Abstract
Monitoring microbiological water quality is important for protecting water resources and the health of swimmers. Routine monitoring relies on cultivating fecal indicator bacteria (FIB), frequently using defined substrate technology. Defined substrate technology is designed to specifically enrich for FIB, but a complete understanding of the assay microbiology requires culture-independent analysis of the enrichments. This study aimed to identify bacteria in positive wells of Colilert and Enterolert Quanti-Tray/2000 (IDEXX Laboratories) FIB assays in environmental water samples and to quantify the degree of false-positive results for samples from an urban creek by molecular methods. Pooled Escherichia coli- and Enterococcus-positive Quanti-Tray/2000 enrichments, either from urban creek dry weather flow or municipal sewage, harbored diverse bacterial populations based on 16S rRNA gene sequences and terminal restriction fragment length polymorphism analyses. Target taxa (coliforms or enterococci) and nontarget taxa (Vibrio spp., Shewanella spp., Bacteroidetes, and Clostridium spp.) were identified in pooled and individual positive Colilert and Enterolert wells based on terminal restriction fragments that were in common with those generated in silico from clone sequences. False-positive rates of between 4 and 23% occurred for the urban creek samples, based on the absence of target terminal restriction fragments in individual positive wells. This study suggests that increased selective inhibition of nontarget bacteria could improve the accuracy of the Colilert and Enterolert assays.
Quantifying fecal pollution in recreational waters is important for protecting the health of swimmers. Current standards for microbiological water quality in the United States and elsewhere are based on culturable fecal indicator bacteria (FIB), i.e., total coliforms, fecal coliforms, or Escherichia coli, and enterococci (21, 53, 57). Various methods are used to quantify FIB, including multiple-tube fermentation, membrane filtration, and defined substrate technologies (9, 16, 45). All rely on temperature, substrate, and selective growth inhibitors to select for FIB (23, 29). The commercially available defined substrate technologies Colilert and Enterolert (IDEXX Laboratories, Westbrook, ME) are accepted by the U.S. Environmental Protection Agency as alternatives to the multiple-tube fermentation and membrane filtration methods for fresh, marine, and estuarine surface waters (54). Specific enzyme-substrate relationships are the basis of both assays (45). In the Colilert assay, total coliforms and E. coli are indicated by a yellow or a yellow and a fluorescent metabolite, respectively. Positive Enterolert assays are indicated by a fluorescent metabolite. The manufacturer-supplied tray format is more convenient than multiple-tube fermentation and membrane filter techniques (20, 45). However, there is evidence that the Colilert and Enterolert assays are not specific (2, 6, 17, 43, 51).
False-positive Colilert or Enterolert readings could occur when there is sufficient abundance of nontarget bacteria, i.e., non-FIB, that enzymatically cleave the chromogenic or fluorogenic substrates. Beta-galactosidase activity, which yields the yellow color in Colilert, has been described for nontarget species in the genera Aeromonas, Vibrio, Pseudomonas, and Flavobacterium (17, 19, 51). Beta-glucuronidase activity, which yields the fluorescence in Colilert, has been found in Shigella, Salmonella, and Yersinia strains, in Flavobacteria, and in some streptococci, clostridia, Bacteroides spp., and Corynebacterium spp. (17, 35, 45). In the Enterolert assay, several nontarget bacteria were identified that yielded false-positive reactions, such as Proteus vulgaris, Serratia marcescens, Sphingomonas spp., and Flavobacterium spp. (2, 9). The basis for defined substrate technology is that nontarget bacterial growth should be excluded, and thus, high concentrations of nontarget bacteria would have to be initially present in a water sample to yield false-positive results (12, 17, 20). However, previous assessments of nontarget bacteria in defined substrate technology assays involved two cultivation steps, the first with the initial assay and the second with bacterial isolation from assay enrichments. Since many environmental bacteria resist cultivation (55), relying on isolate cultivation to characterize assay enrichments could incompletely describe nontarget populations in defined substrate technology assays.
In this study, we used culture-independent methods to identify bacterial taxa in the positively scored wells of Colilert and Enterolert enrichments for selected water samples. Our objectives were to more thoroughly describe bacterial assemblages within the completed assays and to quantify the rates of false positives. The bacterial assemblages in pooled positive Colilert and Enterolert wells of creek water and sewage samples were analyzed for terminal restriction fragment length polymorphism (TRFLP) of genes encoding 16S rRNA. Clone libraries based on 16S rRNA genes were developed for the same pooled positive wells. Bacterial community structure and false-positive rates were determined in individual positive Colilert and Enterolert wells of additional creek water samples by quantifying the occurrence of terminal restriction fragments (TRFs) not associated with target coliform and enterococcus bacteria. Taxa that had not been previously associated with IDEXX assays were found, and the apparent growth of nontarget bacteria in IDEXX assay enrichments resulted in quantifiable false-positive rates for these samples.
MATERIALS AND METHODS
Study sites and sampling design.
The study site was Arroyo Burro Creek in Santa Barbara, CA, which drains a semiurban watershed and is listed under the Clean Water Act Section 303(d) as impaired for recreation because of high FIB concentrations. A prior study revealed sources of human waste entering the creek via storm drains that flow year-round (47). The watershed is 23 km2 with 16% urban and suburban land use in the lower watershed and 84% natural land, including mixed forest, chaparral, and rural residential land use, in the upper watershed (13). Arroyo Burro Creek discharges into a brackish lagoon just upstream of a popular beach that is frequently posted with swimming advisories based on weekly FIB monitoring. The creek sampling site, which was just upstream of the lagoon, was furthest downstream in the urban and suburban contaminated reach described previously (47).
