Abstract
Most Enterococcus faecalis isolates carry gelE, but many are gelatinase nonproducers due to the lack of fsrC (EF_1820) to EF_1841 (fsrC-EF_1841; 23.9 kb in strain V583), including most of the locus encoding Fsr, which activates gelE expression. Analysis of 22 accessible E. faecalis genomes revealed the identity of the 53-amino-acid propeptide of fsrD across multiple MLSTs (multilocus sequence types), although 12 distinctly different variations were found in the EF_1814-to-EF_1902 region. Diversity was seen in fsrABC, in the region EF_1814 to EF_1902, and in a 700-kb region surrounding fsrC-EF_1841. However, analysis of five sequenced strains carrying the fsrC-EF_1841 deletion and the putative integrative conjugative element efaB5 showed almost identical single nucleotide polymorphisms (SNPs) in gelE and an identical junction sequence, despite their unrelated MLSTs, in contrast to those shown by strains without the deletion. Further analysis confirmed the conserved gelE SNPs in 6 additional strains (11 in total) with the deletion. While we were unable to detect evidence of spontaneous deletion using OG1RF and 8 other strains, we were able to engineer a deletion of the 37-kb fsrC-EF_1841 region of OG1RF without deleterious effects, and the 37-kb mutant showed changes in biofilm and chaining similar to those shown by fsr-gelE mutants. In conclusion, we describe the identity of fsrD despite high plasticity within the fsrC-EF_1841 region and the surrounding sequence. However, strains lacking the fsrC-EF_1841 region show a distinct conservation of the sequence surrounding this deletion and in gelE, suggesting that the deletion may result from horizontal transfer and recombination.
Enterococci are Gram-positive commensal bacteria of humans, animals, and insects but can also cause a wide range of diseases, including urinary tract infections, bloodstream infections, wound infections, and endocarditis (26). Nosocomial enterococcal infections are frequently caused by hospital-acquired strains rather than by strains from the patient's own community-derived indigenous flora (13, 46). These strains often belong to specific clones which are thought to have acquired factors important for virulence, colonization, and/or for fitness in the hospital environment (46).
Gelatinase is produced by approximately 60% of clinical Enterococcus faecalis isolates (7). The Fsr system, encoded by the fsrABDC operon, and gelatinase production are intrinsically linked since the Fsr system is the main activator of gelE expression (4, 36, 37). The fsrABDC operon, a homologue of the agrABCD operon in Staphylococcus aureus, encodes the elements of a quorum-sensing system (29, 30). While the presence of a functional Fsr system and/or gelatinase production increases the severity of disease in animal, plant, and nematode models of E. faecalis infection (10, 16, 28, 37, 44, 45), neither are required for the organism to cause disease (7, 45).
When testing E. faecalis isolates for gelatinase activity, Qin et al. observed that isolates that did not produce gelatinase but had the gelE gene often lacked the fsrB gene (37). This study was followed by a report by Nakayama et al. which found that the majority of gelatinase nonproducer but gelE+ strains lacked a region present in strain V583 between the 5′-end portion of fsrC and the 3′-end portion of EF_1841, including the fsrABD genes; this missing region corresponds to a 23.9-kb fragment in V583 (31). Strains lacking this region are gelatinase nonproducers in standard assays due to the absence of a functional Fsr system (31, 37, 40). The possible benefit of not having the fsrC (EF_1820)-to-EF_1841 (fsrC-EF_1841) region, a situation that occurs in approximately 28% of reported strains, remains unclear (37, 40). Two groups have reported the presence of gelatinase nonproducer colonies within a culture of a gelatinase producer isolate (9, 31), leading to the hypothesis of ongoing spontaneous deletion among gelatinase producer strains. However, in both cases, no further experimentation to confirm that the gelatinase nonproducer isolates were related to their gelatinase-positive parent isolate was reported.
In a previous study (36), the last 150 bp of fsrB (now fsrD) homologous to agrD were shown to be identical in five clinical isolates, while three isolates had single silent base pair changes. Due to the availability of multiple E. faecalis genome sequences, in the present study we set out to investigate the chromosomal diversity of 22 isolates of E. faecalis from multiple multilocus sequence types (MLSTs) in the region from EF_1814 to EF_1902 (based on V583 annotation). Although we found considerable variability in this region, we did indeed observe identity in fsrD, and in clonally unrelated isolates lacking the fsrC-EF_1841 region, we also found unique and highly conserved nucleotide changes in gelE and the region close to the fsrC-EF_1841 junction, raising the possibility that this deletion may have been introduced horizontally by recombination. We were also able to delete the fsrC-EF_1841 region using allelic replacement in strain OG1RF and found that growth, biofilm, and chaining phenotypes were similar to those in fsr-gelE mutants of OG1RF, demonstrating that this region is not needed for in vitro growth.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Tables 1 and 2. Twenty-two E. faecalis-accessible genomes were analyzed for their genomic organization. An additional 38 isolates were chosen to represent diverse multilocus sequence types (MLSTs), while an additional 10 isolates were selected based on previous studies showing the lack of fsrB but the presence of gelE (40). Insertion mutants TX5240 (insertion in fsrA), TX5241 (insertion in fsrB), TX5242 (insertion in fsrC) (36), and TX5128 (insertion in gelE) (45) were used for biofilm and chaining assays. All strains were grown routinely in brain heart infusion (BHI) broth (Difco laboratories, Detroit, MI) at 150 to 200 rpm or on BHI agar at 37°C. Gelatinase production was measured as previously described after overnight incubation at 37°C (37).
TABLE 1.
