Abstract
A combinatorial biosynthetic approach was used to interrogate the donor substrate flexibility of GilGT, the glycosyltransferase involved in C-glycosylation during gilvocarcin biosynthesis. Complementation of gilvocarcin mutant Streptomyces lividans TK24 (cosG9B3-U−), in which the biosynthesis of the natural sugar donor substrate was compromised, with various deoxysugar plasmids led to the generation of six gilvocarcin analogues with altered saccharide moieties. Characterization of the isolated gilvocarcin derivatives revealed five new compounds, including 4-β-C-d-olivosyl-gilvocarcin V (d-olivosyl GV), 4-β-C-d-olivosyl-gilvocarcin M (d-olivosyl GM), 4-β-C-d-olivosyl-gilvocarcin E (d-olivosyl GE), 4-α-C-l-rhamnosyl-gilvocarcin M (polycarcin M), 4-α-C-l-rhamnosyl-gilvocarcin E (polycarcin E), and the recently characterized 4-α-C-l-rhamnosyl-gilvocarcin V (polycarcin V). Preliminary anticancer assays showed that d-olivosyl-gilvocarcin and polycarcin V exhibit antitumor activities comparable to that of their parent drug congener, gilvocarcin V, against human lung cancer (H460), murine lung cancer (LL/2), and breast cancer (MCF-7) cell lines. Our findings demonstrate GilGT to be a moderately flexible C-glycosyltransferase able to transfer both d- and l-hexopyranose moieties to the unique angucyclinone-derived benzo[d]naphtho[1,2b]pyran-6-one backbone of the gilvocarcins.
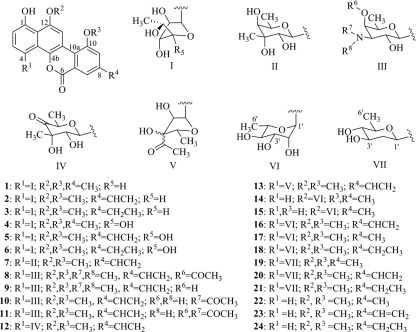
Gilvocarcin V (GV) (structure 2; see Fig. 1 for numbered structures), the principal product of Streptomyces griseoflavus Gö 3592 and other Streptomyces spp., is the most prominent member of a distinct class of antitumor antibiotics that share a polyketide-derived coumarin-based benzo[d]naphtho[1,2-b] pyran-6-one moiety. This small family of antitumor drugs is often referred to as gilvocarcin-type anticancer drugs (5, 10, 26, 41). Most of these natural products possess C-glycosidically linked 6-deoxy-d-hexose moieties in the 4-position, while variants within this group exist as either furanose (structures 1 to 6, structure 13) or pyranose sugars (structures 7 to 12 and 14 to 16) (5, 27, 38, 43, 45). Notably, BE-12406A (structure 14) and B (structure 15) (15, 25), as well as polycarcin V (structure 16) (16), are the only examples of 6-deoxy-l-sugars found in gilvocarcin-type compounds, with structures 14 and 15 representing the only O-glycosidically linked analogues reported so far.
FIG. 1.
Glycodiversity of gilvocarcin-type anticancer drugs. Gilvocarcins (structures 1 to 3), 4′-OH gilvocarcins (structures 4 to 6), chrysomycin V (structure 7), ravidomycin V (structure 8), deacetylravidomycin V (structure 9), FE35A (structure 10) and B (structure 11), Mer1020 dC (structure 12) and dD (structure 13), BE-12406A (structure 14) and B (structure 15), polycarcins (structures 16 to 18), d-olivosyl-gilvocarcins (structures 19 to 21), and defucogilvocarcins M, V, and E (structures 22 to 24).
GV's most likely mechanism of action is a photoactivated [2 + 2] cycloaddition of its vinyl side chain with thymine residues of DNA caused by near-UV or visible blue light which results in single-strand scissions leading to covalent binding with DNA (1, 3, 9, 14, 24). In addition, GV's activity is also attributed to a unique selective cross-linking of DNA and histone H3, a core component of the histone complex that plays an important role for DNA replication and transcription (2, 8, 22, 23, 30). The saccharide moiety, d-fucofuranose, of GV is essential for this activity, as it is believed to facilitate binding of histone H3 (17, 18).
Recently, we identified and characterized largely the deoxysugar biosynthetic pathway that leads to d-fucofuranose (structure 26), the sugar moiety of GV (18). It was possible to shut down the biosynthesis of gilvocarcin's natural sugar donor through the inactivation of gilU, the 4-ketoreductase involved in the biosynthesis of GV's deoxysugar moiety. The inactivation of gilU afforded a more active gilvocarcin analogue, 4′-hydroxy gilvocarcin V (structure 5, 4′-OH GV), illustrating an inherent substrate flexibility of GilGT, the glycosyltranferase responsible for C-glycosylation in GV biosynthesis (17, 18). Additionally, the improved bioactivity of 4′-OH GV over that of GV clearly demonstrated the importance of further investigations into modifying the glycosylation pattern of gilvocarcins.