The study design was comprised of field sampling and analysis to (i) characterize bacterial assemblages from within creek waters and from a contaminated reference sample (sewage), (ii) characterize bacterial assemblages recovered from Colilert and Enterolert assays of the samples, (iii) identify taxa in source samples and FIB enrichments using clone library analysis, and (iv) quantify the rate of false-positive reactions due to nontarget bacteria in additional samples from the same site. Three creek samples were acquired during dry weather, and one additional wet weather sample was acquired for the clone library development. Accordingly, one creek (CR-1) and one sewage (SEW-1) sample were taken on 24 October 2005, when there had been no rainfall for at least a month preceding. The wet weather creek sample was acquired on 9 November 2005 after a first seasonal rainfall event. The same creek location was sampled again during dry weather on 6 November 2007 (CR-2) and 18 December 2007 (CR-3).
Sampling and FIB quantification.
Creek water was sampled (2 liters) approximately 10 cm beneath the creek water surface. Municipal sewage was sampled from the influent of the El Estero Wastewater Treatment Plant (Santa Barbara, CA). Samples were filtered through 22- to 25-μm-pore-size sterile Miracloth (Calbiochem-Novabiochem, La Jolla, CA) to remove large debris and stored on ice until processing in the laboratory (<3 h). Total coliforms, E. coli, and Enterococcus spp. were quantified using the Colilert and Enterolert Quanti-Tray/2000 (IDEXX Laboratories, Westbrook, ME) according to the manufacturer's instructions. Samples were diluted to either 1:10 (creek water) or 1:100,000 (sewage) in sterile Nanopure water. The results were scored using the IDEXX MPN Generator 3.2, a software tool calculating the most probable number (MPN) based on the statistical analysis proposed by Hurley and Roscoe (28).
DNA extraction from environmental samples and Quanti-Tray wells.
DNA was extracted from water and sewage samples, pooled positive Colilert/Enterolert wells, and individual positive Colilert/Enterolert wells. For the creek water and sewage, the samples were vacuum filtered (0.22 μm) to the point of refusal to collect bacteria, and the filters were frozen (−20°C) prior to DNA extraction using an UltraClean water DNA kit (Mo Bio, Carlsbad, CA) according to the manufacturer's instructions. The extracted DNA was ethanol precipitated and resuspended in 50 μl of 0.1× WD5 solution from the kit. The volumes filtered were approximately 1,500 ml (creek samples) and 135 ml (sewage sample).
Individual large (∼1.9 ml) Quanti-Tray wells were sampled through the sterilized (70% ethanol swabbed) paper backing by using a sterile syringe and needle or sterile pipette tips. Small wells (∼0.19 ml) were not sampled, as the small culture volume was not expected to yield sufficient DNA for analysis. To recover the DNA from the FIB enrichments of CR-1 and SEW-1, yellow/fluorescent Colilert and fluorescent Enterolert Quanti-Tray well contents for a sample were each pooled from 18 to 49 positive wells (28 to 43 ml) and were filtered and extracted using an UltraClean water DNA kit as described above. This DNA was used for TRFLP and clone library analyses.
To recover the DNA from CR-2 and CR-3 FIB enrichments, the contents of individual yellow Colilert, yellow/fluorescent Colilert, and fluorescent Enterolert wells were collected into sterile 2-ml screw-cap microcentrifuge vials (BioSpec Products, Bartlesville, OK). Each sample was centrifuged (10 min at 16,000 × g), and the pellet was stored (−20°C) until DNA extraction. DNA extraction was performed following a previously published protocol using bead beating in CTAB (cetyltrimethylammonium bromide) buffer followed by purification using phenol-chloroform (24) but with some modifications. First, the diethyl pyrocarbonate step to remove RNase was not used. Second, the pellet was lysed by mixing three times for 30 s (with 20 s of cooling between intervals) in a BioSpec MiniBeadbeater-8 (BioSpec Products, Bartlesville, OK) after adding 0.5 g of 0.1-mm zirconia/silica beads (BioSpec Products, Bartlesville, OK) plus published reagents. Third, the pellet was extracted again after adding 0.5 ml CTAB buffer, and the aqueous phases of the extractions were pooled. Total nucleic acids were precipitated overnight using 0.1 volume of 3 M sodium acetate (pH 5.5) and 1 volume of isopropanol. After centrifugation (30 min at 16,000 × g), the pellet was washed with ice-cold 70% (vol/vol) ethanol, centrifuged again, and air-dried prior to resuspension in 50 μl of sterile 0.1× Tris buffer. The final DNA concentrations were determined with fluorimetry, using a Quant-iT Picogreen double-stranded DNA assay kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions.
Terminal restriction fragment length polymorphism analysis.
The bacterial community structure was investigated using TRFLP and clone library analysis for CR-1, SEW-1, and the wet weather sample and for the pooled positive Quanti-Tray enrichments of CR-1 and SEW-1. The dominant TRFs in the pooled positive wells of the IDEXX assay enrichments were identified by in silico restriction digest (as described below). TRFLP analysis was also performed for CR-2 and -3.
The amplification of 16S rRNA genes by PCR and subsequent TRFLP analysis of HhaI restriction digests were based on published protocols (33). PCR was performed using the primers 8F hex (fluorescently labeled forward primer, 5′-AGAGTTTGATCCTGGCTCAG-3′) and 1389R (5′-ACGGGCGGTGTGTACAAG-3′), using reaction conditions as previously described (33) but with the addition of bovine serum albumin (0.2 mg/ml) and Q-Solution (Qiagen, Valencia, CA) to the PCR master mix. PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA), and 200 to 300 ng of purified DNA was digested with HhaI (New England Biolabs, Ipswich, MA). After inactivation of the restriction enzyme by heating (65°C for 20 min), the lengths of the fluorescently labeled fragments were determined with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA) at the Genomics Technology Support Facility (Michigan State University). TRFs with lengths between 50 and 1,000 bp were aligned and normalized to total peak height with Excel software, using a macro developed by C. Walsh (http://www.wsc.monash.edu.au/∼cwalsh/treeflap.xls). Peaks with a relative peak height of less than 1% were discarded (44). The automatic alignment was verified manually.