Sequenced strains analyzed in this studyb
| Genotype | Straina | MLST | Relevant characteristic |
Serverc | Reference(s) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| fsrB | gelE | Gel+ | 23.9-kb region | 14.8-kb region | efaB5 | |||||
| A | V583 | 6 | + | + | + | + | − | + | TIGR | 35, 42 |
| Merz96 | 103 | + | + | + | + | − | + | BI | 34 | |
| TX0104 | 2 | + | + | + | + | − | + | BCM | This study | |
| HH22 | 6 | + | + | + | + | − | + | BCM | 27 | |
| A′ | CH188 | 9 | + | + | − | + | − | + | BI | 19, 39 |
| B′ | ATCC 4200 | 105 | − | + | − | − | − | + | BI | 23 |
| B | HIP11704 | 4 | − | + | − | − | − | + | BI | 24 |
| T2 | 11 | − | + | − | − | − | + | BI | 20 | |
| T8 | 8 | − | + | − | − | − | + | BI | 20 | |
| JH2-2 | 8 | − | + | − | − | − | + | 15 | ||
| TX1322 | 64 | − | + | − | − | − | + | BCM | This study | |
| C | OG1RF | 1 | + | + | + | + | + | − | BCM | 8, 14 |
| T1 | 21 | + | + | + | + | + | − | BI | 20 | |
| T3 | 67 | + | + | + | + | + | − | BI | 20 | |
| C′ | E1Sol | 93 | + | + | + | + | + | − | BI | 12 |
| T11 | 65 | + | + | + | + | + | − | BI | 20 | |
| D | Fly1 | 101 | + | + | + | + | P | − | BI | 23 |
| E | JH1 | 40 | + | + | − | + | − | − | BI | 2, 15 |
| F | ARO1/DG | 108 | + | + | + | P | − | − | BI | 22 |
| G | X98 | 19 | + | + | − | − | − | − | BI | 5, 43 |
| H | D6 | 16 | − | − | − | P | − | − | BI | 23 |
| I | ATCC 29200 | 21 | − | P | − | P | − | − | BCM | 1 |
| —d | TX5599 | 1 | − | + | − | − | − | − | This study | |
The strains are listed by the genotype of the EF_1814-to-EF_1902 region. The three genotypes (A, B, and C) encompassing the greatest number of strains are listed first, followed by the genotypes with one strain each. Variations of genotypes A, B, and C were detected in the strains CH188, ATCC 4200, and E1Sol and T11 and designated A′, B′, or C′, respectively. Sequences of these isolates were analyzed for the fsrC-EF_1841 region (23.9 kb), the 14.8-kb insertion region described in OG1RF, and the putative integrative conjugative element mobile element efaB5. Strains with genotype A (V583, Merz96, TX0104, and HH22) are within CC2.
Information on the MLST and the presence (+) or absence (−) of fsrB (EF_1820), gelE (EF_1818), and gelatinase production (Gel+) was either acquired from the literature (23) and confirmed or tested by us. A “P” indicates the presence of the part of the locus/gene mentioned.
TIGR, the Institute for Genomic Research; BI, Broad Institute; BCM, Baylor College of Medicine.
TX5599 is the mutant constructed in this study by deleting 37 kb (the 23.9-kb region and the 14.8-kb region) in OG1RF.
TABLE 2.
fsrC-EF_1841 region genotypes and gelatinase phenotypes of clonally diverse nonsequenced isolates tested in this study
| Straina | STb | Genotypec | Relevant characteristic |
|
|---|---|---|---|---|
| efaB5d | Gelatinase | |||
| TX0066* | T501 | Deletion | + | − |
| TX0079* | T502 | Deletion | + | − |
| TX0085* | T503 | Deletion | + | − |
| TX0047* | T517 | Deletion | − | − |
| TX0017* | T520 | Deletion | + | − |
| TX0063* | T521 | Deletion | + | − |
| TX0108* | 4 | Deletion | + | − |
| TX0855 (A0220) | 4 | Deletion | + | − |
| TX0860 (A0217) | 11 | Deletion | + | − |
| TX2141 (E1825) | 25 | Deletion | + | − |
| TX0083* | 30 | Deletion | + | − |
| TX2134 (E1052) | 30 | Deletion | + | − |
| TX4249 (A0203) | 54 | Deletion | + | − |
| TX0044* | 62 | Deletion | + | − |
| TX0103* | 62 | Deletion | + | − |
| TX4239 (A0825) | 97 | Deletion | + | − |
| TX4260 (A1008) | 137 | Deletion | + | − |
| TX2486 (A0219) | 2 | fsrC+ EF_1841+ | + | + |
| TX2621 (A0218) | 2 | fsrC+ EF_1841+ | + | + |
| TX2783 (A0221) | 5 | fsrC+ EF_1841+ | − | + |
| TX0052 (A0225) | 6 | fsrC+ EF_1841+ | −† | + |
| TX0614 (A0222) | 6 | fsrC+ EF_1841+ | + | + |
| TX0630 (A0214) | 9 | fsrC+ EF_1841+ | + | + |
| TX0635 (A0215) | 9 | fsrC+ EF_1841+ | + | + |
| TX0645 (A0216) | 10 | fsrC+ EF_1841+ | −† | + |
| TX2137 (E1798) | 16 | fsrC+ EF_1841+ | −† | + |
| TX4244 (E1022) | 27 | fsrC+ EF_1841+ | −† | + |
| TX4246 (E1873) | 29 | fsrC+ EF_1841+ | − | + |
| TX2147 (E1845) | 36 | fsrC+ EF_1841+ | − | + |
| TX2144 (E1840) | 40 | fsrC+ EF_1841+ | −† | + |
| TX4242 (A0834) | 47 | fsrC+ EF_1841+ | − | + |
| TX4251 (A0206) | 58 | fsrC+ EF_1841+ | − | + |
| TX2146 (E1844) | 61 | fsrC+ EF_1841+ | − | + |
| TX4254 (A0802) | 82 | fsrC+ EF_1841+ | + | + |
| TX4255 (A0808) | 88 | fsrC+ EF_1841+ | + | + |
| TX4238 (A0823) | 96 | fsrC+ EF_1841+ | −† | + |
| TX4241 (A0828) | 99 | fsrC+ EF_1841+ | − | + |
| TX4257 (A1001) | 130 | fsrC+ EF_1841+ | −† | + |
| TX4259 (A1006) | 135 | fsrC+ EF_1841+ | −† | + |
| TX4247 (E1876) | 20 | fsrC only | − | − |
| TX2139 (E1802) | 35 | fsrC only | − | − |
| TX4248 (E1877) | 40 | fsrC only | −† | + |
| TX2135 (A1795) | 44 | fsrC only | −† | − |
| TX4245 (E1872) | 16 | EF_1841 only | − | − |
| TX4243 (E0252 | 23 | EF_1841 only | −† | − |
| TX2140 (E1803) | 38 | EF_1841 only | −† | − |
| TX2138 (E1801) | 48 | EF_1841 only | −† | − |
| TX4240 (A0826) | 98 | EF_1841 only | − | − |
Parentheses indicate an alternative name used in the literature, while asterisks indicate the 10 additional isolates chosen due to previous data (38) indicating the absence of the fsrC-EF_1841 region. Isolates without asterisks are the initial 38 isolates chosen for study.