In our initial attempt to probe the donor substrate flexibility of GilGT and advance the glycodiversity of gilvocarcin-type anticancer drugs, we complemented the aforementioned GilU mutant, Streptomyces lividans TK24 (cosG9B3-U−), with various deoxysugar plasmids directing the biosynthesis of neutral deoxysugars, namely, l-olivose (pLN2) (32), l-rhamnose (pRHAM) (33), l-mycarose (pFL942) (19), l-digitoxose (pLNBIV) (32), 4-keto-l-olivose (pKOLV), and d-oliose (pOLO). These plasmids are designed for producing specific activated deoxysugars through the combination of genes from one or more deoxysugar biosynthetic pathways, and they have been utilized in successfully altering the saccharide moieties of mithramycin, steffimycin, elloramycin, and rebeccamycin/staurosporin (4, 28, 31, 35, 36). We were particularly interested in the attachment of l-hexose sugars, as these are rarely found in gilvocarcin-type anticancer drugs. Of the six deoxysugar plasmids tested, complementation with pLN2 and pRHAM produced new gilvocarcin analogues with d-olivosyl and l-rhamnosyl moieties, respectively.
MATERIALS AND METHODS
Microorganisms and culture conditions.
All complementation experiments were carried out in the heterologous host S. lividans TK24 (13). The mutant cosmid cosG9B3-U− was introduced into S. lividans TK24 through conjugal transfer according to standard protocols (13), producing S. lividans TK24 (cosG9B3-U−). Conjugation was carried out on MS agar (13) supplemented with 10 mM MgCl2 and overlaid with nalidixic acid with appropriate antibiotics after 18 h. Exconjugates were grown on solid M2 medium (4 g/liter glucose, 10 g/liter malt extract, 4 g/liter yeast extract, 1g/liter CaCO3, 15 g/liter agar) supplemented with appropriate antibiotics. S. lividans TK24 (cosG9B3-U−) was transformed via protoplast transformation with deoxysugar plasmids (pLN2, pRHAM, pLNBIV, pFL942, pKOLO, and pOLO) according to standard protocols (13). Protoplasts were regenerated on R2YE agar medium and overlaid after 18 h with R3 soft agar (171 g/liter sucrose, 10 g/liter glucose, 4 g/liter peptone, 0.5 g/liter K2SO4, 8.1 g/liter MgCl2·6H2O, 2.2 g/liter CaCl2, 8.8 g/liter agar) supplemented with appropriate antibiotics (13). Regenerated protoplasts were transferred to solid M2 agar supplemented with appropriate antibiotics. Initially, each strain was grown in 250-ml baffled Erlenmeyer flasks containing 100 ml of liquid SG medium (20 g/liter glucose, 10 g/liter soy peptone, 2 g/liter CaCO3, 0.001 g/liter cobalt-II chloride, pH 7.2) with appropriate antibiotics and screened for gilvocarcin analogue production using high-performance liquid chromatography-mass spectrometry (HPLC-MS) as described previously (11). Escherichia coli XL1-Blue (Stratagene) was used as the subcloning host and was grown at 37°C in lysogeny broth medium, while E. coli ET12567/pUZ8002 (21, 29) was used for conjugation. When antibiotics were required for strain selection, 50 μg/ml of kanamycin, 100 μg/ml of ampicillin, 50 μg/ml of apramycin, and 25 μg/ml of thiostrepton were used.
DNA manipulation and PCR amplification.
Plasmid DNA isolations were carried out using the GeneJet plasmid miniprep kit (Fermentas). All restriction endonuclease digestions, alkaline phosphatase treatments, ligations, and other DNA manipulations were performed according to standard protocols (37). PCR was used to amplify mtmU from Streptomyces argillaceus ATCC 12596 (mithramycin producer) using the specific oligonucleotide primer mtmU_F (5′-ACTAGTAGAAGGAGCGCCGTGCCCG) (SpeI site is underlined). PCR conditions were as follows: 50 ng of template DNA was mixed with 50 ng of each primer with 1.25 units of Native PFU DNA polymerase (Stratagene) in a total volume of 50 μl containing 0.4 mM each deoxynucleoside triphosphate (dNTP), 2 mM MgCl2, and 5 μl dimethyl sulfoxide (DMSO). The normal PCR thermocycler conditions were 60 s at 98°C followed by 30 cycles of 60 s at 98°C, 45 s at 65°C, and 90 s at 76°C and then 10 min at 76°C. The PCR product was gel purified using a QIAquick gel extraction kit (Qiagen), subcloned into PCR-Blunt II-TOPO (Invitrogen), and sequenced (SeqWright, Houston, TX).
Plasmid constructs.
Deoxysugar plasmid pKOLV was constructed by the deletion of oleU, as a SpeI-NheI fragment, from pLN2 and then self-ligated (Table 1). To produce pOLO, oleU from pLN2 was replaced with mtmU as a SpeI-NheI fragment.
TABLE 1.
Plasmid constructs generated in this work
| Plasmid | Genes | Description |
|---|---|---|
| pKOLO | oleVoleWoleYoleLoleSoleE | pLN2 was digested with SpeI and NheI and then religated |
| pOLO | oleVoleWmtmUoleYoleLoleSoleE | pLN2 was digested with SpeI and NheI and replaced with mtmU with the same flanking restriction sites |
Production, isolation, and purification of gilvocarcin analogues.
Strains producing novel gilvocarcin analogues were grown as seed cultures in 250-ml baffled Erlenmeyer flasks containing 100 ml liquid SG medium, as described above, and allowed to grow at 28°C for 48 h at 250 rpm. The seed cultures of S. lividans TK24 (cosG9B3-U−/pLN2) and S. lividans TK24 (cosG9B3-U−/pRHAM) were used to inoculate 60 liters and 80 liters of liquid SG medium, respectively. After 5 days of fermentation at 28°C with reciprocal shaking (250 rpm), the flasks were removed and combined with celite (100 g/liter) and filtered. The mycelial cake was extracted with acetone by sonication and again filtered. The acetone was removed under vacuum, and the aqueous portion was combined with the culture broth filtrate and passed through C18 reverse-phase silica. The column was eluted with 1-liter fractions of 0, 25, 50, 75, and 100% methanol in water. Fractions containing gilvocarcin analogues were dried and lyophilized. Further purification was obtained through high-performance liquid chromatography (HPLC) according to previously described procedures (11).