For CR-1, -2, and -3 and SEW-1, the richness (S) and Shannon diversity (H) indices were estimated using PRIMER version 6 software (Primer-E Ltd., Plymouth, United Kingdom), based on TRFLP patterns. Hierarchical cluster analysis was performed using TRFLP data based on Bray-Curtis similarities (15). Significant results were analyzed using the similarity profile routine (SIMPROF) (14), which tests for random clustering (8). The SIMPROF test works by ordering similarities from a group of a priori unstructured samples from smallest to largest and plotting similarities against their rank. The observed profile is compared with that expected under the null hypothesis of no meaningful structure within that group, using permutation. Repeated application of this test generates a stopping rule for a posteriori division of the samples into ever smaller subgroups, as in hierarchical cluster analysis (14).
The in silico PCR and Restriction program of the web-based tool Microbial Community Analysis (MiCA) (48) was used to suggest possible phylogenetic affiliations of peaks from the electropherograms in relationship to cloned sequences. A size window of ±3 bp was used to account for the possible differences between real and predicted TRF lengths (36, 39). Only taxa that were found in the clone library analysis (below) were further evaluated.
Clone library analysis and TRF identification.
Partial 16S rRNA genes were amplified using primers 8F (not labeled) and 1398R and cloned into the pCR2.1 vector (Invitrogen, Carlsbad CA). Blue/white screening and further processing and sequencing were performed by Agencourt Bioscience (Beverly, MA). The primers used for sequencing were the same as for PCR amplification. Sequencing was performed using the BigDye terminator version 3.1 chemistry, which is optimized for longer reads, uniform peak heights, and robustness. Ninety-six clones were sequenced for each sample, which was expected to yield a sufficient number of different clones for adequate sampling of the IDEXX assay enrichments and for identifying the dominant TRFLP peaks. The occurrence of chimeras was determined using Bellerophon (27), Check_Chimera (34), Pintail (4), and manual comparison of putative chimeric sequences. Excluding sequences of insufficient quality and chimeras, the final numbers of clones analyzed were 62 (CR-1), 73 (CR-1, yellow/fluorescent Colilert), 73 (CR-1, fluorescent Enterolert), 56 (SEW-1), 88 (SEW-1, yellow/fluorescent Colilert), and 89 (SEW-1, fluorescent Enterolert). Based on the sequence identity matrix, clones sharing ≥97% identity were grouped into one operational taxonomical unit (OTU). The BLAST algorithm (3) was used to determine the phylogenetic affiliations of all OTUs. Rarefaction analysis and estimation of the clone library richness (SChao1) were performed using a web interface (http://www.aslo.org/lomethods/free/2004/0114a.html) (31). SChao1 is an abundance-based richness estimator well suited for estimating phylotype richness from prokaryotic libraries of 16S rRNA genes (31).
The clone libraries were used to identify the dominant TRFs from the pooled positive Colilert and Enterolert wells and from the source samples. The MiCA web tool was used for in silico restriction by HhaI of all clone library taxa. The resulting in silico TRFs, with known phylogenetic affiliations, were then compared to the sample TRFs.
Identification of false-positive wells.
To determine the rates of false positives in the Colilert and Enterolert assays, individual positive wells arising from CR-2 and CR-3 were assessed as described above for TRFs associated with nontarget taxa. False-positive wells were defined as those that produced color or fluorescence in the absence of target TRFs.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) under accession numbers EF658766 to EF659414.
RESULTS
Fecal indicator bacteria and bacterial community composition in creek and sewage samples.
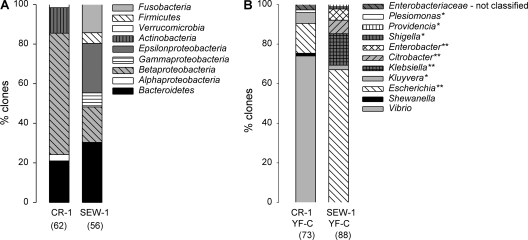
FIB concentrations were highest in the sewage influent sample and much lower in the creek samples (Table 1). The majority of the clones from CR-1 belonged to the Betaproteobacteria and, significantly, to the phyla Bacteroidetes and Actinobacteria (Fig. 1 A). Cloned sequences from SEW-1 mostly belonged to the phyla Bacteroidetes, Betaproteobacteria, Epsilonproteobacteria, and Fusobacteria, with a minority belonging to the phyla Gammaproteobacteria and Firmicutes (Fig. 1A). The TRFLP profiles across CR-1, -2, and -3 were fairly similar and differed from that of SEW-1 (Fig. 2 A), which was confirmed by hierarchical clustering and SIMPROF analysis (data not shown). Bacterial diversity estimates based on TRFLP patterns and clone library analysis showed the highest diversity in the sewage sample and lower but similar diversities in the creek samples (Table 1).
TABLE 1.
Concentrations of fecal indicator bacteria, TRFLP richness and Shannon diversity indices, and clone library rarefaction-based Chao richness estimation for sewage and dry weather creek samplesa
| Expt | Sample | Concna (MPN/100 ml) of: |
Sb | Hc | SChao1d | ||
|---|---|---|---|---|---|---|---|
| TC | EC | ENT | |||||
| Pooled | SEW-1 | 2.8 × 107 | 1.2 × 107 | 7.4 × 105 | 16 | 2.2 | 124e |
| CR-1 | 1.6 × 104 | 2.8 × 102 | 2.6 × 102 | 8 | 1.4 | 25 | |
| Individual | CR-2 | 3.4 × 103 | 1.6 × 102 | 2.2 × 102 | 10 | 1.9 | NA |
| CR-3 | 3.2 × 104 | 2.7 × 102 | 2.8 × 102 | 12 | 1.9 | NA | |
TC, total coliform; EC, E. coli; ENT, Enterococcus spp.