STs were done by MLST, except those designated with a “T,” which are trilocus sequence types (6).
The fsrC-EF_1841 genotypes were detected using a multiplex PCR, as described in Materials and Methods, where AB83 and AB84 amplify EF_1841, AB85 and AB86 amplify fsrC (EF_1820), and AB83 and AB85 amplify the fsrC::EF_1841 junction sequence, indicating the deletion.
The efaB5 mobile element was also detected using a multiplex PCR, which would amplify three bands if present (see Materials and Methods). The absence of the element would produce a 689-bp band between the noncoding region after EF_1845 and EF_1898. “†” indicates that none of these bands were produced.
Computer analyses and annotation.
In addition to V583 and OG1RF, 20 E. faecalis genomes accessible in June 2009 were analyzed for their genomic organization between EF_1814 and EF_1902. For each genome, the sequences were recovered from the NCBI server or the Broad Institute server and analyzed with resources described elsewhere (3). Each gene is denoted either with its characterized name, by an EF_ tag based on the V583 annotation (35), or with an OG1RF tag if it was first described as a unique sequence in OG1RF (3).
Detection of fsrC, EF_1841, or the junction sequence.
To detect the presence or lack of the fsrC-EF_1841 region, we designed a multiplex PCR based on the following four primers. AB83 (outside the deletion) and AB84 would amplify a 575-bp fragment of EF_1841, while AB85 (outside the deletion) and AB86 would amplify a 940-bp fragment of fsrC (EF_1820) when the genes are intact (see Table S1 and Fig. S1 in the supplemental material). In strains lacking the fsrC-EF_1841 region, AB83 and AB85 would amplify a 739-bp fragment of the junction sequence. Additional primers AB74, AB75, AB244, and AB245 were used to confirm the presence of fsrC (see Table S1).
Conjugation experiments.
The possibility of transposition of efaB5 with the fsrC-EF_1841 deletion was investigated using cross-streak and filter mating between JH2-2::Tn916 as the donor (11) and OG1RF tagged with kanamycin as the recipient (32). Mating plates were incubated overnight at 37°C. Growth present in the area of the cross or on the filter was resuspended in 1.0 ml of sterile saline (0.9% NaCl), and serial 10-fold dilutions were made. Aliquots were plated onto Todd-Hewitt agar plates containing 2 mg/ml kanamycin (to select for the recipient OG1RF cells) and 10 μg/ml tetracycline (to select for Tn916). OG1RF transconjugants were then replica plated on 3% gelatin to detect the possible loss of the fsrC-EF_1841 region.
Detection of the efaB5 element in relation to the fsrC-EF_1841 region deletion.
A total of 17 strains (7 from the original 38 strains plus the 10 previously described [40]) from diverse sequence types (by multilocus sequence typing or trilocus sequence typing [6, 41]) known to lack fsrB but to have gelE (40) were tested for the 739-bp PCR product indicating the lack of the fsrC-EF_1841 region (using AB83 and AB85 as described above) and for the presence of the efaB5 mobile element in conjunction with the deletion using primers EF_1877efaB5F, EF_1877efaB5R, 5′efaB5F, 5′efaB5R, 3′efaB5F, and 3′efaB5R in a multiplex PCR (see Table S1 and Fig. S1 in the supplemental material). The EF_1877efaB5F and EF1877efaB5R primers should amplify a 338-bp region of EF1877, a gene conserved within the efaB5 element, if the efaB5 element is present. 5′efaB5F and 5′efaB5R should amplify a 616-bp region if the 5′ region of the efaB5 element is inserted before EF_1846, and the 3′efaB5F and 3′efaB5R primers should amplify a 517-bp region if the 3′ region of efaB5 is inserted before EF_1898. Therefore, in JH2-2, which contains the deletion and efaB5 inserted between EF_1844 and EF_1898, all three regions amplified. In OG1RF, which does not contain the deletion or the efaB5 element, 5′efaB5F and 3′efaB5R amplified a 689-bp region between the noncoding regions before EF_1846 and EF_1898.
Generation and detection of a large chromosomal deletion.
To detect the possible spontaneous occurrence of the deletion of the fsrC-EF_1841 region, primers AB83 and AB85 were used. To create a deletion mutant of OG1RF, we used the PheS* system as described previously (18), using the primers AB152 and AB153 (see Table S1 in the supplemental material) to amplify a 2.8-kb fragment overlapping the junction (Jct) between fsrC and EF_1841 from JH2-2 (reference strain for the “23.9-kb” deletion) for the purpose of deleting the fsrC-EF_1841 region in OG1RF. This 2.8-kb fragment was cloned into pCJK47, creating pCJK47::Jct. We transformed pCJK47::Jct into OG1RF and selected for erythromycin-resistant colonies (resulting from a single crossover between pCJK47::Jct and OG1RF). We then selected for the excision of the plasmid on MM9YEG agar medium containing p-Cl-Phe (catalog no. 6506; Sigma), resulting in colonies which carried either the parent or the deletion genotype. The colonies with deletion of the fsrC-EF_1841 region were then scored on gelatinase plates after incubation overnight at 37°C. One Gel− colony was selected and named TX5599. The deletion was confirmed by pulsed-field gel electrophoresis (PFGE) with NotI digestion, lack of gelatinase activity, and sequencing after amplification with AB152 and AB153.
Biofilm formation assays.
A biofilm density formation assay was carried out as described previously (25). The absorbance of each well was measured at an optical density at 570 nm (OD570). Each assay was performed in triplicate in eight independent experiments.
RESULTS AND DISCUSSION
Genomic diversity between EF_1814 and EF_1902.
