 (Table 1) by Southern hybridization, essentially as described by Roig and Amaro (24). Briefly, DNA samples were subjected to electrophoresis in 0.7% (wt/vol) agarose gels (molecular grade; Roche) and transferred to the membranes (zeta-probe blotting membrane; Bio-Rad) with a vacuum blotter (model 785; Bio-Rad) following the manufacturer's instructions. The samples were genomic DNA extractions (3) and plasmid extractions (24). As molecular weight markers (MWM) Generuler DNA ladder mix and plasmids of the E. coli strain 39R861 were used. Probes of 1,087 nucleotides (nt) were generated from PCR products obtained with primers 2F and 2R (Table 2 ) labeled with digoxigenin (DIG)-11-dUTP (Roche, Switzerland), and the probes against the MWM were derived from the MWM using a DIG High-Prime kit (Roche, Switzerland. Hybridization was performed using a DIG Easy Hyb Granules kit (Roche, Switzerland), and color was developed with anti-digoxigenin-alkaline phosphatase (AP) Fab fragments together with blocking reagent (Roche, Switzerland), which were used following the manufacturer's instructions.
TABLE 2.
Primer pairs for sequencing rtxA1 in various strains
| CECT 4999 (C), A14 (P), and 95-8-161 (P)a |
CECT 529T |
11028 |
94-9-119 |
Riu-1 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Primer | Sequence (5′-3′) | Primer | Sequence (5′-3′) | Primer | Sequence (5′-3′) | Primer | Sequence (5′-3′) | Primer | Sequence (5′-3′) |
| 0F | GTGACGCTCTTTGGTGCTC | 0F | GTGACGCTCTTTGGTGCTC | 0F | GTGACGCTCTTTGGTGCTC | 0F | GTGACGCTCTTTGGTGCTC | 0F | GTGACGCTCTTTGGTGCTC |
| 0R | CAGTCGTTTGTCTTTGGTG | 0R | CAGTCGTTTGTCTTTGGTG | 0R | CAGTCGTTTGTCTTTGGTG | 0R | CAGTCGTTTGTCTTTGGTG | 0R | CAGTCGTTTGTCTTTGGTG |
| 1F | CCACACGGTTGAAGTTGCCA | 1F | CCACACGGTTGAAGTTGCCA | 1F | CCACACGGTTGAAGTTGCCA | 1F | CCACACGGTTGAAGTTGCCA | 1F | CCACACGGTTGAAGTTGCCA |
| 1R | ACGCCAGAAGATTTGCCTCGT | 1R | ACGCCAGAAGATTTGCCTCGT | 1R | ACGCCAGAAGATTTGCCTCGT | 1R | ACGCCAGAAGATTTGCCTCGT | 1R | ACGCCAGAAGATTTGCCTCGT |
| 2F | GACGAGGCAAATCTTCTGGCG | 2F | GACGAGGCAAATCTTCTGGCG | 2F | GACGAGGCAAATCTTCTGGCG | 2F | GACGAGGCAAATCTTCTGGCG | 2F | GACGAGGCAAATCTTCTGGCG |
| 2R | GATGGATGCGAATGGTCTT | 2R | GATGGATGCGAATGGTCTT | 2R | GATGGATGCGAATGGTCTT | 2R | GATGGATGCGAATGGTCTT | 2R | GATGGATGCGAATGGTCTT |
| 3F_Bt2 | CGTCTACAGCCAGTTCAG | 3F_Bt1 | GATGTCGCCTTCCGCAATACC | 3F_Bt1 | GATGTCGCCTTCCGCAATACC | 3F_Bt1 | GATGTCGCCTTCCGCAATACC | 3F_Bt2 | CGTCTACAGCCAGTTCAG |
| 3R_Bt2 | GGTGGATGCGAAAGCGACT | 3R_Bt2 | GGTGGATGCGAAAGCGACT | 5R_Bt1 | CTGAGCAATCCAACCCTTTAC | 3R_Bt1 | GGAGTCACAGTCATTGGC | 3R_Bt2 | GGTGGATGCGAAAGCGACT |
| 4F_Bt2 | CTGTCTGAGTAAAGCGTTGG | 4F_Bt2 | CTGTCTGAGTAAAGCGTTGG | 6F_Bt1 | CCTGCCACACTTTCATC | 4F2_Bt1 | AATGTTGTTCGCAGCACCAG | 4F_Bt2 | CTGTCTGAGTAAAGCGTTGG |
| 4R_Bt2 | CAACCTGCGTGGCTATGG | 6R_Bt1 | GCGGTGTCTGGCTTATT | 6R_Bt1 | GCGGTGTCTGGCTTATT | 4R2_Bt1 | CAGGGCGTAATCAAGCG | 4R_Bt2 | CAACCTGCGTGGCTATGG |
| 5F_Bt2 | CGCTTTGGTCATACTTGGC | 7F_Bt1 | CGTTAGCAGCAGAATCGGCGT | 7F_Bt1 | CGTTAGCAGCAGAATCGGCGT | 5F_Bt1 | CGCATCAAAACCAAGGTCAC | 5F_Riu1 | CGGTCAAGCAATAAGCCAGA |
| 5R_Bt2 | ATGCGAAGAACCCAATG | 7R_Bt1 | TCTGAAGGCTATGCGAG | 7R_Bt1 | TCTGAAGGCTATGCGAG | 5R_Bt1 | CTGAGCAATCCAACCCTTTAC | 5R_Riu1 | GTATCTGGACGGAAACGG |
| 6F_Bt2 | GGTATCAAGAACATCAGTG | 8F_Bt1 | CTTCATCTGCTTGGCTGGC | 6F_Riu1 | GACAAGGCATCAAGGAG | 6F_Bt1 | CCTGCCACACTTTCATC | 6F_Riu1 | GACAAGGCATCAAGGAG |
| 6R_Bt2 | TGCTCGTCTCATTGCTTTC | 8R_Bt1 | GGATAACAAGGTGCTCTCGC | 8R_Bt1 | GGATAACAAGGTGCTCTCGC | 6R_Bt1 | GCGGTGTCTGGCTTATT | 8F_Bt1 | CTTCATCTGCTTGGCTGGC |
| 7F_Bt2 | GTCTTCCTGTAGTCCTTTC | 8F1_Bt1 | GATACTCCGTCAGGGCGT | 8F1_Bt1 | GATACTCCGTCAGGGCGT | 7F_Bt1 | CGTTAGCAGCAGAATCGGCGT | 6R_Riu1 | GTGGTATGTTCCTTGAC |