TRFLP richness index (number of peaks).
TRFLP Shannon diversity index.
Clone library rarefaction-based Chao richness estimation for sewage and dry weather creek samples. NA, not analyzed because no clone library data were available.
Based on rarefaction analysis, diversity was underestimated.
FIG. 1.
Composition of the clone libraries, with the total numbers of high-quality nonchimera clones analyzed indicated in parentheses. (A) Creek (CR-1) and sewage (SEW-1) samples. Phylogenetic affiliations indicated are at phylum/class level. (B) Pooled yellow/fluorescent Colilert (YF-C) wells of CR-1 and SEW-1 samples. Phylogenetic affiliations are at genus level, except for clones that could not be classified. Genera belonging to the Enterobacteriaceae are indicated with one asterisk; genera belonging to the coliform group within the Enterobacteriaceae are indicated with two asterisks.
FIG. 2.
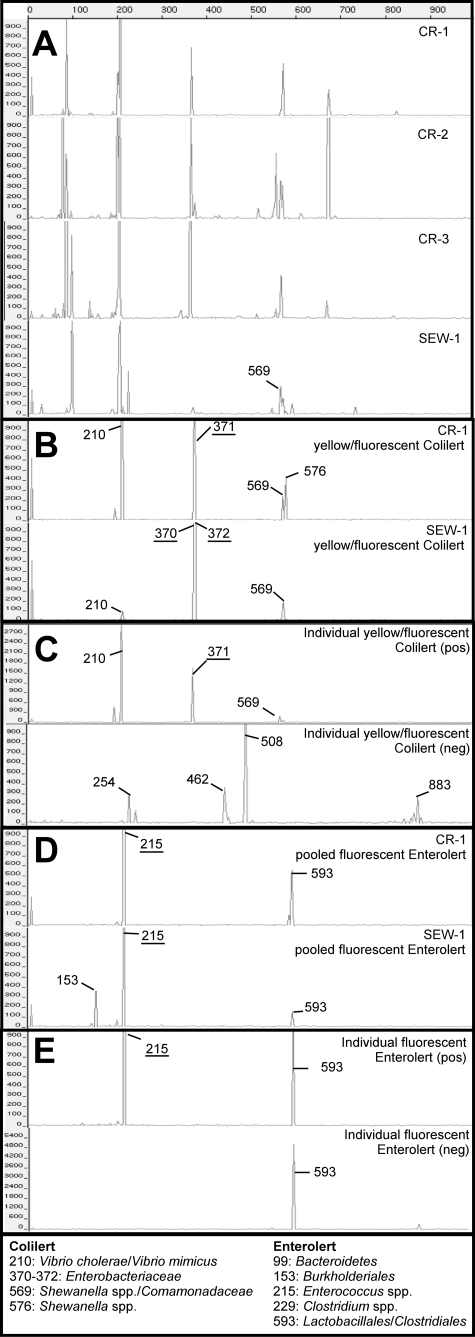
TRFLP electropherograms for creek and sewage samples (A), pooled yellow/fluorescent Colilert wells (B), representative individual yellow/fluorescent Colilert wells containing (pos) or not containing (neg) the target terminal restriction fragments (TRFs) (C), pooled fluorescent Enterolert wells (D), and representative individual fluorescent Enterolert wells containing (pos) or not containing (neg) the target TRFs (E). The dominant peaks in the Colilert and Enterolert enrichments are indicated by their fragment lengths (bp) and are also shown in panel A when they were detected. TRFs corresponding to the target organisms for Colilert and Enterolert assays are underlined. Lengths (bp) and putative phylogenetic affiliations of the dominant TRFs are listed at the bottom.
Bacterial community composition in pooled positive Quanti-Tray/2000 wells.
Multiple TRFLP peaks (≤8) were present in the pooled yellow/fluorescent Colilert and fluorescent Enterolert enrichments (Fig. 2B and D). The most abundant TRFs for each enrichment were similar for CR-1 and SEW-1. No TRFs were shared between environmental samples and IDEXX assay enrichments, except for one (569 bp in SEW-1 and pooled yellow/fluorescent Colilert wells of SEW-1). All clones from the SEW-1 pooled yellow/fluorescent Colilert wells belonged to the Enterobacteriaceae, and the majority (67%) were most closely related to E. coli (Fig. 1B). However, clones from CR-1 pooled yellow/fluorescent Colilert wells mostly belonged to the genus Vibrio (73%) and some to the genus Shewanella (1%). All clones from SEW-1 pooled fluorescent Enterolert and CR-1 pooled fluorescent Enterolert wells belonged to the target genus Enterococcus (data not shown).
The identities of the major TRFLP peaks in the electropherograms of the pooled Quanti-Tray/2000 enrichments were delineated using the MiCA web tool. The list of putative taxa was culled to include only genera belonging to taxonomic groups found in the enrichment clone libraries. The latter included a library from a wet weather sample in order to enable identification of all TRFs. These taxonomic groups were as follows: for yellow/fluorescent Colilert wells, Enterobacteriaceae, Comamonadaceae, Vibrio spp., and Shewanella spp., and for fluorescent Enterolert wells, Enterococcus spp., Bacteroidetes, Clostridium spp., and Burkholderiales. Most importantly, the analysis allowed the assignment of TRFs to the target bacterial groups Enterobacteriaceae (370 to 372 bp) and Enterococcus spp. (215 bp).
