Abstract
Phosphatidylinositol 3-kinase (PI3K) regulates several vital cellular processes, including signal transduction and membrane trafficking. In order to study the intracellular localization of the PI3K product, phosphatidylinositol 3-phosphate [PI(3)P], we constructed a probe consisting of two PI(3)P-binding FYVE domains. The probe was found to bind specifically, and with high affinity, to PI(3)P both in vitro and in vivo. When expressed in fibroblasts, a tagged probe localized to endosomes, as detected by fluorescence microscopy. Electron microscopy of untransfected fibroblasts showed that PI(3)P is highly enriched on early endosomes and in the internal vesicles of multivesicular endosomes. While yeast cells deficient in PI3K activity (vps15 and vps34 mutants) were not labelled, PI(3)P was found on intralumenal vesicles of endosomes and vacuoles of wild-type yeast. vps27Δ yeast cells, which have impaired endosome to vacuole trafficking, showed a decreased vacuolar labelling and increased endosome labelling. Thus PI(3)P follows a conserved intralumenal degradation pathway, and its generation, accessibility and turnover are likely to play a crucial role in defining the early endosome and the subsequent steps leading to multivesicular endosome formation.
Keywords: electron microscopy/membrane traffic/multivesicular body/phosphoinositide/PI 3-kinase
Introduction
The temporal and spatial regulation of complex cellular processes such as signal transduction and membrane trafficking relies on the reversible recruitment of proteins to restricted intracellular membranes. Phosphorylated derivatives of phosphatidylinositol (PI), known as phosphoinositides, play a key role in this recruitment by binding (and sometimes activating) specific proteins (Corvera et al., 1999). Since phosphoinositides function to recruit proteins to specific membranes, it is clearly of great interest to determine their intracellular localization. To date, this has been attempted mainly through the expression of green fluorescent protein (GFP) fused with pleckstrin homology (PH) domains that show a phosphoinositide-selective binding profile in vitro. In this manner, the PH domain from phospholipase C δ1 (PLC-δ1) has been employed as a probe for PI(4,5)P2, and the PH domains of ARNO or Bruton’s tyrosine kinase have been used in order to detect PI(3,4,5)P3 (Stauffer et al., 1998; Várnai and Balla, 1998; Venkateswarlu et al., 1998; Hirose et al., 1999; Várnai et al., 1999). While such studies have indicated the presence of both PI(4,5)P2 and PI(3,4,5)P3 at the plasma membrane, their interpretations are complicated by several issues. First, it has been argued that the recruitment of PH domains to membranes may depend on binding to other determinants in addition to phosphoinositides. For instance, while the PLC-δ1 PH domain is targeted to the plasma membrane, the PH domain of oxysterol-binding protein, which shows a comparable affinity and specificity for PI(4,5)P2 in vitro, is targeted to the Golgi apparatus (Levine and Munro, 1998). Secondly, the high level expression required in order to detect a GFP–PH fusion protein by fluorescence microscopy may by itself affect phosphoinositide distribution. Thirdly, the resolution at the light microscopic level is insufficient to yield a precise localization of any phosphoinositide. Fourthly, fluorescence microscopy does not allow a quantitative estimation of phosphoinositide levels.
The products of phosphatidylinositol 3-kinases (PI3Ks) have received special attention due to their importance in signal transduction, membrane trafficking, cytoskeletal regulation and apoptosis (Leevers et al., 1999; Rameh and Cantley, 1999). PI(3,4,5)P3 is produced upon agonist stimulation of mammalian cells and interacts with a number of proteins containing PH domains (Rameh and Cantley, 1999). In contrast, PI(3)P is constitutively produced in both yeast and mammalian cells, and binds to proteins containing FYVE finger domains (Stenmark and Aasland, 1999). In order to understand the functions of FYVE finger proteins, several of which have been shown to play crucial roles in physiology and cell biology (Stenmark and Aasland, 1999), it is essential to localize PI(3)P within the cell. Fluorescence microscopy has been used to show that the C-terminal part of the early endosomal autoantigen EEA1 (Mu et al., 1995), which contains a FYVE finger domain, is targeted to early endosomes (Stenmark et al., 1996; Burd and Emr, 1998; Gaullier et al., 1998; Patki et al., 1998). However, since this targeting also requires an adjacent domain that binds to the early endosomal GTPase Rab5 (Stenmark et al., 1996; Simonsen et al., 1998b; Lawe et al., 2000), this provides only circumstantial evidence for the presence of PI(3)P on early endosomes and no information about the absence or presence of PI(3)P on other membranes. Here we have used biochemical and morphological assays to characterize a new probe with unique specificity for PI(3)P. We have used this probe to determine the localization of PI(3)P in both mammalian and yeast cells. We now show that PI(3)P is found on the limiting membranes of early endosomes and, in addition, on the internal vesicles of multivesicular endosomes and the yeast vacuole.