| 7R_Bt2 | CCAGTCAAACCACCAAAGC | 8R1_Bt1 | CGATTATGACTGGGACAAACGC | 8R1_Bt1 | CGATTATGACTGGGACAAACGC | 7R_Bt1 | TCTGAAGGCTATGCGAG | 6R2_Riu1 | GCTGGTGGTTTGTTAGA |
| 8F_Bt2 | GCCCATCATCACTCACC | 8F2_Bt1 | CCAGACCAGAAGGAAACAATGC | 8F2_Bt1 | CCAGACCAGAAGGAAACAATGC | 8F_Bt1 | CTTCATCTGCTTGGCTGGC | 9F | GTGTGCGTCAGATTCGG |
| 8R_Bt2 | GTCCGTTGCGAAGTCTGAAGC | 8R2_Bt1 | GAAATCTGAAGCGGGTGTGG | 8R2_Bt1 | GAAATCTGAAGCGGGTGTGG | 8R_Bt1 | GGATAACAAGGTGCTCTCGC | 9R | GAAGAAGTGGGCTCGGAC |
| 9F | GTGTGCGTCAGATTCGG | 9F | GTGTGCGTCAGATTCGG | 9F | GTGTGCGTCAGATTCGG | 8F1_Bt1 | GATACTCCGTCAGGGCGT | 10F | CCTTCACCATTCACGCCATA |
| 9R | GAAGAAGTGGGCTCGGAC | 9R | GAAGAAGTGGGCTCGGAC | 9R | GAAGAAGTGGGCTCGGAC | 8R1_Bt1 | CGATTATGACTGGGACAAACGC | 10R | GGTGTTGTGGCGGCAGC |
| 10F | CCTTCACCATTCACGCCATA | 10F | CCTTCACCATTCACGCCATA | 10F | CCTTCACCATTCACGCCATA | 8F2_Bt1 | CCAGACCAGAAGGAAACAATGC | 11F | CAACAACCCTACCGTGG |
| 10R | GGTGTTGTGGCGGCAGC | 10R | GGTGTTGTGGCGGCAGC | 10R | GGTGTTGTGGCGGCAGC | 8R2_Bt1 | GAAATCTGAAGCGGGTGTGG | 11R | CCGCAGGTATGTTGGGC |
| 11F | CAACAACCCTACCGTGG | 11F | CAACAACCCTACCGTGG | 11F | CAACAACCCTACCGTGG | 9F | GTGTGCGTCAGATTCGG | 12F | CACCGCCCATCACCGCAA |
| 11R | CCGCAGGTATGTTGGGC | 11R | CCGCAGGTATGTTGGGC | 11R | CCGCAGGTATGTTGGGC | 9R | GAAGAAGTGGGCTCGGAC | 12R | AGTGTGCCATTGCCGTAGG |
| 12F | CACCGCCCATCACCGCAA | 12F | CACCGCCCATCACCGCAA | 12F | CACCGCCCATCACCGCAA | 10F | CCTTCACCATTCACGCCATA | 13F | CACCGCTTGAACATCG |
| 12R | CCTACGGCAATGGCACACT | 12R | CCTACGGCAATGGCACACT | 12R | CCTACGGCAATGGCACACT | 10R | GGTGTTGTGGCGGCAGC | 13R | AAGTGGCGATGGCAATGTG |
| 13F | CACCGCTTGAACATCG | 13F | CACCGCTTGAACATCG | 13F | CACCGCTTGAACATCG | 11F | CAACAACCCTACCGTGG | 14F | CCTTGGTTGGTTTCAT |
| 13R | AAGTGGCGATGGCAATGTG | 13R | AAGTGGCGATGGCAATGTG | 13R | AAGTGGCGATGGCAATGTG | 11R | CCGCAGGTATGTTGGGC | 14R | CTACTACGAAGATGAGCAC |
| 14F | CCTTGGTTGGTTTCAT | 14F | CCTTGGTTGGTTTCAT | 14F | CCTTGGTTGGTTTCAT | 12F | CACCGCCCATCACCGCAA | ||
| 14R | CTACTACGAAGATGAGCAC | 14R | CTACTACGAAGATGAGCAC | 14R | CTACTACGAAGATGAGCAC | 12R | CCTACGGCAATGGCACACT | ||
| 13F | CACCGCTTGAACATCG | ||||||||
| 13R | AAGTGGCGATGGCAATGTG | ||||||||
| 14F | CCTTGGTTGGTTTCAT | ||||||||
| 14R | CTACTACGAAGATGAGCAC | ||||||||
C, rtxA1 copy located on a chromosome; P, rtxA1 copy located on a plasmid.
PCR and DNA sequencing.
To sequence the whole rtxA1 gene in the strains CECT 529T, 94-9-119, Riu-1, A14, 95-8-161, 11028, and CT218 and the common fragment (an 828-bp fragment from the 5′ region of the gene obtained with the primers 12F and 12R) in the V. vulnificus collection of strains, PCRs were performed using the primers listed in Table 2. These primers were designed from the published genome sequences of two human biotype 1 strains (YJ016 and CMCP6) (7, 15) and of the plasmids of two biotype 2 strains (CECT 4999 and CECT 4602) (18). Reaction mixtures for PCRs (120 μl) contained 0.48 mM forward and reverse primer, 2.5 units of Taq DNA polymerase (5 U/μl GoTaq; Promega), 18 μl of 5× Taq reaction buffer (Gotaq Green; Promega), 1.3 mM MgCl2, 0.75 mM deoxynucleoside triphosphate (dNTP) mix (Promega), and 1 μl of DNA. PCRs were performed in a Techne thermocycler (TC-412). The reaction started with 10 min of denaturation at 94°C and was followed by 45 cycles of 40 s of denaturation at 94°C, 45 s of annealing at 52°C, and 90 s of extension at 72°C. An additional extension at 72°C for 10 min completed the reaction. A negative (no template DNA) and a positive (purified DNA of positive isolates) control were included in each PCR batch. Amplified products were separated by electrophoresis at 100 V for 30 min in a 2% (wt/vol) agarose (Low EEO; Pronadisa) gel with Tris-acetate-EDTA (TAE) buffer (Real, Spain) and were visualized by staining with ethidium bromide.
Highly conserved regions, repetitive regions, and protein domain analysis.