Bacterial community composition in individual positive Quanti-Tray/2000 wells.
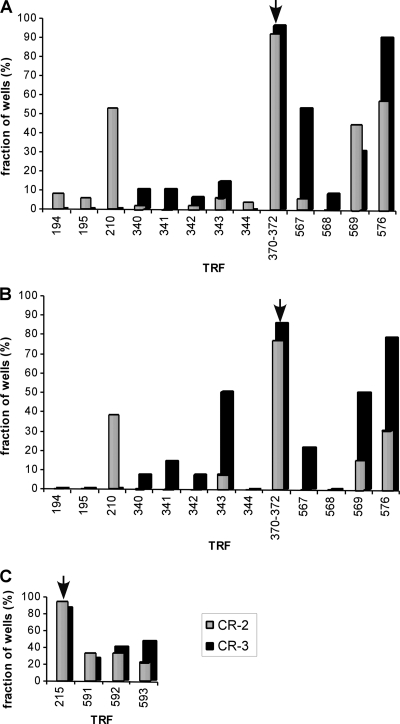
Most individual yellow Colilert wells contained the TRFs of 370 to 372 bp for Enterobacteriaceae (Fig. 3A). Only 4 and 2 wells out of 49 for CR-2 and CR-3, respectively, did not contain those TRFs. Other TRFs were typically also present, such as a 210-bp fragment corresponding to Vibrio spp., an unidentified 567-bp fragment, a 569-bp fragment corresponding to Shewanella spp. and the Comamonadaceae, and a 576-bp fragment corresponding to Shewanella spp. Most individual yellow/fluorescent Colilert wells also contained the TRFs of 370 to 372 bp for Enterobacteriaceae (Fig. 3B). Only 3 of 13 wells for CR-2 and 2 of 14 wells for CR-3 did not contain those TRFs. Individual yellow/fluorescent Colilert wells also contained the 210-, 569-, and 576-bp TRFs. In addition, the unidentified 343-bp TRF occurred frequently in CR-3 yellow/fluorescent Colilert wells. Finally, most individual fluorescent Enterolert wells contained the 215-bp TRF corresponding to Enterococcus spp. (Fig. 3C). Only 1 of 19 CR-2 wells and 3 of 16 CR-3 wells did not contain this TRF. Almost all individual fluorescent Enterolert wells that contained the 215-bp TRF also contained unidentified 591- or 592-bp TRFs and a 593-bp TRF corresponding to unidentified Lactobacillales or Clostridiales.
FIG. 3.
Distribution of terminal restriction fragments (TRFs) across positively scored individual large Quanti-Tray/2000 wells for CR-2 and CR-3 FIB enrichments. The x axis shows TRF bp length. The y axis is the percentage of positive wells containing the TRFs for 49 yellow Colilert wells each for CR-2 and CR-3 (A), 13 and 14 yellow/fluorescent Colilert wells for CR-2 and CR-3, respectively (B), and 18 and 16 fluorescent Enterolert wells for CR-2 and CR-3, respectively (C). Samples were diluted 1:10 before addition to the Quanti-Trays. TRFs expected for total coliforms, E. coli, or Enterococcus spp. are indicated with an arrow.
False-positive well rates.
TRFs of target and nontarget bacteria were usually observed simultaneously in individual positive Colilert and Enterolert wells (Fig. 3). In those cases, the individual wells were deemed true positives. However, in some of the individual wells presumed positive for total coliforms, E. coli, or Enterococcus, only TRFs associated with nontarget bacteria were detected. Based on the absence of target TRFs in positively scored individual Colilert wells, the false-positive rates for CR-2 and CR-3 water samples under dry weather conditions were 4 to 8% for total coliform, 14 to 23% for E. coli, and 6 to 20% for Enterolert (Fig. 3; Table 2).
TABLE 2.
Calculation of false-positive rates based on the number of positive wells before or after subtraction of false-positive large wells
| Phylogenetic affiliationa | Sample | FPb | No. of positive wells (L, S)c | FP rate (%) |
|---|---|---|---|---|
| TC | CR-2 | + | 49, 22 | 8 |
| − | 45, 22 | |||
| CR-3 | + | 49, 48 | 4 | |
| − | 47, 48 | |||
| EC | CR-2 | + | 13, 3 | 23 |
| − | 10, 3 | |||
| CR-3 | + | 14, 0 | 14 | |
| − | 12, 0 | |||
| ENT | CR-2 | + | 18, 3 | 6 |
| − | 17, 3 | |||
| CR-3 | + | 15, 2 | 20 | |
| − | 12, 2 |
TC, total coliform; EC, E. coli; ENT, Enterococcus spp.
FP, false positive; +, positive wells before subtraction of the number of false-positive large wells; −, positive wells after subtraction of the number of false-positive large wells.
L, large wells; S, small wells.
DISCUSSION
FIB assays are a requisite component of microbiological water quality monitoring, and thus, understanding the specificity of defined substrate technology for FIB quantification is important. Prior studies of defined substrate technology enrichment microbiology relied upon phenotypic profiling of enrichment isolates to reveal false positives (1, 9, 11, 17, 19, 23, 43, 51). In this study, culture-independent bacterial community analyses were used to reduce potential biases due to selective culture media. While TRFLP can have its own biases, e.g., originating during DNA extraction, PCR amplification, enzymatic restriction, and electrophoresis (22, 25, 41), relative changes in community structures are reliably reflected using TRFLP (25, 46). Further, dominant taxa can be detected, even if the dominant taxa are not represented by the dominant TRFs (22, 40). TRFLP biases were minimized in this study by equalizing PCR and restriction template DNA concentrations, using high DNA template concentrations (∼250 ng) before restriction, limiting the number of PCR cycles to 28, and using relative fluorescence intensities for data analysis. Finally, the absence of target TRFs in water samples—but the dominance of target TRFs in the positive wells—indicated that positive or negative biases, e.g., due to primer mismatches (49), were unlikely to be significant for the target TRFs.