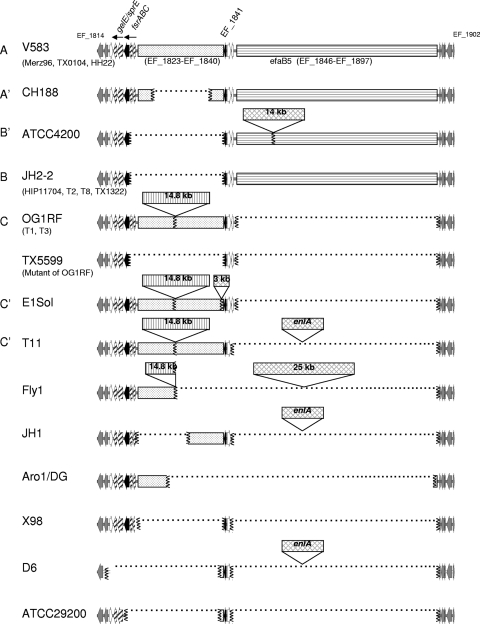
Since we had previously noted the conservation of fsrD in a small subset (36) and had previously observed that isolates that did not produce gelatinase but had the gelE gene often lacked the fsrB gene (37) (which was later described to be the “23.9-kb” deletion [31]), we set out to analyze the fsr-gelE genomic region to see how diverse or conserved this region is and if this could be related to MLST or strains from specific clones. The 22 accessible genomes that were analyzed come from different multilocus sequence types (MLSTs) (Table 1), sources (clinical and commensal), and times of isolation (from 1925 to 2005) (23). We first blasted each genome with gelE (EF_1818) and EF_1842 to determine the location of the fsrC-EF_1841 region on the chromosome/contig and the size of the fragment. The analysis of this region was performed with genes EF_1814 to EF_1902, which were present in all genomes. A total of 12 different genetic organizations, or genotypes, were detected based on variation in the region between EF_1814 and EF_1902 (Fig. 1), with three genotypes (A, B, and C) containing more than one isolate each. Genotype A carries the longest DNA sequence between EF_1814 and EF_1902, due to the presence of the previously described mobile element efaB5 (consisting of EF_1846 to EF_1897, which is ∼49.5 kb in strain V583 [35]), in addition to the presence of the fsrC-EF_1841 region. This genotype is present in strains within clonal complex 2 (CC2), as follows: V583 (sequence type 6 [ST6]), HH22 (ST6), TX0104 (ST2), and Merz96 (ST103). Isolates within genotype B/B′ lack the fsrC-EF_1841 region but possess an efaB5 element (the B′ genotype carries an additional 14 kb within the efaB5 element). The isolates with genotype B are found in diverse lineages by MLST (Fig. 2) and include JH2-2 (ST8), HIP11704 (ST4), T2 (ST11), T8 (ST8), and TX1322 (ST64). Finally, genotype C isolates carry an additional 14.8-kb fragment (encoding OG1RF_0128 to OG1RF_0140) in the fsrC-EF_1841 region; none of the three isolates with this genotype (OG1RF [ST1], T1 [ST21], and T3 [ST67]) carry an efaB5 element or are related by ST. For the 10 remaining EF_1814 to EF_1902 genotypes, the length of the sequence between EF_1814 and EF_1902 was found to range from 12.1 kb (ATCC 29200) to more than 70 kb for CH188. In summary, 13 strains were found to carry a complete fsr-gelE locus, while 9 strains were found to have an incomplete locus, with 6 of which having a conserved deletion (5 having an identical fsrC-EF_1841 deletion junction and 1 having a single base change, as seen in Fig. S2 in the supplemental material).
FIG. 1.
Genomic diversity of E. faecalis between EF_1814 and EF_1902. Twenty-two of the sequenced strains plus the 37-kb deletion mutant of OG1RF are shown. Redasoft Visual Cloning was used to find open reading frames (ORFs) representing EF_1814 to EF_1902 in sequences downloaded from either the NCBI server or the Broad Institute server. The first two ORFs (EF_1814 and EF_1815) and the last five ORFs (EF_1898 to EF_1902) of this region, which are common between all strains, are shown in gray. The fsrC (EF_1820) and EF_1841 genes are indicated by black arrows. When a region is missing, the junction areas are indicated with vertical squiggly lines, and the space between is filled with a dotted line. The fsrAB (EF_1821 and EF_1822), gelE (EF_1818), and sprE (EF_1817) genes are labeled with arrows filled with diagonal lines. The immediately adjacent loci, EF_1816, EF_1843, and EF_1844, are represented with white arrows. The area from EF_1823 to EF_1840 is represented with a spotted background, while the one representing the 14.8-kb region contains vertical lines. The efaB5 region from EF_1847 to EF_1896 is represented with horizontal lines. All other insertions are represented with crosshatching. Letters on the left side of the diagram indicate the genotypes based on EF_1814 to EF_1902, corresponding to Table 1.
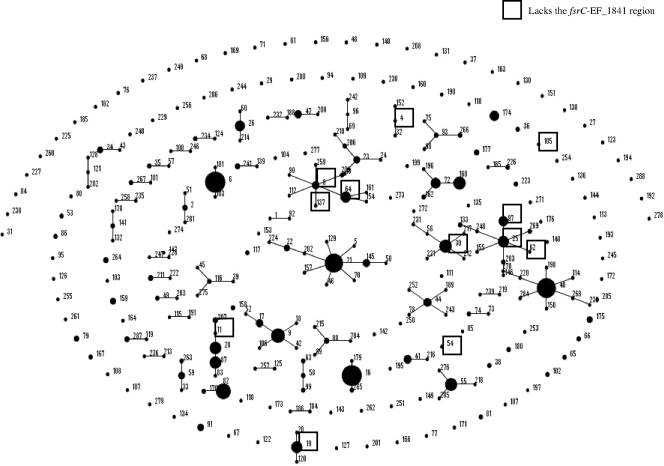
FIG. 2.
Strains lacking the fsrC-EF_1841 region are found in diverse MLST lineages. In this diagram, an eBURST of all MLSTs in the E. faecalis MLST database (http://efaecalis.mlst.net/) are depicted. Single-locus variants are linked by a line between them. The larger the middle circle, the more abundant this ST is in the database. The STs of 17 strains (6 sequenced and 11 from our collection) within this study lacking the fsrC-EF_1841 region are boxed.
Genetic variation of E. faecalis in a 700-kb region, including EF_1814 to EF_1902.