The highly conserved terminal regions of the rtxA1 genes sequenced in this work were identified by alignment of the amino acid sequences of the following: MARTXVc toxin of strains M66-2, AM19226, MO10, O395, and N16961 (YP_002810174.1, YP_002175744.1, ZP_05237921.1, NC_009457.1, NC_002505.1, respectively); MARTX toxin of L. anguillarum (MARTXLa) of isolate M93 (EU_155486.1); MARTX toxin of V. splendidus (MARTXVs) of strain 12B01 (ZP_00989505.1); and MARTXVv toxin of YJ016, CMCP6, CECT 4999, and CECT 4602 (YP_001393222.1, YP_001393065.1, NP_937086.1, and NP_762440.1, respectively) using the program AlignX from Invitrogen. Repetitive sequences were identified by searching the consensus sequences in the alignments using the MEGA program as sequence editor (33) and the AlignX (Invitrogen) and Expasy programs (12). Finally, the domains were determined as described by Satchell (27) by direct alignment by using InterProScan Sequence Search, Superfamily, Psi-BLAST, and SBase Web programs.
Phylogeny reconstruction.
Phylogenetic trees were reconstructed using the neighbor-joining algorithm (25) and the maximum-likelihood method. Neighbor-joining trees were obtained using MEGA, version 4 (33), with the maximum-composite-likelihood distance (34) in the case of nucleotide sequences and the Poisson correction for amino acid alignments (37). Support for the groupings derived in these reconstructions was evaluated by bootstrapping using 1,000 replicates (10). Maximum-likelihood trees were obtained using PhyML (13). The best evolutionary model for the sequences according to jModelTest (23) and considering the Akaike information criterion (AIC) turned out to be the generalized time-reversible model of substitution with a gamma distribution (GTR+G) accounting for heterogeneity in evolutionary rates among sites. This model was used to derive the maximum-likelihood trees obtained in this work. The whole or partial rtxA1 genes of V. cholerae (strains M66-2, MO10, N16961, O395, and AM19226), V. splendidus (strain 12B01), L. anguillarum (strain M93), and A. hydrophila (strain ATCC 7966) were used as outgroups.
The congruence among phylogenetic reconstructions obtained with different alignments was checked using Shimodaira-Hasegawa (SH) (31) and expected-likelihood weight (ELW) tests as implemented in TreePuzzle, version 5.2 (29, 32).
A maximum-parsimony network using the Wagner method (9, 16) was constructed using the program MOVE in the PHYLIP package (PHYLIP, Phylogeny Inference Package, version 3.6[http://www.evolution.genetics.washington.edu]) from the presence/absence of the main domains in the rtx gene to show the relationships among the above genes.
Nucleotide sequence accession numbers.
Sequences of the whole rtxA1 genes determined in this study were deposited in the GenBank under accession numbers FJ002578 to FJ002581 and GU452642 to GU452644. Sequences of the common fragment of the rtxA1 gene were deposited under accession numbers GU452542 to GU452641 and GU452645 to GU452660.
RESULTS
Location of the gene rtxA1.
All V. vulnificus isolates tested of the three biotypes carried a copy of the rtxA1 gene in the chromosome (described in the literature as chromosome II or “small chromosome” [7, 15]), and those of biotype 2 had an additional copy in the plasmid of 68 to 70 kb (see Fig. S1 in the supplemental material).
MARTXVv toxin domain organization.
Figure 1 shows the comparison of conserved and nonconserved domains of the predicted MARTX toxin for V. vulnificus (four sequences previously published and seven obtained in this work) compared with those of MARTXVc toxin (strains M66-2, AM19226, MO10, O395, and N16961), MARTXLa toxin (L. anguillarum) (strain M93), MARTXVs toxin (strain 12B01), and the MARTX toxin of A. hydrophila (MARTXAh) (strain ATCC 7966). Tables 3 and 4 summarize the main structural characteristics of these proteins in V. vulnificus. The predicted proteins vary in length, ranging from 4,596 aa for biotype 2 isolates to 5,207 aa for three biotype 1 isolates (Fig. 1). The multiple alignment of MARTXVv toxins showed that they could be divided in three regions, two corresponding to conserved sequences among biotypes, located at the N- and C-terminal ends, respectively, and another of variable sequence, located in the central portion of the protein (Fig. 1).
FIG. 1.
Domain organization of the MARTX toxin structures from the analyzed genomes. The external regions, the repeats (vertical lines), and the internal domains for each toxin are color coded as indicated at the bottom of the figure. Diagrams are drawn to scale.
TABLE 3.
Repeat sequences in MARTXVv toxins
| Strain (location of rtxA1gene copy)a | Profile of repeat sequences by type |
|||||||
|---|---|---|---|---|---|---|---|---|
| A type A.1 |
B type B.1 |
B type B.2 |
C type C.1 |
|||||
| No. of repeats | Location (aa) | No. of repeats | Location (aa) | No. of repeats | Location (aa) | No. of repeats | Location (aa) | |
| V. vulnificus | ||||||||
| YJ016 | 9 | 30-1483 | 29 | 729-1330 | 18 | 742-1287 | 9 | 4584-5063 |
| CECT 529T | 9 | 30-1480 | 29 | 726-1328 | 18 | 739-1284 | 9 | 4078-4557 |
| Riu-1 | 9 | 30-1483 | 30 | 724-1325 | 18 | 756-1301 | 9 | 4096-4572 |
| CMCP6 | 9 | 30-1483 | 29 | 729-1330 | 18 | 742-1287 | 9 | 4584-5063 |
| 94-9-119 | 9 | 30-1483 | 29 | 729-1330 | 18 | 742-1287 | 9 | 4584-5063 |
| A14 (P) | 9 | 30-1485 | 29 | 731-1332 | 18 | 744-1289 | 9 | 3973-4452 |
| 95-8-161 (P) | 9 | 30-1485 | 29 | 731-1332 | 18 | 744-1289 | 9 | 3973-4452 |
| CECT 4999 (C) | 9 | 30-1485 | 29 | 731-1332 | 18 | 744-1289 | 9 | 3973-4452 |
| CECT 4602 (P) | 9 | 30-1485 | 29 | 731-1332 | 18 | 744-1289 | 9 | 3973-4452 |
| CECT 4999 (P) | 9 | 30-1485 | 29 | 731-1332 | 18 | 744-1289 | 9 | 3973-4452 |
| 11028 | 9 | 30-1483 | 29 | 729-1330 | 18 | 742-1287 | 9 | 4081-4560 |
| V. cholerae M66-2 | 9 | 30-1478 | 29 | 726-1328 | 18 | 739-1284 | 9 | 3923-4402 |
| A. hydrophila ATCC 7966 | 10 | 30-3012 | 27 | 704-1286 | 17 | 717-1224 | 9 | 4072-4542 |
| L. anguillarum M93 | 9 | 30-1483 | 29 | 729-1330 | 18 | 742-1287 | 9 | 3783-4256 |
C, chromosome; P, plasmid.