A variety of nontarget taxa were detected in positive Colilert and Enterolert wells, including taxa that were not detected in previous studies using culture-based techniques. Nontarget TRFs were mainly associated with Shewanella spp., Comamonadaceae, and Vibrio spp. in positive Colilert assays and with unidentified Lactobacillales or Clostridiales in positive Enterolert assays. Vibrio spp. have been recovered from yellow and yellow/fluorescent Colilert wells associated with marine (42, 43) and freshwater (51) samples before, but this study also suggested that Vibrio spp. can be among the dominant populations in these wells. Noncoliforms such as Salmonella spp. have been isolated from positive Colilert tubes or wells from freshwater samples before, although only in a minority of the samples (17, 43). A larger diversity of noncoliform genera was isolated from tropical freshwaters (11). For Enterolert, growth of members of the class Flavobacteria (but no other Bacteroidetes bacteria) and the order Lactobacillales was previously shown (9), but growth within the orders Clostridiales and Burkholderiales (in SEW-1 pooled fluorescent Enterolert wells) is newly reported with this study. As both Enterococcus spp. and Clostridiales are considered fastidious organisms (7, 56), it is reasonable that the Enterolert medium can support the growth of both groups of bacteria. It is known that some Clostridiales exhibit significant beta-glucosidase activity (37). However, since enzyme activity can differ substantially, even within a genus or species (37), direct proof of the ability of Lactobacillales or Clostridiales to cause false-positive results in the Enterolert assay should involve isolation of bacteria.
The relative abundances of target and nontarget TRFs in water samples versus IDEXX assay enrichments suggested growth of nontarget bacteria in the IDEXX assays. The growth of coliforms and Enterococcus spp. during incubation in Quanti-Trays increased the relative abundance of the target TRFs from nondetectable (in water samples) to dominant (in individual positive Quanti-Tray wells). The absence of these TRFs in the source water samples is not surprising, given the FIB concentrations in this study and published detection limits for TRFLP (∼107 CFU per 100 ml) (46). Therefore, it is reasonable to infer that where Vibrio and Lactobacillales/Clostridiales were the dominant nontarget TRFs in IDEXX enrichments, the cause was growth.
The increased detection of nontarget organisms in this study compared to their detection in previous studies is probably rooted in the differences between culture-independent versus culture-dependent approaches. Previous culture-based studies have used either general purpose culture media (e.g., tryptic soy agar and R2A agar) or selective media (MacConkey agar, m-ENDO agar, and m-TEC agar) to isolate bacteria from yellow/fluorescent Colilert wells/tubes (10, 17, 19, 43). Selective media chemistry will bias the detection of nontarget organisms, as previously shown for the detection of Vibrio spp. in positive Colilert tubes (42). However, even general purpose media are selective and can underestimate culture-independent bacterial diversity (5, 30). Accordingly, the culture media (bile-esculine agar and MacConkey agar) and aerobic incubation conditions used previously for isolation and phenotypic profiling of bacteria growing in fluorescent Enterolert wells (1, 9, 23) were not suitable for detecting either Clostridiales or Burkholderiales. Still, it is possible that differences between this study and prior studies originate with the samples analyzed. Even within this study, some sample-to-sample variability was observed, as no Vibrio spp. were detected in the individual yellow/fluorescent Colilert wells of CR-3.
False-positive wells were identified as yellow Colilert, yellow/fluorescent Colilert, and fluorescent Enterolert wells with no target TRFs present. Consequently, at least one of the nontarget TRFs was associated with bacterial taxa causing the false-positive signals. Since DNA yields from the false-positive wells were sufficiently high for reliable PCR and TRFLP, target TRFs were dominant in other (true) positive wells, and TRFLP is generally very reproducible (25, 46), the absence of target TRFs in some wells was unlikely to be caused by TRFLP biases. The false-positive rates in this study, based on TRFLP, are very similar to those reported previously for total coliforms but slightly higher for E. coli (1, 9, 23) and higher for Enterolert (6 to 20% here versus 2.4 to 5.1% previously) (1, 9, 23, 26). The TRF of 370 to 372 bp cannot distinguish between E. coli and other coliforms and can also be associated with noncoliform Enterobacteriaceae. Therefore, the false-positive rates for E. coli and total coliforms based on TRFLP are conservative estimates.
Based on the results from this study, a more general assessment of the impact of false positives on the overestimation of FIB concentrations in environmental water samples is recommended. In this study, a limited number of samples, representative for dry weather flow at one urban creek location, were analyzed in order to support a detailed analysis using clone libraries. Additional research is needed to determine false-positive rates in IDEXX assays for a larger number and wider range of samples (e.g., storm flow). Nonetheless, the results of this study, including the identification of cloned sequences and TRFs, can be applied to detailed false-positive assessments of other locations, samples, and types of FIB assays. Lastly, an implication of our study is that future efforts aimed at reducing false positives in IDEXX assays could constructively focus on reducing the growth of nontarget bacteria, e.g., by testing a variety of selective growth-inhibiting compounds available for members of the Lactobacillales and Clostridiales and for Vibrio spp. (18, 32, 38, 50, 52).
Acknowledgments
This work was supported by Measure B funding from the Creeks Division of the City of Santa Barbara and by a Leadership Grant from the Switzer Foundation. Additional support during the preparation of the manuscript was via the UC Marine Council.