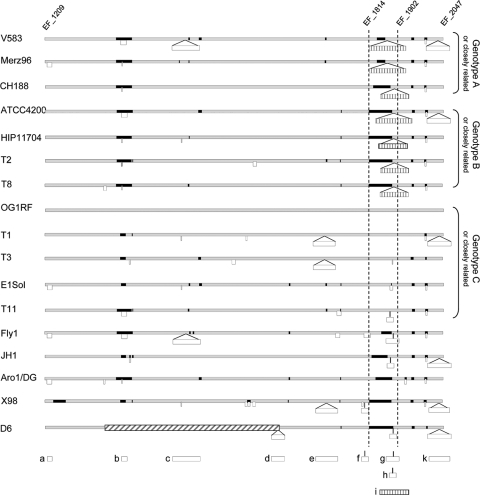
Since the genomes with genotype B presented such a high degree of similarity between each other in the EF_1814-to-EF_1902 region, we investigated the similarities of a larger region to see if all the strains were conserved or if the similarities were limited to the EF_1814-to-EF_1902 region. As mentioned previously, only the V583 and OG1RF genomes are complete and closed, while the other genomes are in a less finished stage, ranging from simple shotgun sequences to large assembled contigs. OG1RF was used as a reference since it contains a limited number of putative mobile elements. For 17 of the genomes, we could identify a 700-kb region in common, encompassing a sequence homologous to that of the EF_1814-to-EF_1902 region, corresponding to position 1.02 to 1.72 Mb in OG1RF (EF_1209 to EF_2047 in V583). As shown in Fig. 3, the 700-kb regions differ considerably among the strains, mainly due to deletion/insertion events. The three genotype B strains presented 7 major differences from each other, indicating that although highly similar in the EF_1814-to-EF_1902 region, those strains are not as highly conserved in other areas of the genome.
FIG. 3.
Genetic diversity of E. faecalis in an ∼700-kb region, including EF_1209 to EF_2047. Seventeen of the genomes which contain ∼700 kb in common corresponding to nucleotide positions 1.02 Mb to 1.72 Mb in OG1RF (EF_1209 to EF_2047 in V583) were analyzed for genomic differences using OG1RF as a reference. The gray bars indicate the sequences corresponding to OG1RF, deletions are indicated with black bars, and insertions of >1 kb are indicated with white bars. The black and white bars are to scale with the size of the deletion/insertion. Smaller white bars are insertions of >1 kb but <5 kb, while larger white bars designated with letters are insertions of >5 kb or insertional elements that have been previously described; these are positioned directly below where they appear on the diagram for reference. “a” carries a WxL locus, “b” carries a probable ABC transporter and a two-component system, “c” is phage 3, “d” is a probable multidrug resistance transposon, “e” is a probable 38.5-kb phage, “f” is a probable 11-kb fragment that does not appear to be mobile, “g” is a probable 25-kb phage, “h” carries enlA, “i” (bar with vertical lines) is the efaB5 element, and “k” is phage 4. The bar with diagonal lines indicates an inversion in the D6 genome compared to OG1RF. There were 5 to 16 variations of >1 kb between each genome and OG1RF, with an average of 9 variations per genome.
Strains lacking the fsrC-EF_1841 region represent diverse MLSTs and do not produce gelatinase.
While Nakayama et al. found that 56% of the 46 isolates obtained from a local hospital lacked the fsrC-EF_1841 region (31), Roberts et al. reported that 28% of 215 isolates lacked the fsrC-EF_1841 region in a more diverse collection of isolates (40). However, in both studies, the clonality of those isolates was unknown. We therefore initially studied 38 strains (Table 2) belonging to diverse MLSTs (in addition to the sequenced strains), used a set of multiplex primers to determine the absence or presence of this region, and correlated this with their ability to produce gelatinase. As shown in Table 2, only 7 of the initial 38 strains (18%) from our collection generated a 739-bp PCR product characteristic of the deletion of the fsrC-EF_1841 region, and they were all gelatinase nonproducers; 6 of these strains were unrelated by MLST, while 2 were double-locus variants of each other (Fig. 2). A total of 22 of the 38 isolates (58%) resulted in a PCR product, indicative of the presence of the full fsrC and EF_1841 genes, all of which produced gelatinase. Of the remaining 9 strains, 4 strains were positive for only the fsrC region, and 3 of those 4 strains did not produce gelatinase. The other 5 strains were negative for the fsrC region but positive for EF_1841 by PCR, and none produced gelatinase.
Through a combination of previously published data on the sequenced strains (23), sequence analysis, and confirmation of these results through multiplex PCR and gelatinase production assays (Table 1), we verified that all sequenced genotype B/B′ strains (negative for the fsrC-EF_1841 region) were gelatinase nonproducers although gelE positive, while all genotype A and genotype C/C′ strains were gelatinase producers. Of the remaining 7 strains, Fly1 and ARO1/DG are gelatinase producers, as to be expected since they have the fsrABDC and gelE-sprE operons intact. However, CH188 and JH1 were gelatinase nonproducers, despite complete fsrABDC and gelE-sprE loci. X98, D6, and ATCC 29200 are gelatinase nonproducers since some or all of the fsrABDC operon is absent. In sum, 14 of the 60 initial isolates (23%) examined (38 tested by PCR and the 22 sequenced genomes analyzed) lacked the fsrC-EF_1841 region, and these strains do not produce gelatinase.
Limited variability in the Fsr system and none in FsrD.
In a previous study (36), the conservation of fsrD in a limited group of isolates had been noted. Since the 13 E. faecalis strains in this study with complete Fsr systems were more diverse by MLST, we also compared their fsr-gelE loci for possible sequence diversity. It is known that among the agr loci in Staphylococcus aureus, which has the best described cyclic peptide-mediated quorum-sensing system among Gram-positive bacteria and is a homologue of the fsrABDC system, sequence variation is particularly evident in the autoinducer precursor AgrD (only a cysteine conserved of the 7 to 9 amino acids), AgrB (only 68 of the 187 amino acids are conserved), and AgrC (only 177 of the 423 amino acids are conserved) due to the presence of a hypervariable region which results in at least four agr specificity groups (17, 33). Unlike what has been found in staphylococci (30), we observed perfect conservation, not only of the 11 amino acids that are the basis for the autoinducer cyclic peptide but also of the full FsrD polypeptide (53 amino acids) of the 13 diverse strains of E. faecalis evaluated. When looking at the other constituents of the fsr operon, we found that the 13 strains carrying the complete fsrABC genes had up to 6 amino acid variations for fsrA, 7 amino acid variations for fsrB, and 9 amino acid variations for fsrC, relative to V583. Among the 20 strains carrying gelE and sprE, we found up to 12 amino acid variations for gelE and 15 for sprE, relative to V583. None of the proteins of the fsr-gelE locus were truncated as a consequence of a frameshift or point mutation.
A conserved junction sequence and SNPs in strains lacking the fsrC-EF_1841 region suggest that this deletion may occur by homologous recombination.