NMR analysis.
The 1H and 13C nuclear magnetic resonance (NMR) data (Tables 2 and 3; see Table S7 in the supplemental material) were recorded on a Varian Vnmr 500 spectrometer, operating on 500 MHz (1H) and 125 MHz (13C), respectively, using the indicated deuterated solvents.
TABLE 2.
1H (500 MHz) and 13C (125 MHz) NMR data for d-olivosyl-gilvocarcin M (structure 19) DMSO-d6
| Position | δH(J in Hz) | δC, mult.d |
|---|---|---|
| 1 | 152.9, C | |
| 1-OH | 9.69, s | |
| 2 | 6.90, d (8.5) | 111.9, CH |
| 3 | 7.71, d (8.0) | 129.4, CH |
| 4 | 126.6, C | |
| 4a | 122.3, C | |
| 4b | 141.0, C | |
| 6 | 159.3, C | |
| 6a | 121.1, C | |
| 7 | 7.58, s | 120.4, CH |
| 8 | 140.2, C | |
| 8-CH3 | 2.39, s | 21.1, CH3 |
| 9 | 7.20, s | 118.5, CH |
| 10 | 156.6, C | |
| 10-OCH3 | 3.96, s | 56.0, CH3 |
| 10a | 121.0, C | |
| 10b | 113.4, C | |
| 11 | 8.22, s | 101.6, CH |
| 12 | 151.6, C | |
| 12-OCH3 | 4.02, s | 56.2, CH3 |
| 12a | 114.6, C | |
| 1′ | 5.63, d (10.5) | 75.0, CH |
| 2′-Ha | 1.30, dd (11.5, 11.5)a | 43.2, CH2 |
| 2′-He | 2.36, mc | |
| 2′-OH | ||
| 3′ | 3.80, m | 72.0, CH |
| 3′-OH | 4.73, d (4.5)b | |
| 4′ | 2.91, dd (9.0, 8.5)a | 77.8, CH |
| 4′-OH | 4.96, d (5.0)b | |
| 5′ | 3.49, m | 76.0, CH |
| 6′-H3 | 1.27, d (6.0) | 18.7, CH3 |
| 1″ | ||
| 2″-He | ||
| 2″-Hz |
Coupling constants calculated after D2O exchange.
Signal lost after D2O exchange.
Signal partially obscured.
mult., multiplicity.
TABLE 3.
1H (500 MHz) and 13C (125 MHz) NMR data for d-olivosyl-gilvocarcin V (structure 20) DMSO-d6
| Position | δH(J in Hz) | δC, mult.c |
|---|---|---|
| 1 | 153.0, C | |
| 1-OH | 9.75, s | |
| 2 | 6.96, d (8.5) | 112.4, CH |
| 3 | 7.76, d (8.5) | 129.6, CH |
| 4 | 126.9, C | |
| 4a | 122.8, C | |
| 4b | 141.7, C | |
| 6 | 159.4, C | |
| 6a | 122.5, C | |
| 7 | 7.97, s | 119.2, CH |
| 8 | 138.9, C | |
| 8-CH3 | ||
| 9 | 7.71, s | 114.6, CH |
| 10 | 157.4, C | |
| 10-OCH3 | 4.15, s | 56.3, CH3 |
| 10a | 122.2, C | |
| 10b | 113.4, C | |
| 11 | 8.45, s | 101.8, CH |
| 12 | 151.9, C | |
| 12-OCH3 | 4.10, s | 56.7, CH3 |
| 12a | 115.0, C | |
| 1′ | 5.73, d (11.0) | 75.0, CH |
| 2′-Ha | 1.30, dd (11.5, 11.5)a | 43.1, CH2 |
| 2′-He | 2.33, m | |
| 2′-OH | ||
| 3′ | 3.81, m | 72.0, CH |
| 3′-OH | 4.73, d (5.0)b | |
| 4′ | 2.89, dd (9.0, 8.5)a | 77.8, CH |
| 4′-OH | 4.94, d (5.0)b | |
| 5′ | 3.47, m | 76.0, CH |
| 6′-H3 | 1.26, d (6.0) | 18.6, CH3 |
| 1″ | 6.93, dd (17.5, 11.0) | 135.2, CH |
| 2″-He | 6.14, d (18.0) | 122.0, CH2 |
| 2″-Hz | 5.50, d (11.0) |
Coupling constants calculated after D2O exchange.
Signal lost after D2O exchange.
mult., multiplicity.
RESULTS
In vivo coexpression of S. lividans TK24 (cosG9B3-U−) and pLN2.
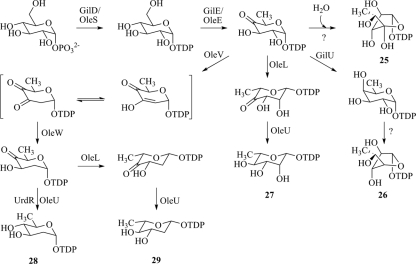
The well-established deoxysugar plasmid pLN2 is responsible for producing the TDP-activated 2,6-dideoxy-hexopyranose, TDP-l-olivose (structure 29) (32). The biosynthesis of TDP-l-olivose involves two unique 2,6-dideoxy-4-ketosugar intermediates, one in d- and the other in l-configuration, which can further be reduced to TDP-d- or TDP-l-olivose. In addition, the ketosugars themselves may also serve as acceptable donor substrates for GilGT (Fig. 2), as was shown previously for the 4-keto-d-fucofuranose (18).