We acknowledge the contributions of Christopher Ehrhardt of the UCSB, Jill Zachary of the City of Santa Barbara, and Scott Coombs, Blair Goodridge, and Jonathan Fram, associated with the Santa Barbara Coastal LTER (project numbers NSF OCE 9982105 and OCE 0620276).
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Abbott, S., B. Caughley, and G. Scott. 1998. Evaluation of Enterolert for the enumeration of enterococci in the marine environment. N. Z. J. Mar. Freshw. Res. 32:505-513. [Google Scholar]
- 2.Adcock, P. W., and C. P. Saint. 2001. Development of glucosidase agar for the confirmation of water-borne Enterococcus. Water Res. 35:4243-4246. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ball, C. L., and R. L. Crawford. 2006. Bacterial diversity within the planktonic community of an artesian water supply. Can. J. Microbiol. 52:246-259. [DOI] [PubMed] [Google Scholar]
- 6.Berger, S. A. 1994. Increased protection afforded by the defined substrate technology Colilert system by its ability to detect Shigella beta-glucuronidase. Lett. Appl. Microbiol. 19:53-56. [Google Scholar]
- 7.Bergey, D. H., and J. G. Holt. 1994. Bergey's manual of determinative bacteriology. Williams & Wilkins, Baltimore, MD.
- 8.Bradford, A., R. D. Handy, J. W. Readman, A. Atfield, and M. Muhling. 2009. Impact of silver nanoparticle contamination on the genetic diversity of natural bacterial assemblages in estuarine sediments. Environ. Sci. Technol. 43:4530-4536. [DOI] [PubMed] [Google Scholar]
- 9.Budnick, G. E., R. T. Howard, and D. R. Mayo. 1996. Evaluation of Enterolert for enumeration of enterococci in recreational waters. Appl. Environ. Microbiol. 62:3881-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao, K. K., C. C. Chao, and W. L. Chao. 2004. Evaluation of Colilert-18 for detection of coliforms and Escherichia coli in subtropical freshwater. Appl. Environ. Microbiol. 70:1242-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao, W. L. 2006. Evaluation of Colilert-18 for the detection of coliforms and Escherichia coli in tropical fresh water. Lett. Appl. Microbiol. 42:115-120. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. M., K. Doherty, H. Gu, G. Dichter, and A. Naqui. 1996. Enterolert: a rapid method for the detection of Enterococcus spp., abstr. Q.-448, p. 464. Abstr. 96th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 13.City of Santa Barbara. 2005. Existing conditions study of the Arroyo Burro, Mission, Sycamore, and Laguna Creek watersheds. Creeks Restoration and Water Quality Improvement Division, Santa Barbara, CA.
- 14.Clarke, K. R., P. J. Somerfield, and R. N. Gorley. 2008. Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J. Exp. Mar. Biol. Ecol. 366:56-69. [Google Scholar]
- 15.Clarke, K. R., and R. M. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd ed. Primer-E, Plymouth, United Kingdom.
- 16.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton. 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 17.Covert, T. C., L. C. Shadix, E. W. Rice, J. R. Haines, and R. W. Freyberg. 1989. Evaluation of the autoanalysis Colilert test for detection and enumeration of total coliforms. Appl. Environ. Microbiol. 55:2443-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.del Castillo, C. S., I. Wahid, T. Yoshikawa, and T. Sakata. 2008. Isolation and inhibitory effect of anti-Vibrio substances from Pseudoalteromonas sp. A1-J11 isolated from the coastal sea water of Kagoshima Bay. Fish. Sci. 74:174-179. [Google Scholar]
- 19.Edberg, S. C., M. J. Allen, and D. B. Smith. 1988. National field evaluation of a defined substrate method for the simultaneous enumeration of total coliforms and Escherichia coli from drinking water: comparison with the standard multiple tube fermentation method. Appl. Environ. Microbiol. 54:1595-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edberg, S. C., and M. M. Edberg. 1988. A defined substrate technology for the enumeration of microbial indicators of environmental pollution. Yale J. Biol. Med. 61:389-399. [PMC free article] [PubMed] [Google Scholar]
- 21.European Union. 2006. Directive 2006/7/EC of the European Parliament and of the council of 16 February 2006 concerning the management of bathing water quality and repealing directive 76/160/EEC. Off. J. Eur. Union L64:37-51. [Google Scholar]
- 22.Frey, J. C., E. R. Angert, and A. N. Pell. 2006. Assessment of biases associated with profiling simple, model communities using terminal-restriction fragment length polymorphism-based analyses. J. Microbiol. Methods 67:9-19. [DOI] [PubMed] [Google Scholar]
- 23.Fricker, E. J., and C. R. Fricker. 1996. Use of defined substrate technology and a novel procedure for estimating the numbers of enterococci in water. J. Microbiol. Methods 27:207-210. [Google Scholar]
- 24.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartmann, M., and F. Widmer. 2008. Reliability for detecting composition and changes of microbial communities by T-RFLP genetic profiling. FEMS Microbiol. Ecol. 63:249-260. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez, J. F., A. M. Pourcher, J. M. Delattre, C. Oger, and J. L. Loeuillard. 1993. MPN miniaturized procedure for the enumeration of fecal enterococci in fresh and marine waters: the must procedure. Water Res. 27:597-606. [Google Scholar]
- 27.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 28.Hurley, M. A., and M. E. Roscoe. 1983. Automated statistical analysis of microbial enumeration by dilution series. J. Appl. Bacteriol. 55:159-164. [Google Scholar]
- 29.IDEXX Laboratories. 2010. Colilert brochure. IDEXX Laboratories, Carlsbad, CA.