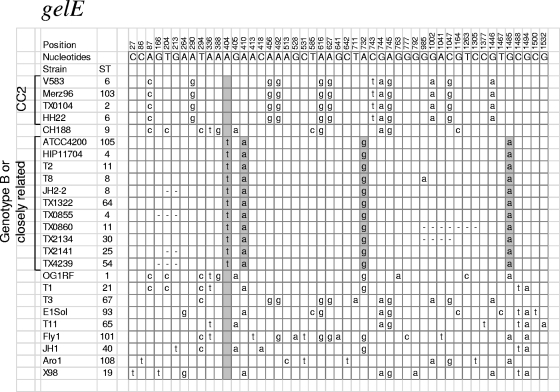
A detailed analysis of the 6 genotype B/B′ strains lacking the fsrC-EF_1841 region found an identical sequence (∼600 bp, with 300 bp of fsrC and 300 bp of EF_1841) overlapping the junction (except for T8, which had a single base difference). Most of the sequences from the junction through gelE between these sequenced strains were identical; however, some of the strains do not have complete sequence coverage within this region. Furthermore, 11 strains (6 sequenced strains and 5 from our collection) lacking the fsrC-EF_1841 region also shared a nearly identical gelE gene profile with 4 identical single nucleotide polymorphisms (SNPs) over the 1,533 nucleotides within the gene (0.26% variation), compared to up to 3.2% variation among the 12 other gelE sequences from strains carrying the fsrC-EF_1841 region (Fig. 4). We had speculated that there might be an even higher level of differences in strains lacking the fsrC-EF_1841 region since the gelE gene is not expressed, at least under standard in vitro conditions, and thus, there might be more degeneration of the sequence.
FIG. 4.
SNP analysis of gelE. SNP analysis was done on the entire gelE open reading frame of 25 strains (20 sequenced strains with intact gelE genes and 5 strains from our collection). The position of a SNP is indicated with the numbers at the top. The letters at the top indicate the nucleotide at each position that is shared by the most isolates (consensus). All other letters indicate the SNPs found in the corresponding strain. Only variations from the consensus are indicated. A dash indicates that sequence was not available for that region or nucleotide due to lack of sequence coverage. This figure includes all sequenced strains plus isolates from our collection lacking the fsrC-EF_1841 region for which we had the gelE sequence.
The fact that the strains lacking the fsrC-EF_1841 region are scattered among various MLSTs (some other members of which have this region) (Fig. 2) suggests that the deletion of the fsrC-EF_1841 region is not an ancient event that occurred before those strains evolved into distinct clones. Instead, the presence of a highly conserved and distinctive junction sequence suggests a common mechanism for the deletion; moreover, the presence of a virtually identical gelE gene SNP profile suggests that the region surrounding the deletion was introduced by horizontal transfer from a common or conserved source and recombined into the chromosome.
Rice and Carias reported that insertion of Tn5385 into a recipient's chromosome likely occurred by recombination across flanking regions homologous between donors and recipients, since identical deletions present in the donor sequences flanking Tn5385 were found in the transconjugants (38). If there is frequent recombination in this region, it is possible that the deletion could be transferred via homologous recombination through the movement of a mobile element, such as efaB5, as postulated previously (31). EfaB5 is a Tn916-like element containing several genes related to virulence and flanked by site-specific recombinases, indicative of a mobile element. We reasoned that the “23.9-kb” deletion could be occurring in a scenario similar to that described for Tn5385 by Rice and Carias, where the deletion would be cotransferred with efaB5 and incorporated into the genome via homologous recombination, leading to deletion of the fsrC-EF_1841 region. Using a multiplex PCR, we also screened for the presence of the efaB5 element in relation to the deletion in the 7 strains (for which gelE was sequenced and the presence of the deletion known) plus 10 additional isolates for which previous findings indicated the deletion (40). We found that 16 out of these 17 strains lacking the fsrC-EF_1841 region (94%) had the efaB5 element, compared to 7 out of 22 strains which contain the fsrC-EF_1841 region (32%) (Table 2). An additional 8 isolates did not give a PCR product indicative of the presence of the efaB5 element nor the absence of the efaB5 element (PCR positive for the junction between EF_1846 and EF_1898, the region flanking the efaB5 element) (see Fig. S1 in the supplemental material). One possibility is that these isolates contain a large insertion, such as the enlA element or the 25-kb insertion found in Fly1, in which case, although efaB5 is not present, the junction would not amplify due to these large insertions.
We next attempted to show transfer of the deletion. Since the efaB5 element does not carry any marker, we used a JH2-2::Tn916 (Tet) donor strain to mediate conjugation. After 10,000 tetracycline-resistant transconjugant colonies on gelatinase plates were screened, all were found to be gelatinase producers, indicating that none had the deletion. In a recent report, Manson et al. (21) reported that the frequency of transfer of chromosomal regions mediated by conjugative plasmids was 10−9; our assay conditions were not sufficiently sensitive to detect transconjugants that arrive at this frequency. This mechanism could also mediate horizontal transfer of the deletion and surrounding regions.
No spontaneous occurrence of a gelatinase nonproducer phenotype.
It was suggested by two groups that gelatinase nonproducer colonies may happen by spontaneous loss of the “23.9-kb” region (9, 31). To estimate the probability of a natural occurrence of this phenotype, we spread dilutions of overnight cultures of OG1RF and 7 strains from CC2 (all gelE positive) on gelatinase plates but failed to observe gelatinase nonproducer colonies. Then, we grew OG1RF for 60 generations, extracted genomic DNA (gDNA), and used primers AB83 to AB85 to screen for the junction if the deletion had occurred spontaneously. With a mixture of OG1RF (which has the fsrC-EF_1841 region) and JH2-2 (which lacks the fsrC-EF_1841 region), our level of detection was 1 CFU of JH2-2 for 106 CFU of OG1RF. We repeated this experiment 5 times with the same outcome: we never detected a PCR product, indicating the presence of a spontaneous deletion of the fsrC-EF_1841 region. Since it was proposed that the efaB5 element may be implicated in the deletion of the fsrC-EF_1841 region (31), we also tested the 7 CC2 strains (genotype A carrying the efaB5 element from various origins) for the natural occurrence of the deletion by using the same conditions as the ones described above for OG1RF but, again, failed to detect the incidence of a deletion by PCR.
Generation and analysis of an isogenic mutant of OG1RF with a deletion of the fsrC-EF_1841 region.