TABLE 4.
Putative functional domains in MARTXVv toxins
| Strain (location of rtxA1 gene copy)a | Domain nameb | Protein |
Gene |
||
|---|---|---|---|---|---|
| Position (aa) | % Identityc | Position (aa) | % Identityc | ||
| YJ016 | DUF | 1957-2345 | ND | 5871-7035 | ND |
| RID | 2368-2915 | 87.6 | 7081-8719 | 82.4 | |
| α/β-Hydrolase | 2918-3277 | 85.8 | 8709-9609 | 89.7 | |
| Efa1/LifA | 3276-3519 | 54.1 | 9844-10569 | 55.5 | |
| rtxAPA | 3604-4080 | 57 | 10789-12201 | 61.4 | |
| CPD | 4082-4188 | 75.7 | 12202-12525 | 70.6 | |
| CMCP6 | DUF | 1957-2345 | ND | 5871-7035 | ND |
| RID | 2368-2915 | 87.6 | 7081-8719 | 82.2 | |
| α/β-Hydrolase | 2918-3277 | 85.8 | 8709-9609 | 89.1 | |
| Efa1/LifA | 3276-3519 | 54.1 | 9844-10569 | 55.6 | |
| rtxAPA | 3604-4080 | 57 | 10789-12201 | 61.5 | |
| CPD | 4082-4188 | 75.7 | 12202-12525 | 70 | |
| 94-9-119 | DUF | 1957-2345 | ND | 5871-7035 | ND |
| RID | 2368-2915 | 86.5 | 7081-8719 | 81.1 | |
| α/β-Hydrolase | 2918-3277 | 85.2 | 8709-9609 | 89.5 | |
| Efa1/LifA | 3276-3519 | 53.4 | 9844-10569 | 55.6 | |
| rtxAPA | 3604-4080 | 57 | 10789-12201 | 61.4 | |
| CPD | 4082-4188 | 76.6 | 12202-12525 | 70.3 | |
| CECT 529T | DUF | 1954-2342 | ND | 5862-7026 | ND |
| RID | 2365-2912 | 86.3 | 7072-8710 | 80,9 | |
| α/β-Hydrolase | 2915-3224 | 86.1 | 8700-9600 | 89.6 | |
| Efa1/LifA | 3273-3516 | 53.4 | 9839-10548 | 56.2 | |
| CPD | 3576-3682 | 86.9 | 10687-11007 | 75.5 | |
| Riu-1 | DUF | 1971-2359 | ND | 5913-7077 | ND |
| RID | 2382-2929 | 80.1 | 7123-8761 | 78.9 | |
| α/β-Hydrolase | 2932-3241 | 86.8 | 8751-9651 | 90 | |
| Efa1/LifA | 3290-3533 | 54.3 | 9870-10599 | 57.0 | |
| CPD | 3593-3699 | 86 | 10738-11058 | 75.8 | |
| CECT 4999 (C) | ACD | 1958-2421 | 89 | 5856-7225 | 82.5 |
| CECT 4999 (P) | Efa1/LifA (copy 1) | 2492-2735 | 53.9 | 7495-8205 | 56.7 |
| CECT 4602 (P) | α/β-Hydrolase | 2810-3119 | 86.5 | 8385-9285 | 97.1 |
| A14 (P) | Efa1/LifA (copy 2) | 3168-3411 | 54.3 | 9523-10233 | 56.7 |
| 94-9-161 (P) | CPD | 3471-3577 | 86 | 10372-10692 | 76.1 |
| 11028 | DUF | 1957-2345 | ND | 5871-7035 | ND |
| RID | 2368-2915 | 83.6 | 7081-8719 | 81.5 | |
| α/β-Hydrolase | 2918-3227 | 86.1 | 8709-9609 | 90.2 | |
| Efa1/LifA | 3276-3519 | 53.9 | 9844-10569 | 56.3 | |
| CPD | 3579-3685 | 86 | 10695-11016 | 76.1 | |
C, chromosome; P, plasmid.
CPD, autocatalytic cysteine protease domain; Efa/LifA, LifA-like protein of Escherichia coli O127:H6 E2348/69; RID, Rho-GTPase inactivation domain; DUFVv, domain with an unknown function; rtxPA, rtxA of P. asymbiotica; ACD, actin cross-linking domain.
Percent identity with the V. cholerae N16961 rtxA gene for ACD, RID, CPD, and α/β-hydrolase domains, with the E. coli Efa/LifA gene for the Efa1/LifA domain, and with the P. asymbiotica rtxA gene for rtxAPA domain. ND, no similarity detected.
Within MARTXVv toxin, the N- and C-terminal ends were very similar (93 to 100% and 90.5 to 100%, respectively) (see Table S1 in the supplemental material). The similarity of MARTXVv toxin to the MARTXVc, MARTXVs, and MARTXLa toxins was also high (61.1 to 100%) (with the exception of the strain O395, which lacks nearly all of the N-terminal region although similarity was even lower with MARTXAh toxin [46.5 to 57.2%]) (see Table S1 in the supplemental material). According to Satchel (27), the MARTX toxin subfamily carries three different types of repetitive sequences in the N- and C-terminal ends, which share a G-7X-GXXN central motif. We were unable to find these repeats in the same number and positions described by Satchell (27) in the strains we analyzed, which showed the following repetitive sequences: A-type repeat, GXXGXXXXXG; B-type repeat B.1, TXVGXGXX; B-type repeat B.2, GXANXXTXX; and C-type repeat, GGXGXDXXX. The results obtained for the redefined repeat sequences are shown in Table 3 and Fig. 1. The new repeats, their positions as well as the entire N and C termini, were highly conserved across the species and even within the genus (Fig. 1; see also Tables S2 to S5 in the supplemental material).
The internal region of the MARTXVv toxin was very variable, showing a global identity and similarity of 18% and 44.5%, respectively (see Table S1). However, these values rose to over 95% for the group formed by the human biotype 1 isolates (YJ016, CMCP6, and 94-9-119) and to 100% for biotype 2 isolates (see Table S1).