- 30.Kawai, M., et al. 2002. 16S ribosomal DNA-based analysis of bacterial diversity in purified water used in pharmaceutical manufacturing processes by PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 68:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp, P. F., and J. Y. Aller. 2004. Estimating prokaryotic diversity: when are 16S rDNA libraries large enough? Limnol. Oceanogr. Methods 2:114-125. [Google Scholar]
- 32.Kurdi, P., K. Kawanishi, K. Mizutani, and A. Yokota. 2006. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J. Bacteriol. 188:1979-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaMontagne, M. G., and P. A. Holden. 2003. Comparison of free-living and particle-associated bacterial communities in a coastal lagoon. Microbiol. Ecol. 46:228-237. [DOI] [PubMed] [Google Scholar]
- 34.Maidak, B. L., et al. 1994. The Ribosomal Database project. Nucleic Acids Res. 22:3485-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manafi, M., W. Kneifel, and S. Bascomb. 1991. Fluorogenic and chromogenic substrates used in bacterial diagnostics. Microbiol. Rev. 55:335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto, M., M. Sakamoto, H. Hayashi, and Y. Benno. 2005. Novel phylogenetic assignment database for terminal-restriction fragment length polymorphism analysis of human colonic microbiota. J. Microbiol. Methods 61:305-319. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, J., et al. 2002. Comparison of four microbial enzymes in clostridia and Bacteroides isolated from human feces. Microbiol. Immunol. 46:487-490. [DOI] [PubMed] [Google Scholar]
- 38.Nath, J., and A. G. Datta. 1970. Studies on the mechanism of inhibition of growth of Vibrio cholerae by erythrose. J. Gen. Microbiol. 62:17-25. [DOI] [PubMed] [Google Scholar]
- 39.Nyman, J. L., et al. 2006. Heterogeneous response to biostimulation for U(VI) reduction in replicated sediment microcosms. Biodegradation 17:303-316. [DOI] [PubMed] [Google Scholar]
- 40.Orcutt, B., B. Bailey, H. Staudigel, B. M. Tebo, and K. J. Edwards. 2009. An interlaboratory comparison of 16S rRNA gene-based terminal restriction fragment length polymorphism and sequencing methods for assessing microbial diversity of seafloor basalts. Environ. Microbiol. 11:1728-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborn, A. M., E. R. B. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 42.Palmer, C. J., Y. L. Tsai, A. L. Lang, and L. R. Sangermano. 1993. Evaluation of Colilert-marine water for detection of total coliforms and Escherichia coli in the marine environment. Appl. Environ. Microbiol. 59:786-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pisciotta, J. M., D. F. Rath, P. A. Stanek, D. M. Flanery, and V. J. Harwood. 2002. Marine bacteria cause false-positive results in the Colilert-18 rapid identification test for Escherichia coli in Florida waters. Appl. Environ. Microbiol. 68:539-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rees, G. N., D. S. Baldwin, G. O. Watson, S. Perryman, and D. L. Nielsen. 2004. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie Van Leeuwenhoek 86:339-347. [DOI] [PubMed] [Google Scholar]
- 45.Rompre, A., P. Servais, J. Baudart, M.-R. de-Roubin, and P. Laurent. 2002. Detection and enumeration of coliforms in drinking water: current methods and emerging approaches. J. Microbiol. Methods 49:31. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, J. K., B. Konig, and U. Reichl. 2007. Characterization of a three bacteria mixed culture in a chemostat: evaluation and application of a quantitative terminal-restriction fragment length polymorphism (T-RFLP) analysis for absolute and species specific cell enumeration. Biotechnol. Bioeng. 96:738-756. [DOI] [PubMed] [Google Scholar]
- 47.Sercu, B., L. C. Van De Werfhorst, J. Murray, and P. A. Holden. 2009. Storm drains are sources of human fecal pollution during dry weather in three urban Southern California watersheds. Environ. Sci. Technol. 43:293-298. [DOI] [PubMed] [Google Scholar]
- 48.Shyu, C., T. Soule, S. J. Bent, J. A. Foster, and L. J. Forney. 2007. MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. Microb. Ecol. 53:562-570. [DOI] [PubMed] [Google Scholar]
- 49.Sipos, R., et al. 2007. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targetting bacterial community analysis. FEMS Microbiol. Ecol. 60:341-350. [DOI] [PubMed] [Google Scholar]
- 50.Sklenickova, O. J., et al. 2010. Selective growth inhibitory effect of biochanin A against intestinal tract colonizing bacteria. Molecules 15:1270-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, M. M. 1999. Total coliform methods are not equal: sensitivity differences to specific genera of bacteria, abstr. 50358. In Proceedings of the 1999 Water Quality Technology Conference. American Water Works Association, Denver, CO.
- 52.Sundaram, S., and K. V. Murthy. 1983. Occurrence of 2,4-diamino-6,7-diisopropyl-pteridine (0/129) resistance in human isolates of Vibrio cholerae. FEMS Microbiol. Lett. 19:115-117. [Google Scholar]
- 53.U.S. Environmental Protection Agency. 2003. Bacterial water quality standards for recreational waters (fresh and marine waters). EPA/823.R-03/008. Office of Water, U.S. Environmental Protection Agency, Washington, DC.
- 54.U.S. Environmental Protection Agency. 2003. Guidelines establishing test procedures for the analysis of pollutants: analytical methods for biological pollutants in ambient water, final rule. Fed. Regist. 68:43272-43283. [Google Scholar]
- 55.Wagner, M., R. Amann, H. Lemmer, and K. H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for Proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiegel, J., R. Tanner, and F. A. Rainey. 2006. An introduction to the family Clostridiaceae, p. 654-678. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 4. Springer, New York, NY. [Google Scholar]
- 57.World Health Organization. 2003. Guidelines for safe recreational water environments, vol. 1. Coastal and fresh waters. World Health Organization, Geneva, Switzerland.