FIG. 2.
Biosynthesis of TDP-4′-OH-d-fucofuranose (structure 25), TDP-d-fucofuranose (structure 26), TDP-l-rhamnose (structure 27), TDP-d-olivose (structure 28), and TDP-l-olivose (structure 29).
Fermentation of the recombinant mutant strain S. lividans TK24 (cosG9B3-U−/pLN2) accumulated three new peaks, structures 19, 20, and 21 (Fig. 3b). Large-scale fermentation and preparative HPLC were used to isolate the new compounds, which were structurally characterized through NMR spectroscopy and mass spectrometry. A 60-liter fermentation of S. lividans TK24 (cosG9B3-U−/pLN2) afforded 4 mg of the major peak, d-olivosyl-gilvocarcin M (structure 19; ∼0.07 mg/liter). MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) high-resolution mass spectrometry of structure 19 gave m/z = 466.1590 ([M+H]+ C26H25O8, requires 466.1549). The 1H and 13C NMR of structure 19 clearly showed the presence of the gilvocarcin chromophore; however, new sugar signals were observed (Tables 2 and 3). We expected the sugar to be l-olivose; however, two-dimensional (2D) NMR (correlation spectroscopy [COSY], nuclear Overhauser enhancement spectroscopy [NOESY], heteronuclear multiple-bond correlation [HMBC], and heteronuclear single quantum correlation [HSQC]) experiments confirmed the sugar residue to be d-olivose (Fig. 4A). From HMBC correlations, we found clear evidence of C-glycosidic attachment at the C-4 position (2JC-H coupling between 1′-H and C-4), while the typical 4C1-conformation of d-sugars was confirmed through NOESY correlations between 1′-H, 3′-H, and 5′-H and between 2′-Ha and 4′-H (Fig. 4B). Assuming GilGT belongs to the GT-1 family of inverting glycosyltransferases (6, 7, 42), the use of TDP-d-olivose as a donor substrate should result in a glycosidic bond with β-d-configuration, as evident from the large coupling constant of C-1′ (δ 5.63, J = 10.5 Hz).
FIG. 3.
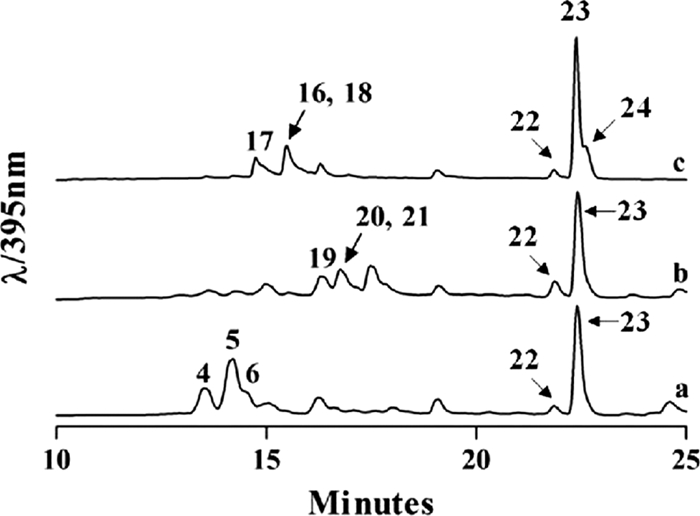
HPLC trace of S. lividans TK24 (cosG9B3-U−) (a), S. lividans TK24 (cosG9B3-U−/pLN2) (b), and S. lividans TK24 (cosG9B3-U−/pRHAM) (c).
FIG. 4.
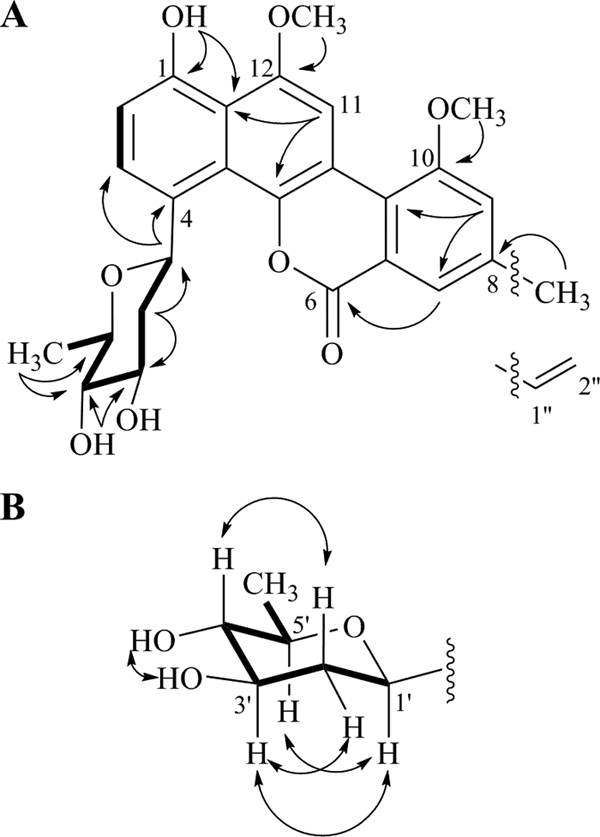
2D NMR results for d-olivosyl-gilvocarcin M highlighting COSY (thick line) and selected HMBC (→) (A) as well as NOESY (↔) (B) correlations.