The functional domains and their positions are presented in Fig. 1 and Table 4. Three different modular types of MARTXVv toxins (called types I, II, and III) (Fig. 1) were found, all of which possessed a CPD domain, an α/β-hydrolase domain, and a newly reassigned domain, denoted Efa/LifA (common domains). This new domain corresponds to Mcf-DUF (27) and was renamed because of its very high similarity to the Efa/LifA-like protein of Escherichia coli O127:H6 E2348/69 (YP_002328637.1) (E-values between 2e−64 and 3e−65 for the domains of the different MARTXVv toxins). MARTXVv toxin type I was similar to the MARTXVvbt1 toxin described by Satchell (27). This type was present in human isolates of biotype 1, strains YJ016, CMCP6, and 94-9-119, and contained the common domains plus a DUF domain, with no similarity to any known protein (V. vulnificus DUF [DUFVv]), a RID domain, and a newly reassigned domain called rtxA of Photorhabdus asymbiotica (rtxAPA). This new domain corresponds to PMT C1/C2 (27) and was renamed because of its similarity to the rtxA protein of P. asymbiotica (access number YP_003040934.1) (E-values of 3e−158 to 1e−170 for the domains of the different MARTXVv toxins). MARTXVv toxin type II was represented by the type strain of the species, isolate CECT 529T of biotype 1, an environmental isolate of biotype 1, Riu-1, and the isolate of biotype 3, 11028. Type II differed from type I in that it lacked the rtxAPA domain, and, consequently, it was smaller (around 4,704 aa). MARTXVv toxin type III was identical to MARTXVvbt2 toxin, a plasmidic variant described by Lee et al. (18), which, in addition to the common domains, had an ACD domain and two copies of the Efa/LifA domain flanking the common α/β-hydrolase domain.
MARTXVc toxin of the five strains analyzed showed the same domain organization although MARTXVc toxin of strain O395 lacked around 1,910 aa in the N-terminal region (Fig. 1). MARTXLa toxin showed an organization similar to that of MARTXVv toxin type II except that it lacked the DUFVv domain, and MARTXVs toxin showed an organization similar to that of MARTXAh toxin and different from that of the MARTXVv and MARTXVc toxins with a replacement of the ACD domain by a domain of unknown function different from that of V. vulnificus (V. splendidus DUF [DUFVs]).
Phylogenetic reconstruction.
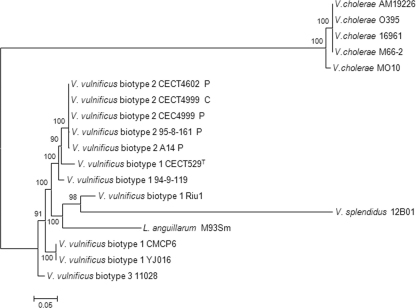
The different phylogenetic trees derived from the complete nucleotide sequences from V. vulnificus and the outgroup species were coincident in revealing a topology in which V. cholerae strains formed a well-supported, monophyletic group quite distinct from its sister group, which included V. vulnificus and two other outgroups, V. splendidus and L. anguillarum, which were associated with V. vulnificus strain Riu-1 (Fig. 2). As a result of the variable patterns of modular organization among the different strains and species analyzed (Fig. 1), the phylogenetic reconstruction derived from the multiple alignment of the complete gene may not be an accurate representation of the evolutionary relationships among the different sequences considered. In fact, it was not possible to obtain a reliable alignment including the outgroup species A. hydrophila. Hence, alternative strategies were used to decipher the evolutionary history of rtxA1 in these taxa. To this end, the phylogenetic trees from the external and internal regions of rtxA1 were analyzed separately. As can be seen in Fig. 3, they were not congruent with one another or with the sequence from the whole gene (Fig. 2 and 3). Thus, all V. vulnificus isolates grouped together with L. anguillarum and separately from V. cholerae and V. splendidus when only the external 5′-terminal regions were considered (Fig. 3A); they formed a well-supported monophyletic group highly related to the V. cholerae group, with V. splendidus and L. anguillarum occupying external positions, when only the external 3′-terminal regions were considered. In contrast, V. vulnificus strains were divided into two groups, one formed by biotype 1 and 3 isolates together with L. anguillarum and V. splendidus as a basal taxon to this group and the other formed by biotype 2 isolates when only the internal region was considered (Fig. 3C). The phylogenetic reconstruction from the common fragment of 828 nt corresponding to the 5′ end of the rtxA1 gene is shown in Fig. S2 in the supplemental material. The fragment was highly similar (95 to 100%) although there were differences between strains that were not related to biotype, with isolate Riu-1 presenting the largest number of substitutions (see Fig. S2). Nevertheless, the similarity between fragments was too high to infer any conclusive results from the evolutionary history of this portion of the gene.
FIG. 2.
Maximum-likelihood tree derived from the aligned regions from complete rtxA1 gene sequences (20,394 nt) using the GTR+G model of evolution. Bootstrap support values higher than 80% are indicated in the corresponding nodes.
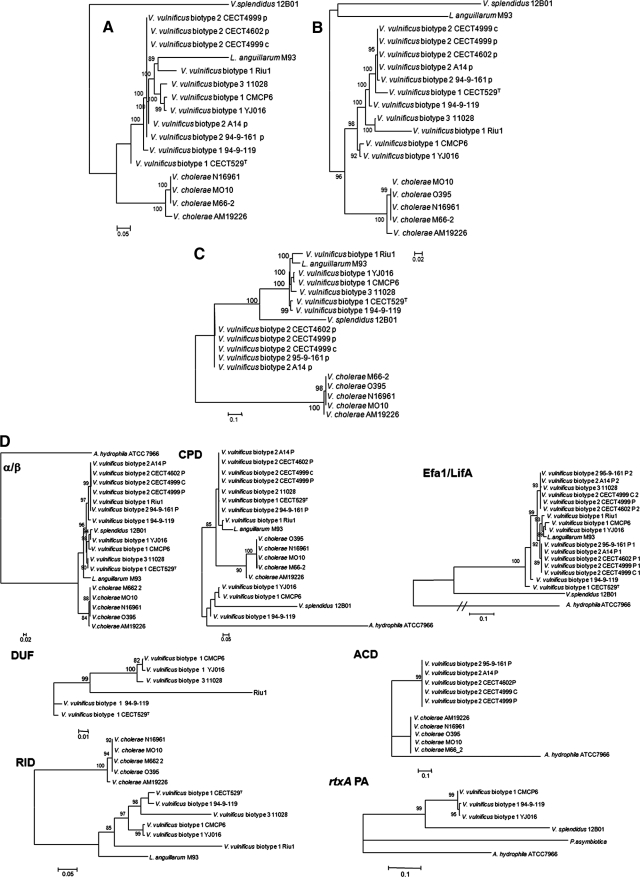
FIG. 3.