The fsrC-EF_1841 region in OG1RF is very similar to the one present in V583, except for an additional 14.8-kb fragment inserted between the corresponding EF_1826 and EF_1827 loci (4), leading to a fragment of 37 kb from fsrC to EF_1841. Since most naturally occurring Gel− E. faecalis isolates lack this region, unlike laboratory-generated fsr or gelE mutants, we sought to see if the rest of this region was important for the phenotypes, i.e., changes observed in biofilm and chaining with laboratory-generated fsr-gelE insertion mutants. Toward this end, we replaced the fsrC-EF_1841 region with the JH2-2 junction sequence by allelic replacement using the PheS* system (18). One mutant was selected and designated TX5599. Analysis of the sequence of the 2-kb region overlapping the junction sequence in TX5599 and JH2-2 compared to that in OG1RF showed that one crossover event occurred between 756 and 550 bp upstream of the junction and the second occurred between 938 and 1,134 bp downstream of the junction. The deletion was also confirmed by PFGE after digestion with NotI (Fig. 5). Therefore, an isogenic mutant of OG1RF with a deletion of the fsrC-EF_1841 region was easily obtained, indicating that major adaptations were not needed and that this region is not needed for growth in vitro in OG1RF. We saw very similar biofilm levels (Fig. 6) and chaining phenotypes (data not shown) when comparing the 37-kb deletion mutant of OG1RF and fsr-gelE insertion mutants, also indicating that loss of the rest of this region does not seem to compensate for the decrease in biofilm and chaining seen in fsr-gelE mutants. In addition, these results also demonstrated that a large genomic deletion can be generated in E. faecalis.
FIG. 5.
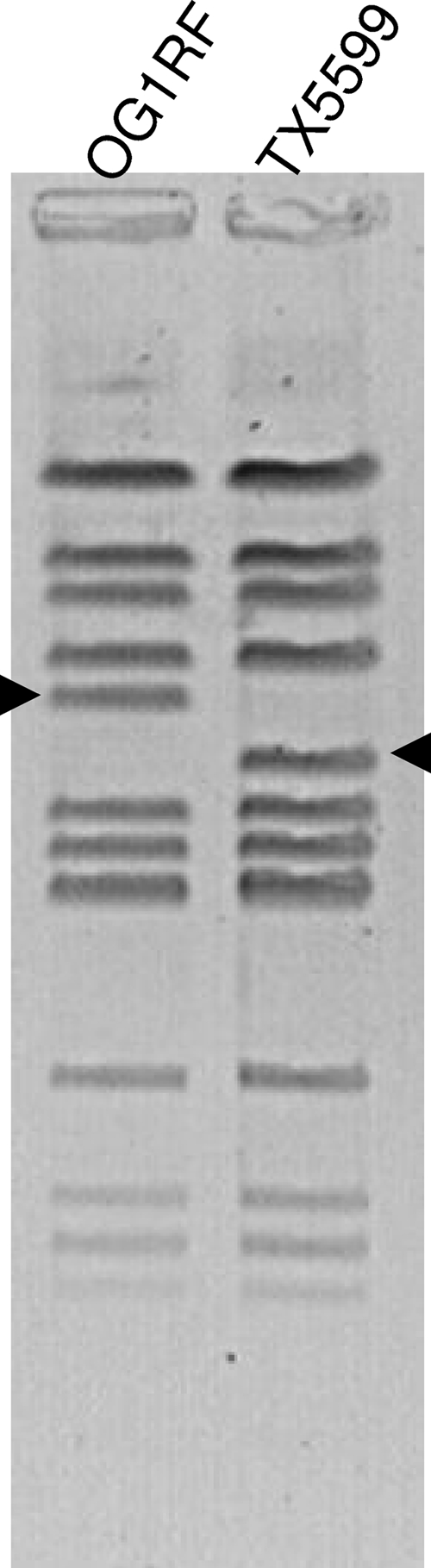
PFGE of NotI digestion of OG1RF and its mutant (TX5599), isogenic for the deletion from fsrC to EF_1841. Digestion with NotI showed the band corresponding to the fsrC-EF_1841 region migrating as ca. 280 kb for OG1RF and ca. 240 kb for TX5599, a difference corresponding to the 37-kb deletion in OG1RF. The band corresponding to the fsrC-EF_1841 region and the fragment after the deletion area is shown by a black arrowhead.
FIG. 6.
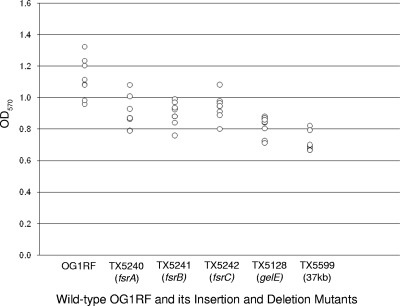
Biofilm formation does not differ between fsr-gelE mutant strains and the 37-kb mutant fsrC-EF_1841 in OG1RF. Biofilm formation in the insertion mutants of fsrA (TX5240), fsrB (TX5241), fsrC (TX5242), and gelE (TX5128) and in the 37-kb deletion mutant (TX5599), all in the OG1RF background, was assayed. Each circle represents the average OD570 of a triplicate of one of eight independent experiments.
Conclusion.
In summary, we analyzed 22 genomes representing 19 different MLSTs with the goal of better characterizing the region that includes the previously designated “23.9-kb” deletion (fsrC-EF_1841). We found 12 unique genotypes based on the sequence surrounding this region, indicating the high plasticity of the genome at this location. We observed that, although the EF_1814-to-EF_1902 region was similar in strains of different MLSTs lacking the fsrC-EF_1841 region, strains differed by insertion and deletion events (Fig. 3) when looking at a larger fragment (corresponding to ∼700 kb in OG1RF).
Despite clonal variability of these genomes by MLST, we found perfect conservation in fsrD, encoding the propeptide of the autoinducer, within all strains and high identity among the fsrA, fsrB, and fsrC genes, unlike what is seen in agr of S. aureus. Including the available sequenced strains, we found that strains from various clonal backgrounds lacking the fsrC-EF_1841 region represent approximately 23% of isolates tested, and none of these strains produced gelatinase. We also found that strains lacking the fsrC-EF_1841 region had a very highly conserved junction sequence (∼600 bp), including an identical SNP profile in gelE, suggesting that the deletion and surrounding region may be horizontally transferred between strains. This is also consistent with the observation that a high percentage (94%) of strains lacking the fsrC-EF_1841 region contain the efaB5 mobile element in its flanking sequence.