The remaining minor compounds were identified as d-olivosyl-gilvocarcin V (structure 20; ∼0.05 mg/liter) and d-olivosyl-gilvocarcin E (structure 21; ∼0.01 mg/liter). As before, structural elucidation of structure 20 was carried out through NMR (Table 3) and mass spectrometry (high-resolution mass spectrometry [HRMS] [MALDI-TOF+] m/z = 478.1490; [M+H]+ C27H25O8, requires 478.1549). Due to the poor production yield of structure 21, we were unable to confirm its structure through NMR and instead relied only on mass spectrometry data (atmospheric pressure chemical ionization [APCI] m/z = 480; [M+H]+ C27H27O8, requires 480.1706) to propose the presence of d-olivosyl-gilvocarcin E. This postulation is indirectly supported by the fact that gilvocarcins (structures 1 to 3) are naturally produced as three congeners differing only in their side chains attached at C-8 (39). This is also illustrated in the host strains' ability to produce compounds 4 to 6, as seen in Fig. 3a.
In vivo coexpression of S. lividans TK24 (cosG9B3-U−) and pRHAM.
The deoxysugar plasmid pRHAM is responsible for producing the TDP-activated 6-deoxy-hexopyranose, TDP-l-rhamnose (structure 27) (33). In comparison with pLN2, pRHAM lacks the genes responsible for 2-deoxygenation (oleV and oleW) and therefore leads to only a single l-4-ketosugar intermediate. Since all gilvocarcin-type compounds discovered thus far contain 2-hydroxy groups in their sugar moieties, pRHAM encoding a TDP-6-deoxyhexopyranose was considered a more conservative attempt to modify the gilvocarcin sugar moiety. It was only after preliminary structural characterization of structure 17 that polycarcin V (structure 16) was reported as a natural secondary metabolite of Streptomyces polyformus (16).
Fermentation of the recombinant mutant strain S. lividans TK24 (cosG9B3-U−/pRHAM) accumulated three new peaks, structures 16, 17, and 18 (Fig. 3c). As before, large-scale fermentation and preparative HPLC were used to isolate all new compounds which were then characterized through NMR and mass spectrometry. Due to poor yields, an 80-liter fermentation of S. lividans TK24 (cosG9B3-U−/pRHAM) was needed to isolate 4 mg of the major peak, l-rhamnosyl-gilvocarcin M (= polycarcin M; structure 17; ∼0.05 mg/liter). To simplify future discussions on the growing collection of gilvocarcin analogues, structures 17 and 18 will be referred to as polycarcin M and polycarcin E, as they are congeners of the recently reported polycarcin V (16). HRMS (MALDI-TOF+) of structure 17 gave m/z = 482.1540 ([M+H]+ C27H27O8, requires 482.1499), which was in agreement with the attachment of l-rhamnose. The structure was confirmed by 1H and 13C NMR (see Fig. S5 and S6 and Table S7 in the supplemental material), as well as various 2D NMR experiments (COSY, NOESY, HMBC, and HSQC). The sugar moiety, l-rhamnose, was further confirmed through comparisons of reported NMR data from polycarcin V (16).
The remaining minor compounds were identified as polycarcin V (l-rhamnosyl-gilvocarcin; structure 16; ∼0.03 mg/liter) and polycarcin E (l-rhamnosyl-gilvocarcin E; structure 18; ∼0.01 mg/liter). The 1H NMR, 13C NMR, and HRMS (MALDI-TOF+) of structure 16 matched those previously reported for polycarcin V (16). Due to the limited quantity of structure 18 isolated (<1 mg) from large fermentations, we relied on APCI mass spectrometry (m/z = 496; [M+H]+ C27H27O8, requires 496.1655) to propose its structure.
The host strain S. lividans TK24 (cosG9B3-U−) as well as the two recombinant strains S. lividans TK24 (cosG9B3-U−/pLN2) and S. lividans TK24 (cosG9B3-U−/pRHAM) also produced significant amounts of the previously described (17) sugar-free defucogilvocarcins, defucogilvocarcin M (structure 22), defucogilvocarcin V (structure 23), and defucogilvocarcin E (structure 24) (Fig. 3).
In vivo coexpression of S. lividans TK24 (cosG9B3-U−) and pLNBIV, pFL942, pKOLV, and pOLO.
The deoxysugar plasmids pLNBIV and pFL942 have been shown to direct biosynthesis toward l-digitoxose and l-mycarose, respectively (19, 32). When these plasmids were used to complement S. lividans TK24 (cosG9B3-U−), no new metabolites were observed. This indicates that GilGT is unable to transfer either l-digitoxose or l-mycarose to the gilvocarcin backbone. Similarly, the newly constructed plasmids pKOLV (4-keto-l-olivose) and pOLO (d-oliose) did not yield new products. We anticipated that the removal of oleU (4-ketoreductase) from pLN2 would result in 4-keto-l-olivose while the replacement of oleU with mtmU (4-ketoreductase that forms the axial 4-OH in d-oliose biosynthesis) would result in d-oliose. The products of plasmids pKOLV and pOLO have not yet been verified; therefore, we cannot claim with certainty that the expected sugars were produced. However, based on their derivation from pLN2, a well-established deoxysugar plasmid, we hypothesize that GilGT cannot utilize 4-keto-l-olivose or d-oliose.