Maximum-likelihood trees derived from 5′-terminal (A), 3′-terminal (B), and internal (C) regions of the rtxA1 gene using the GTR+G model of evolution. The numbers of nucleotide positions considered in the multiple alignments were 5,844, 3,039, and 7,988, respectively. Bootstrap support values higher than 80% are indicated in the corresponding nodes. (D) Maximum-likelihood tree derived from the different modules of the rtxA1 gene using the GTR+G model of evolution. Bootstrap support values higher than 80% are indicated in the corresponding nodes.
A maximum-parsimony network showing the relationships among the genes from the presence/absence of the domains in the rtxA1 gene is shown in Fig. 4. In this analysis, we included A. hydrophila, which was too divergent at the sequence level of the complete gene to be incorporated in the previous maximum-likelihood tree, and excluded V. splendidus because of its different modular organization (Fig. 1). The network suggested a very plastic modular organization in this gene, with a minimum of seven independent gains/losses of domains. Of these, only one homoplasy (the independent gain of domain rtxAPA in A. hydrophila and three strains of biotype 1) was found. Furthermore, V. vulnificus represented the most divergent gene organizations (with four or five gains/losses of domains) among the sequences included in the analysis.
FIG. 4.
Maximum-parsimony reconstruction of the presence/absence of modules in the central portion of rtxA1 genes in the genomes analyzed. Gains or losses of domains are indicated next to each branch.
The next issue we pursued was an individual analysis of each of these domains, for which we used the corresponding multiple nucleotide sequence alignments (Fig. 3D). The only domains common to all the strains and species considered were the α/β-hydrolase and CPD, and their phylogenetic histories were not congruent, again, to one another or with that derived with the complete sequences for this gene (Fig. 2 and 3). The most relevant discrepancies were for the α/β-hydrolase domain, for which the sequence from Riu-1 was identical to sequences from biotype 2 strains, and for the CPD domain, for which the three biotype 1 strains 94-9-119, CMCP6, and YJ016 grouped externally to the rest with V. splendidus, placing V. cholerae and L. anguillarum as members of the ingroup (Fig. 3D). The differences found among biotype 1 sequences for the α/β-hydrolase domain could be explained by an HGT event of this domain from one biotype 2 strain to Riu-1. The simplest explanation for the placement of the three biotype 1 sequences in the CPD domain phylogenetic tree is also their acquisition from an unidentified donor by an HGT event. To further verify these hypotheses, we performed reciprocal tests of congruence for the α/β-hydrolase and CPD domains and the complete gene sequences using the 13 strains common to all three data sets. The results of the Shimodaira-Hasegawa (SH) and expected-likelihood weight (ELW) tests are summarized in Table 5. All the comparisons were highly significant for both tests with only one exception, corresponding to the results of the SH test of the topology obtained with the complete gene sequences versus the CPD domain alignment and its maximum-likelihood tree. This indicates that the phylogenetic reconstructions obtained from each alignment were not congruent with one another, thus providing statistical support for the HGT hypothesis for both domains.
TABLE 5.
Summary of SH and ELW tests for the complete rtxA1 gene sequence and those obtained for the α/β-hydrolase and CPD modules using the 13 common strains and speciesa
| Alignment | Topology | lnL | SH test | ELW test |
|---|---|---|---|---|
| Complete | Complete | −70351.24 | 1.000 | 1.000 |
| α/β-Hydrolase domain | −72657.44 | 0.000 | 0.000 | |
| CPD domain | −76609.58 | 0.000 | 0.000 | |
| α/β-Hydrolase module | α/β-Hydrolase domain | −2165.08 | 1.000 | 1.000 |
| Complete | −2287.23 | 0.000 | 0.000 | |
| CPD domain | −2235.99 | 0.000 | 0.000 | |
| CPD module | CPD domain | −1150.62 | 1.000 | 0.971 |
| Complete | −1159.13 | 0.273 | 0.029 | |
| α/β-Hydrolase domain | −1252.89 | 0.000 | 0.000 |
Each alignment was used to evaluate the log likelihood (lnL) of the ML tree obtained with each of the three data sets. The probability values for each topology and test are shown. Complete, complete rtxA1 gene sequence.
The remaining domains considered in this gene had a less constant distribution in the different strains and species considered. Biotype 2 strains presented two copies of the Efa1/LifA-like domain. The phylogenetic tree derived for the sequences in this domain suggests that the duplication probably occurred before the divergence of biotype 2 strains (Fig. 3D). This was not the only salient feature in the evolution of this domain. Here, the biotype 3 sequence (from strain 11028) grouped with one of the paralogous copies from biotype 2, the one that was not present in biotype 1 strains. Furthermore, two biotype 1 strains (CECT 529T and 94-9-119) did not group with the remaining sequences from this biotype and diverged closer to the root of the V. vulnificus group than they did in the maximum-likelihood tree for the whole-gene alignment. Interestingly, one outgroup sequence, corresponding to L. anguillarum, was very similar to, and grouped with, sequences from V. vulnificus biotype 1 strains. This resemblance was also revealed in the previous analysis of the distribution of domains (Fig. 4) and by the phylogenetic tree from the internal part of the gene (Fig. 3C).
Domains ACD, DUFVv, RID, and rtxAPA were present in only a few strains (Fig. 3D), and their corresponding phylogenetic trees were not too informative on the evolution of the rtxA1 gene. ACD was present in only biotype 2 strains and in the outgroups V. cholerae and A. hydrophila, whereas RID was present in biotype 1 and 3 strains and in the outgroups V. cholerae and L. anguillarum (Fig. 1 and 3D). In both cases, the within-biotype distribution of evolutionary distances was very similar to that observed in the analysis of the complete gene (Fig. 2). Domain rtxAPA was present in only three biotype 1 strains and in three outgroups (P. asymbiotica, V. splendidus, and A. hydrophila) but not in V. cholerae. Finally, DUFVv was present in only V. vulnificus biotypes 1 and 3. In fact, this domain did not show similarity to known proteins by BLAST search. Similar to other portions in this gene, the strain Riu-1 presented a remarkable accumulation of mutations in comparison to other strains of the same subtype.