Despite the frequency of the fsrC-EF_1841 deletion, we did not detect evidence that mutants of OG1RF lacking this region occur spontaneously or after mating with JH2-2::Tn916. This work also demonstrated that the PheS* system can be used to delete a large fragment (37 kb) of chromosomal DNA, and nothing between fsrC and the homologue of EF_1841 is essential for the growth of OG1RF in BHI medium, nor does the deletion of this region seem to compensate for the biofilm and chaining seen in specific fsrA, fsrB, fsrC, and gelE insertion mutants.
Supplementary Material
Acknowledgments
We are grateful to K.V. Singh and S. R. Nallapareddy for helpful discussions.
This work was supported by National Institutes of Health grant R37 AI47923 from the Division of Microbiology and Infectious Diseases to B.E.M. J.R.G.-P. is supported by Molecular Basis of Infectious Diseases Training Grant T32 AI55449.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ackermann, H. W., T. Caprioli, and S. S. Kasatiya. 1975. A large new Streptococcus bacteriophage. Can. J. Microbiol. 21:571-574. [DOI] [PubMed] [Google Scholar]
- 2.Banai, M., and D. J. LeBlanc. 1983. Genetic, molecular, and functional analysis of Streptococcus faecalis R plasmid pJH1. J. Bacteriol. 155:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourgogne, A., et al. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgogne, A., S. G. Hilsenbeck, G. M. Dunny, and B. E. Murray. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 188:2875-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock, T. D., B. Peacher, and D. Pierson. 1963. Survey of the bacteriocines of enterococci. J. Bacteriol. 86:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury, S. A., et al. 2009. A trilocus sequence typing scheme for hospital epidemiology and subspecies differentiation of an important nosocomial pathogen, Enterococcus faecalis. J. Clin. Microbiol. 47:2713-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coque, T. M., J. E. Patterson, J. M. Steckelberg, and B. E. Murray. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171:1223-1229. [DOI] [PubMed] [Google Scholar]
- 8.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. U. S. A. 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton, T. J., and M. J. Gasson. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelbert, M., E. Mylonakis, F. M. Ausubel, S. B. Calderwood, and M. S. Gilmore. 2004. Contribution of gelatinase, serine protease, and fsr to the pathogenesis of Enterococcus faecalis endophthalmitis. Infect. Immun. 72:3628-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner, P., D. H. Smith, H. Beer, and R. C. Moellering, Jr. 1969. Recovery of resistance (R) factors from a drug-free community. Lancet ii:774-776. [DOI] [PubMed] [Google Scholar]
- 13.Gilmore, M. S., and J. J. Ferretti. 2003. Microbiology. The thin line between gut commensal and pathogen. Science 299:1999-2002. [DOI] [PubMed] [Google Scholar]
- 14.Gold, O. G., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 15.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jha, A. K., H. P. Bais, and J. M. Vivanco. 2005. Enterococcus faecalis mammalian virulence-related factors exhibit potent pathogenicity in the Arabidopsis thaliana plant model. Infect. Immun. 73:464-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 18.Kristich, C. J., J. R. Chandler, and G. M. Dunny. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LeBlanc, D. J., and L. N. Lee. 1982. Characterization of two tetracycline resistance determinants in Streptococcus faecalis JH1. J. Bacteriol. 150:835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maekawa, S., M. Yoshioka, and Y. Kumamoto. 1992. Proposal of a new scheme for the serological typing of Enterococcus faecalis strains. Microbiol. Immunol. 36:671-681. [DOI] [PubMed] [Google Scholar]
- 21.Manson, J. M., L. E. Hancock, and M. S. Gilmore. 2010. Mechanism of chromosomal transfer of Enterococcus faecalis pathogenicity island, capsule, antimicrobial resistance, and other traits. Proc. Natl. Acad. Sci. U. S. A. 107:12269-12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manson, J. M., S. Keis, J. M. Smith, and G. M. Cook. 2003. Characterization of a vancomycin-resistant Enterococcus faecalis (VREF) isolate from a dog with mastitis: further evidence of a clonal lineage of VREF in New Zealand. J. Clin. Microbiol. 41:3331-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride, S. M., V. A. Fischetti, D. J. Leblanc, R. C. Moellering, Jr., and M. S. Gilmore. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moellering, R. C., Jr., and A. N. Weinberg. 1971. Studies on antibiotic syngerism against enterococci. II. Effect of various antibiotics on the uptake of 14C-labeled streptomycin by enterococci. J. Clin. Invest. 50:2580-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72:3658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray, B. E. 1990. The life and times of the Enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray, B. E., and B. Mederski-Samaroj. 1983. Transferable beta-lactamase. A new mechanism for in vitro penicillin resistance in Streptococcus faecalis. J. Clin. Invest. 72:1168-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mylonakis, E., et al. 2002. The Enterococcus faecalis fsrB gene, a key component of the fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect. Immun. 70:4678-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama, J., et al. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama, J., et al. 2006. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal AgrD. J. Bacteriol. 188:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama, J., R. Kariyama, and H. Kumon. 2002. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl. Environ. Microbiol. 68:3152-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallapareddy, S. R., et al. 2006. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Invest. 116:2799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 34.Oprea, S. F., et al. 2004. Molecular and clinical epidemiology of vancomycin-resistant Enterococcus faecalis. J. Antimicrob. Chemother. 53:626-630. [DOI] [PubMed] [Google Scholar]
- 35.Paulsen, I. T., et al. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 36.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, L. B., and L. L. Carias. 1998. Transfer of Tn5385, a composite, multiresistance chromosomal element from Enterococcus faecalis. J. Bacteriol. 180:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, L. B., et al. 1991. Chromosomally mediated beta-lactamase production and gentamicin resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 35:272-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts, J. C., K. V. Singh, P. C. Okhuysen, and B. E. Murray. 2004. Molecular epidemiology of the fsr locus and of gelatinase production among different subsets of Enterococcus faecalis isolates. J. Clin. Microbiol. 42:2317-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz-Garbajosa, P., et al. 2006. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 44:2220-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahm, D. F., et al. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharpe, M. E. 1964. Serological types of Streptococcus faecalis and its varieties and their cell wall type antigen. J. Gen. Microbiol. 36:151-160. [DOI] [PubMed] [Google Scholar]
- 44.Sifri, C. D., et al. 2002. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect. Immun. 70:5647-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh, K. V., S. R. Nallapareddy, E. C. Nannini, and B. E. Murray. 2005. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect. Immun. 73:4888-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willems, R. J., et al. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.