Bioactivity study.
Preliminary anticancer assays against human lung (H460), human breast (MCF-7), and murine lung (LL/2) cancer cell lines using the sulforhodamine B assay (34, 40) indicated that structure 16 has anticancer activity that is comparable to those of structures 2 and 5; however, structure 20 showed slightly better anticancer activity toward cancer cell lines H460 and MCF-7 than structures 2 and 5 (Table 4). Additionally, dose-dependent curves showed that structures 16 and 20 have 50% growth-inhibitory (GI50) values comparable to (H460) or higher than (MCF-7 and LL/2) that of parent congener structure 2. It should be noted that in all cell lines structure 5 received the lowest GI50 values (data not shown).
TABLE 4.
Anticancer activity assaysa of gilvocarcins
| Gilvocarcin | Anticancer activity (% T/C at 100 μM)b |
||
|---|---|---|---|
| H460 | MCF-7 | LL/2 | |
| Gilvocarcin V (structure 2) | 5.5 ± 0.3 | 4.5 ± 1.0 | 1.4 ± 0.4 |
| 4′-OH-gilvocarcin V (structure 5) | 2.6 ± 0.5 | 7.0 ± 2.5 | 1.4 ± 0.5 |
| Polycarcin V (structure 16) | 2.3 ± 1.0 | 8.9 ± 0.4 | 4.8 ± 5.3 |
| d-Olivosyl-gilvocarcin V (structure 20) | 1.2 ± 0.2 | 2.5 ± 0.4 | 3.4 ± 2.1 |
DISCUSSION
The bioactivity of glycosylated natural products produced by actinomycetes, in many cases, can be attributed to their saccharide moiety. Recently, glycodiversification studies have garnered much attention, with the overall goal of generating more bioactive compounds or compounds with improved pharmacological properties (20, 42, 44). In this study, we chose to probe the donor substrate flexibility of GilGT, the glycosyltransferase involved in gilvocarcin biosynthesis. The recent generation of S. lividans TK24 (cosG9B3-U−) provided a suitable model host for probing GilGT flexibility, as cosG9B3-U− lacks the genes required for the production of the natural d-fucofuranose moiety of the natural gilvocarcins. By expressing deoxysugar plasmids in S. lividans TK24 (cosG9B3-U−), we were able to supply GilGT with alternative donor substrates for glycosylation.
The complementation of S. lividans TK24 (cosG9B3-U−) with pLN2 and pRHAM resulted in the production of five novel compounds, including d-olivosyl-gilvocarcins M, V, and E as well as polycarcins M and E. The recently reported polycarcin V was isolated as a natural metabolite from S. polyformus; however, its side chain derivatives were not reported (16). Here we report the engineered production of polycarcin V as well as its methyl and ethyl side chain congeners.
The engineered d-olivosyl-gilvocarcins discussed here are the first examples of gilvocarcin-type anticancer drugs with a neutral multiply deoxygenated sugar moiety. The finding that d- and not l-olivose was the attached sugar moiety of these compounds was unexpected and surprising, since pLN2 encodes the biosynthetic pathway to TDP-l-olivose. We assume either that the 4-ketoreductase OleU, encoded by pLN2, can act on the intermediate TDP-4-keto-6-deoxy-d-olivose or that the latter was reduced by a ketoreductase naturally present in S. lividans before it could be converted to 4-keto-6-deoxy-l-olivose by the 3,5 epimerase OleL. Originally, pLN2 was tested with the flexible glycosyltransferase ElmGT from the elloramycin biosynthetic pathway, which resulted in the exclusive production of l-olivosyl-tetracenomycin C (32). In the presence of ElmGT, a GT which naturally transfers l-rhamnose, the biosynthetic pathway favored the production of NDP-l-olivose. However, in the gilvocarcin pathway it is clear that l-olivose cannot be utilized by GilGT, which naturally transfers a d-sugar, and instead it scavenges the limited amount of d-olivose produced by pLN2, generating a small flux toward d-olivose and, therefore, isolable amounts of d-olivosyl-gilvocarcins.
The results presented here provide further examples of successful glycodiversification efforts by utilizing deoxysugar plasmids and relying on the (here limited) flexibility of a glycosyltransferase. We have established GilGT as being a moderately flexible glycosyltransferase able to accept its yet-undetermined natural donor substrate d-fucofuranose or d-fucopyranose as well as d-olivose and l-rhamnose. Additionally, this work demonstrates how substrate preference can shift the biosynthetic flux toward an unexpected outcome, further highlighting the intricacies of engineered biosynthetic approaches to produce new natural product analogues. The fact that several other deoxysugar plasmids failed to trigger the production of new gilvocarcins, however, shows that this method of glycodiversification has its limits when being applied to the gilvocarcin biosynthetic pathway. Besides the limited flexibility of GilGT, the apparently narrow substrate specificity of the terminal enzyme of the gilvocarcin pathway, oxidoreductase GilR (12), may cause problems, and we are currently further investigating this gatekeeper enzyme with the long-term goal to reengineer it toward broader substrate specificity.
Supplementary Material
Acknowledgments
This work was supported by a grant (CA 102102) from the U.S. National Institutes of Health to J.R.