DISCUSSION
In the first part of this work, we have analyzed the structural diversity of MARTX toxins in V. vulnificus biotypes by comparing them to other MARTX toxins. We have found three different modular types according to the domains identified in the internal part of the protein. In contrast, the external regions are highly conserved within the species and even the genus, which correlates with the proposed function for these regions, that is, to interact with the membrane of eukaryotic cells allowing the translocation of effector domains to the cytoplasm. Upon translocation, CPD is activated to process the MARTX toxin, thereby releasing the internal domains, which alter cell function (30). We have redefined two of the internal domains, Mcf-DUF and PMT C1/C2 (originally described by Satchell [27]) as Efa/LifA and rtxPA, respectively, because of the new similarities revealed by BLAST searches. Lymphostatin (Efa/LifA protein) is a common toxin in Gram-negative bacteria with multiple functions, cell adhesion, immunosuppression, and disruption of epithelial barrier function via the modulation of Rho GTPase activities, while no function has been reported for the rtxA of P. asymbiotya. Biotype 1 isolates feature MARTXVv toxin type I, a type already described by Satchell (27), and type II, which is simpler than the former because it lacks the rtxPA domain. In addition, all V. vulnificus biotype 2 isolates, irrespective of their serovar, have MARTXVv toxin type III described by Lee et al. (18) and harbor two copies of the gene, one plasmidic and the other chromosomal. It has been proposed that MARTXVv toxin type I is the main factor for human virulence in V. vulnificus since mutations in this gene practically abolish virulence in mice (19). However, we have found that other human isolates (the biotype 3 isolate) can produce a different type of toxin that lacks the rtxPA domain, which would suggest that this domain is not necessary for virulence. In addition, we have found that fish-pathogenic isolates (biotype 2) show a different type, which could be related to specific virulence for fish. The role of the new types of MARTXVv toxins in virulence as well as the reason for gene duplication in the biotype 2 remains to be elucidated.
In the second part of this work, we have analyzed the rtxA1 gene using different data sets demonstrating that MARTXVv toxins are mosaics of segments with different evolutionary histories. Previous studies of the evolution of V. vulnificus based on sequence-based typing of housekeeping genes suggest that the species can be subdivided into two recent and rapidly expanding phylogenetic clusters (8). One lineage would correspond to biotype 1 isolates, mostly from human sources, including the strains YJO16 and CMCP6 used in this study. The other lineage would be formed by biotype 1 and 2 isolates from various environments, including diseased fish. The phylogenetic position of biotype 3 isolates, which are genetically identical, would be variable depending on the gene and the model and method of analysis (6). An ongoing phylogenetic study of in lab using sequence-based typing of housekeeping and virulence-related genes (26) with the same strains included in the present work suggests that V. vulnificus species are subdivided in three lineages, one including biotype 1 and 2 from the fish farming environment, another including biotype 3 isolates, and a third including biotype 1 strains from human blood and the environment. Interestingly, none of the evolutionary histories inferred from the rtxA1 gene matched the previously hypothesized evolutionary histories of V. vulnificus (6, 8, 26). The phylogenetic tree derived from the complete rtxA1 gene (Fig. 2) presented an unlikely evolutionary scenario: V. cholerae isolates conformed to a well-supported, monophyletic group, but this was not the case for V. vulnificus, which included two outgroups, V. splendidus and L. anguillarum, both related to strain Riu-1. The inclusion of outgroup species within the ingroup can be due to several reasons, which we tried to analyze next by dividing the gene in three different portions. These fragments showed very different evolutionary relationships, which also differed from the previous result with the complete gene. The 5′ end (Fig. 3A) and the internal portion (Fig. 3C) were partially coincident, but the placement of V. splendidus differed in the two trees, being an outgroup to all the sequences in the N-terminal coding portion of the gene and basal to a subgroup of V. vulnificus (including biotypes 1 and 3 and L. anguillarum) in the central portion. The 3′-end coding portion of the gene provided yet another history, but in this case the outgroup species appeared where they were supposed to (Fig. 3B). These results indicated that rtxA1 of V. vulnificus has a complex evolutionary history, and this is also probably true for other closely related species. The modular structure of the gene may be partially responsible for this complexity (Fig. 1) and, in consequence, our next step was to investigate the evolutionary history of these modules.
The maximum-parsimony network for the module composition helped to disentangle the complex history of this protein (Fig. 4). The analysis of gains/losses of modules revealed that V. vulnificus biotype 2 isolates are closer to A. hydrophila than to the other biotypes of the species and that there have been several gain/loss events between biotype 2 and biotypes 1 and 3.
The most likely explanation for this complex evolutionary history involves HGT events from other bacteria and subsequent recombination, yielding new combinations, which might have been favored by selection. The rtxA1 gene belongs to the segmentally variable gene family, previously defined as mosaics of one or more rapidly evolving, variable regions interrupted by conserved regions. Many of the mosaic genes identified encode proteins involved in host-pathogen interactions, defense mechanisms, and intracellular responses to external changes, and this is the case for MARTXVv toxin. We have detected by the bioinformatic analysis evidence of at least two HGT events affecting the α/β-hydrolase and CPD domains and probably also to the Efa/LifA domain a fragment duplicated in biotype 2 isolates. In the cases of the α/β-hydrolase and Efa/LifA domains, the transferred DNA was probably from the same species, whereas in case of the CPD domain, it probably came from a different, unknown donor.
With regard to the domains ACD, DUFVv, RID, and rtxAPA, we can suggest that (i) the DUFVv domain is exclusive of V. vulnificus biotype 1 and does not show similarity to known proteins, (ii) the rtxAPA domain of V. vulnificus is too different from rtxA of P. asymbiotica to have been received directly from this donor by HGT, and (iii) the ACD and RID domains have not been received from the species considered (V. cholerae or V. cholerae/L. anguillarum, respectively) because the distance for these domains is similar to that for the rest of the studied common domains. In the light of these results, the hypothesis of the origin of the ACD domain of MARTXVv toxin type III from the ACD domain of MARTXVc toxin has to be discarded.
V. vulnificus expresses a natural competence system induced by chitin (14) that has also been described in V. cholerae (21), which could work under natural conditions and favor the exchange of genetic material. In addition, the species possesses conjugative plasmids, which can be transmitted between strains and can even mobilize other resident plasmids (such as the virulence one) by forming cointegrates (18). Both genetic exchange mechanisms can result in coinfection with two different but related rtx genes, which could recombine, resulting in a new, different rtx gene. This process is exemplified in the biotype 2 of the species, which has a duplicated rtxA1 gene in the chromosome and in the plasmid and produces a MARTX toxin that is completely different from the toxins of biotypes 1 and 3. The presence of identical duplicate genes in the genome of the biotype 2 isolates suggests that either the acquisition has been recent or a strong purifying selection is acting against mutations that modify gene function.
The phylogenetic study performed in our lab (26) demonstrates that biotype 2 is polyphyletic and suggests that it has emerged by acquisition of a virulence plasmid from V. vulnificus isolates from fish farms. According to this hypothesis, the acquisition of the plasmid could have occurred before or after divergence from the common ancestor for biotype 2 strains. The phylogenetic analysis shown here suggests that the duplication of the Efa/LifA domain likely occurred after the divergence between biotypes 1 and 2 and before the expansion of the biotype 2 clonal complex and, in consequence, more or less contemporaneously to the acquisition of the virulence plasmid mentioned above. This acquisition might have favored recombination events between the chromosomal and plasmidic rtxA1 genes, and the selection of a new combination of the mosaic gene would probably have been advantageous in the fish farming environment and, in consequence, would have spread rapidly throughout suitable habitats throughout the seas, appearing as highly conserved among the different clonal complexes. The reasons for such an advantage will have to be elucidated by obtaining mutants deficient for the different domains and their combinations and then testing these mutants in selected in vitro models and in vivo.
Supplementary Material
Acknowledgments
This work has been financed by grants AGL2008-03977/ACU and BFU2008-03000 and by Programa Consolider-Ingenio CSD2009-00006 from MICINN and project ACOMP/2009/240 from Conselleria d'Educació (Generalitat Valenciana).
We thank the SCSIE of the University of Valencia for technical support in determining the sequences.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amaro, C., and E. G. Biosca. 1996. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl. Environ. Microbiol. 62:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaro, C., E. G. Biosca, B. Fouz, and E. Garay. 1992. Electrophoretic analysis of heterogeneous lipopolysaccharides from various strains of Vibrio vulnificus biotypes 1 and 2 by silver staining and immunoblotting. Curr. Microbiol. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2007. Current protocols in molecular biology. John Wiley, New York, NY.
- 4.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, and J. J. Farmer III. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 5.Bisharat, N., C. Amaro, B. Fouz, A. Llorens, and D. I. Cohen. 2007. Serological and molecular characteristics of Vibrio vulnificus biotype 3: evidence for high clonality. Microbiology 153:846-856. [DOI] [PubMed] [Google Scholar]
- 6.Bisharat, N., D. I. Cohen, M. C. Maiden, D. W. Crook, T. Peto, and R. M. Harding. 2007. The evolution of genetic structure in the marine pathogen, Vibrio vulnificus. Infect. Genet. Evol. 7:685-693. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, A. L., J. D. Oliver, A. DePaola, E. J. Feil, and E. F. Boyd. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eck, R. V., and M. O. Dayhoff. 1966. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152:363-366. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Fouz, B., F. J. Roig, and C. Amaro. 2007. Phenotypic and genotypic characterization of a new fish-virulent Vibrio vulnificus serovar that lacks potential to infect humans. Microbiology 153:1926-1934. [DOI] [PubMed] [Google Scholar]
- 12.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 14.Gulig, P. A., M. S. Tucker, P. C. Thiaville, J. L. Joseph, and R. N. Brown. 2009. User friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl. Environ. Microbiol. 75:4936-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kluge, A. G., and J. S. Farris. 1969. Quantitative phyletics and the evolution of anurans. Syst. Zool. 18:1-32. [Google Scholar]
- 17.Lee, B. C., S. H. Choi, and T. S. Kim. 2008. Vibrio vulnificus RTX toxin plays an important role in the apoptotic death of human intestinal epithelial cells exposed to Vibrio vulnificus. Microbes Infect. 10:1504-1513. [DOI] [PubMed] [Google Scholar]
- 18.Lee, C. T., C. Amaro, K. M. Wu, E. Valiente, Y. F. Chang, S. F. Tsai, C. H. Chang, and L. I. Hor. 2008. A common virulence plasmid in biotype 2 Vibrio vulnificus and its dissemination aided by a conjugal plasmid. J. Bacteriol. 190:1638-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. H., M. W. Kim, B. S. Kim, S. M. Kim, B. C. Lee, T. S. Kim, and S. H. Choi. 2007. Identification and characterization of the Vibrio vulnificus rtxA essential for cytotoxicity in vitro and virulence in mice. J. Microbiol. 45:146-152. [PubMed] [Google Scholar]
- 20.Li, L., J. L. Rock, and D. R. Nelson. 2008. Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 76:2620-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meibom, K. L., M. Blokesch, N. A. Dolganov, C. Y. Wu, and G. K. Schoolnik. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824-1827. [DOI] [PubMed] [Google Scholar]
- 22.Oliver, J. D. 2006. Vibrio vulnificus, p. 349-366. In F. L. Thompson, B. Austin, and J. Swings (ed.), Biology of vibrios. ASM Press, Washington, DC.
- 23.Posada, D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253-1256. [DOI] [PubMed] [Google Scholar]
- 24.Roig, F. J., and C. Amaro. 2009. Plasmid diversity in Vibrio vulnificus biotypes. Microbiology 155:489-497. [DOI] [PubMed] [Google Scholar]
- 25.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 26.Sanjuan, E., F. Gonzalez-Candelas, and C. Amaro. 2011. Polyphyletic origin of Vibrio vulnificus biotype 2 as revealed by sequence-based analysis. Appl. Environ. Microbiol. 77:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satchell, K. J. F. 2007. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect. Immun. 75:5079-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satchell, K. J. F., and B. Geissler. 2009. The multifunctional-autoprocessing RTX toxins of vibrios, p. 113-126. In Thomas Proft (ed.), Microbial toxins: current research and future trends. Caister Academic Press, Auckland, New Zealand.
- 29.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. Tree-Puzzle: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 30.Sheahan, K. L., C. L. Cordero, and K. J. Satchell. 2007. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J. 26:2552-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 32.Strimmer, K., and A. Rambaut. 2002. Inferring confidence sets of possibly misspecified gene trees. Proc. Biol. Sci. 269:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 34.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tison, D. L., M. Nishibuchi, J. D. Greenwood, and R. J. Seidler. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, C., Y. Yang, and A. Y. Jong. 1990. Mini-prep in ten minutes. Biotechniques 8:172-173. [PubMed] [Google Scholar]
- 37.Zuckerkandl, E., and L. Pauling. 1965. Evolutionary divergence and convergence in proteins, p. 97-166. In V. Bryson and H. J. Vogel (ed.), Evolving genes and proteins. Academic Press, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.