The University of Kentucky core facilities for NMR and mass spectrometry are acknowledged for the use of their instruments. We thank Val R. Adams and C. Mattingly for the cytotoxicity testing.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alegria, A. E., L. Zayas, and N. Guevara. 1995. A comparative study of the visible light photochemistry of gilvocarcins V and M. Photochem. Photobiol. 62:409-415. [DOI] [PubMed] [Google Scholar]
- 2.Antczak, A., T. Tsubota, P. Kaufman, and J. Berger. 2006. Structure of the yeast histone H3-ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct. Biol. 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arce, R., R. Oyola, and A. E. Alegria. 1998. The photobiological differences of gilvocarcins V and M are not related to their transient intermediates and triplet yields. Photochem. Photobiol. 68:25-31. [PubMed] [Google Scholar]
- 4.Baig, I., M. Pérez, A. F. Braña, R. Gomathinayagam, C. Damodaran, J. A. Salas, C. Méndez, and J. Rohr. 2008. Mithramycin analogues generated by combinatorial biosynthesis show improved bioactivity. J. Nat. Prod. 71:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balitz, D. M., F. A. O'Herron, J. Bush, D. M. Vyas, D. E. Nettleton, R. E. Grulich, W. T. Bradner, T. W. Doyle, E. Arnold, and J. Clardy. 1981. Antitumor agents from Streptomyces anandii: gilvocarcins V, M and E. J. Antibiot. 34:1544-1555. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326:929-939. (Erratum, 329:719.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 8.Eickbush, T. H., and E. N. Moudrianakis. 1978. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry 17:4955-4964. [DOI] [PubMed] [Google Scholar]
- 9.Elespuru, R. K., and S. K. Gonda. 1984. Activation of antitumor agent gilvocarcins by visible light. Science 223:69-71. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, C., F. Lipata, and J. Rohr. 2003. The complete gene cluster of the antitumor agent gilvocarcin V and its implication for the biosynthesis of the gilvocarcins. J. Am. Chem. Soc. 125:7818-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kharel, M. K., S. E. Nybo, M. D. Shepherd, and J. Rohr. 2010. Cloning and characterization of the ravidomycin and chrysomycin biosynthetic gene clusters. Chembiochem 11:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharel, M. K., P. Pahari, H. Lian, and J. Rohr. 2009. GilR, an unusual lactone-forming enzyme involved in gilvocarcin biosynthesis. Chembiochem 10:1305-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 14.Knobler, R. M., F. B. Radlwimmer, and M. J. Lane. 1992. Gilvocarcin V exhibits both equilibrium DNA binding and UV light induced DNA adduct formation which is sequence context dependent. Nucleic Acids Res. 20:4553-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojiri, K., H. Arakawa, F. Satoh, K. Kawamura, A. Okura, H. Suda, and M. Okanishi. 1991. New antitumor substances, BE-12406A and BE-12406B, produced by a streptomycete. I. Taxonomy, fermentation, isolation, physico-chemical and biological properties. J. Antibiot. (Tokyo) 44:1054-1060. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y. Q., X. S. Huang, K. Ishida, A. Maier, G. Kelter, Y. Jiang, G. Peschel, K. D. Menzel, M. G. Li, M. L. Wen, L. H. Xu, S. Grabley, H. H. Fiebig, C. L. Jiang, C. Hertweck, and I. Sattler. 2008. Plasticity in gilvocarcin-type C-glycoside pathways: discovery and antitumoral evaluation of polycarcin V from Streptomyces polyformus. Org. Biomol. Chem. 6:3601-3605. [DOI] [PubMed] [Google Scholar]
- 17.Liu, T., M. K. Kharel, C. Fischer, A. McCormick, and J. Rohr. 2006. Inactivation of gilGT, encoding a C-glycosyltransferase, and gilOIII, encoding a P450 enzyme, allows the details of the late biosynthetic pathway to gilvocarcin V to be delineated. Chembiochem 7:1070-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, T., M. K. Kharel, L. Zhu, S. A. Bright, C. Mattingly, V. R. Adams, and J. Rohr. 2009. Inactivation of the ketoreductase gilU gene of the gilvocarcin biosynthetic gene cluster yields new analogues with partly improved biological activity. Chembiochem 10:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombó, F., M. Gibson, L. Greenwell, A. F. Braña, J. Rohr, J. A. Salas, and C. Méndez. 2004. Engineering biosynthetic pathways for deoxysugars: branched-chain sugar pathways and derivatives from the antitumor tetracenomycin. Chem. Biol. 11:1709-1718. [DOI] [PubMed] [Google Scholar]
- 20.Luzhetskyy, A., and A. Bechthold. 2008. Features and applications of bacterial glycosyltransferases: current state and prospects. Appl. Microbiol. Biotechnol. 80:945-952. [DOI] [PubMed] [Google Scholar]
- 21.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto, A., Y. Fujiwara, R. K. Elespuru, and P. C. Hanawalt. 1994. Photoactivated gilvocarcin V induces DNA-protein crosslinking in genes for human ribosomal RNA and dihydrofolate reductase. Photochem. Photobiol. 60:225-230. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto, A., and P. C. Hanawalt. 2000. Histone H3 and heat shock protein GRP78 are selectively cross-linked to DNA by photoactivated gilvocarcin V in human fibroblasts. Cancer Res. 60:3921-3926. [PubMed] [Google Scholar]
- 24.McGee, L. R., and R. Misra. 1990. Gilvocarcin photobiology. Isolation and characterization of the DNA photoadduct. J. Am. Chem. Soc. 112:2386-2389. [Google Scholar]
- 25.Nakajima, S., K. Kojiri, H. Suda, and M. Okanishi. 1991. New antitumor substances, BE-12406A and BE-12406B, produced by a streptomycete. II. Structure determination. J. Antibiot. (Tokyo) 44:1061-1064. [DOI] [PubMed] [Google Scholar]
- 26.Nakano, H., Y. Matsuda, K. Ito, S. Ohkubo, M. Morimoto, and F. Tomita. 1981. Gilvocarcins, new antitumor antibiotics. 1. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. 34:266-270. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima, T., T. Fujii, K. Sakai, S. Tomohiro, H. Kumagai, and T. Yoshioka. February 2000. Chrysomycin derivative compounds and use as antitumor agents. U.S. patent 6,030,951.
- 28.Olano, C., M. S. Abdelfattah, S. Gullón, A. F. Braña, J. Rohr, C. Méndez, and J. A. Salas. 2008. Glycosylated derivatives of steffimycin: insights into the role of the sugar moieties for the biological activity. Chembiochem 9:624-633. [DOI] [PubMed] [Google Scholar]
- 29.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peak, M. J., J. G. Peak, C. M. Blaumueller, and R. K. Elespuru. 1988. Photosensitized DNA breaks and DNA-to-protein crosslinks induced in human cells by antitumor agent gilvocarcin V. Chem. Biol. Interact. 67:267-274. [DOI] [PubMed] [Google Scholar]
- 31.Pérez, M., F. Lombó, I. Baig, A. F. Braña, J. Rohr, J. A. Salas, and C. Méndez. 2006. Combinatorial biosynthesis of antitumor deoxysugar pathways in Streptomyces griseus: reconstitution of “unnatural natural gene clusters” for the biosynthesis of four 2,6-d-dideoxyhexoses. Appl. Environ. Microbiol. 72:6644-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez, L., I. Aguirrezabalaga, N. Allende, A. F. Braña, C. Méndez, and J. A. Salas. 2002. Engineering deoxysugar biosynthetic pathways from antibiotic-producing microorganisms: a tool to produce novel glycosylated bioactive compounds. Chem. Biol. 9:721-729. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez, L., C. Oelkers, I. Aguirrezabalaga, A. F. Braña, J. Rohr, C. Méndez, and J. A. Salas. 2000. Generation of hybrid elloramycin analogs by combinatorial biosynthesis using genes from anthracycline-type and macrolide biosynthetic pathways. J. Mol. Microbiol. Biotechnol. 2:271-276. [PubMed] [Google Scholar]
- 34.Rubinstein, L. V., R. H. Shoemaker, K. D. Paull, R. M. Simon, S. Tosini, P. Skehan, D. A. Scudiero, A. Monks, and M. R. Boyd. 1990. Comparison of in vitro anticancer-drug-screening data generated with a tetrazolium assay versus a protein assay against a diverse panel of human tumor cell lines. J. Natl. Cancer Inst. 82:1113-1118. [DOI] [PubMed] [Google Scholar]
- 35.Salas, A. P., L. Zhu, C. Sanchez, A. F. Brana, J. Rohr, C. Mendez, and J. A. Salas. 2005. Deciphering the late steps in the biosynthesis of the anti-tumour indolocarbazole staurosporine: sugar donor substrate flexibility of the StaG glycosyltransferase. Mol. Microbiol. 58:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salas, J. A., and C. Mendez. 2009. Indolocarbazole antitumour compounds by combinatorial biosynthesis. Curr. Opin. Chem. Biol. 13:152-160. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Sehgal, S. N., H. Czerkawski, A. Kudelski, K. Pandev, R. Saucier, and C. Vezina. 1983. Ravidomycin (AY-25,545), a new antitumor antibiotic. J. Antibiot. 35:355-361. [DOI] [PubMed] [Google Scholar]
- 39.Shepherd, M. D., M. K. Kharel, L. L. Zhu, S. G. Van Lanen, and J. Rohr. 2010. Delineating the earliest steps of gilvocarcin biosynthesis: role of GilP and GilQ in starter unit specificity. Org. Biomol. Chem. 8:3851-3856. [DOI] [PubMed] [Google Scholar]
- 40.Skehan, P., R. Storeng, D. Scudiero, A. Monks, J. McMahon, D. Vistica, J. T. Warren, H. Bokesch, S. Kenney, and M. R. Boyd. 1990. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82:1107-1112. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi, K., M. Yoshida, F. Tomita, and K. Shirahata. 1981. Gilvocarcins, new antitumor antibiotics. 2. Structural elucidation. J. Antibiot. 34:271-275. [DOI] [PubMed] [Google Scholar]
- 42.Thibodeaux, C. J., C. E. Melancon III, and H. W. Liu. 2008. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew. Chem. Int. Ed. Engl. 47:9814-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss, U., K. Yoshihira, R. J. Highet, R. J. White, and T. T. Wei. 1982. The chemistry of the antibiotics chrysomycin A and B. Antitumor activity of chrysomycin A. J. Antibiot. 35:1194-1201. [DOI] [PubMed] [Google Scholar]
- 44.Williams, G. J., R. Gantt, and J. S. Thorson. 2008. The impact of enzyme engineering upon natural product glycodiversification. Curr. Opin. Chem. Biol. 12:556-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita, N., K. Shin-ya, K. Furihata, Y. Hayakawa, and H. Seto. 1998. New ravidomycin analogues, FE35A and FE35B, apoptosis inducers produced by Streptomyces rochei. J. Antibiot. 51:1105-1108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.