Results
A double FYVE finger binds with high affinity and specificity to PI(3)P
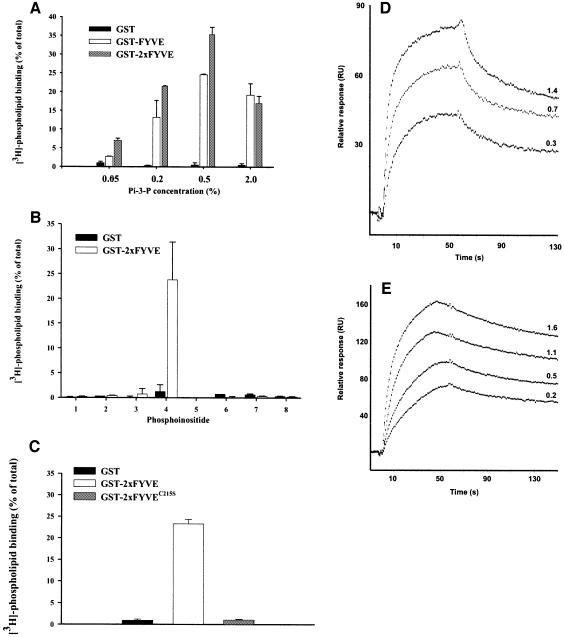
We considered the possibility of using a FYVE finger-derived construct as a probe for PI(3)P. The isolated FYVE finger of EEA1 apparently is unsuitable as a probe for PI(3)P (Stenmark et al., 1996; Lawe et al., 2000), presumably because it binds PI(3)P with too low affinity. We therefore investigated whether the FYVE finger of the receptor tyrosine kinase substrate Hrs (Komada and Kitamura, 1995) could be used for this purpose. However, like the EEA1 FYVE finger, the FYVE finger of Hrs was mainly cytosolic when expressed in cells (data not shown). We therefore constructed a double FYVE finger of Hrs (2×FYVE), with the rationale that this should have a greater affinity for PI(3)P. Indeed, in an in vitro assay, a fusion protein consisting of GST and 2×FYVE bound strongly to PI(3)P-containing liposomes, with maximal binding at ∼0.5% (Figure 1A). Even at a very low PI(3)P concentration (0.05%), a significant binding of 2×FYVE could be detected, in contrast to the single FYVE finger. We also found that 2×FYVE did not bind to liposomes containing any other phosphoinositides (Figure 1B), and a point mutation (C215S) (Gaullier et al., 1998) introduced into both of its constituent FYVE fingers completely abolished PI(3)P binding (Figure 1C). In order to compare the binding kinetics of GST–FYVE and GST–2×FYVE, we studied their binding to PI(3)P using surface plasmon resonance. A sensor surface was coated with mixed phospholipids containing 2% PI(3)P, and sensorgrams were recorded upon addition of the GST fusion proteins. Although the FYVE finger of Hrs has been proposed to interact with PI(3)P as a dimer (Mao et al., 2000), the interaction of GST–FYVE with PI(3)P showed Langmuir association and dissociation kinetics characteristic of a 1:1 binding, with a KD of 38 ± 19 nM (Figure 1D). In contrast, GST–2×FYVE showed more complex association/dissociation kinetics that could be fit into a bivalent binding model (Figure 1E). Because of the complex kinetics of GST–2×FYVE, it is difficult to draw direct comparisons between the two probes, but it is evident that GST–2×FYVE has slower dissociation kinetics than GST–FYVE. The ability of one GST–2×FYVE molecule to interact with two molecules of PI(3)P is thus likely to explain its superior PI(3)P binding compared with GST–FYVE.
Fig. 1. 2×FYVE binds specifically and with high affinity to PI(3)P. (A–C) Portions of GST alone or fused to FYVE, 2×FYVE or 2×FYVE with a double C215S mutation (Gaullier et al., 1998) were immobilized on glutathione–Sepharose beads and then incubated in the presence of 3H-labelled liposomes containing: (A) 0.05–2.0% of PI(3)P, (B) 0.2% of the inositol lipids indicated below or (C) 0.2% PI(3)P. All values are means ± SEM of multiple experiments performed in triplicate. 1, PI; 2, PI(4)P; 3, PI(5)P; 4, PI(3)P; 5, PI(3,4)P2; 6, PI(3,5)P2; 7, PI(4,5)P2; 8, PI(3,4,5)P3. (D) GST–FYVE or (E) GST–2×FYVE were injected into sensor chips that had been loaded with liposomes containing 2% PI(3)P. Sensorgrams at the indicated concentrations (in µM) of the GST fusion proteins are shown.
Transfected 2×FYVE as a specific probe for PI(3)P
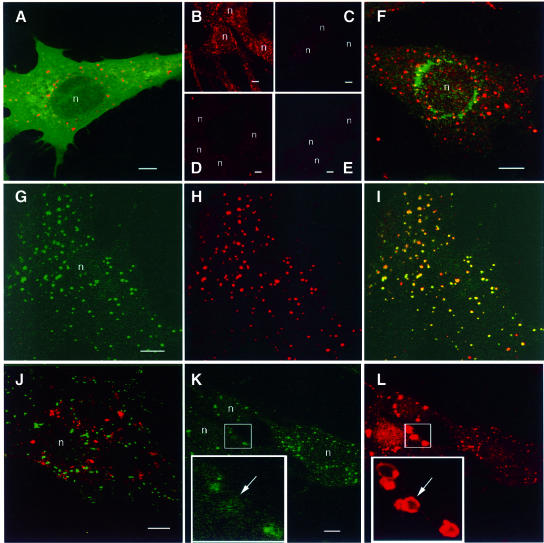
In order to ascertain whether 2×FYVE could be used as a probe for PI(3)P, we examined its distribution in transiently transfected BHK cells (Simonsen et al., 1998a) using confocal immunofluoresence microscopy. When a myc epitope-tagged 2×FYVE was co-expressed with a GFP-tagged PH domain from PLC δ1, 2×FYVE was localized on intracellular vesicular structures, in contrast to GFP–PLC-δ1–PH, which is known to localize mainly to the plasma membrane (Stauffer et al., 1998) (Figure 2A). A similar result was obtained with a 2×FYVE construct derived from EEA1, although the vesicular staining was somewhat fainter (data not shown). By two-hybrid analysis and co-expression experiments, we detected no binding of the 2×FYVE constructs to their native proteins (i.e. EEA1 and Hrs) (data not shown). Incubation of cells with wortmannin, an established inhibitor of PI3K, abolished the localization of 2×FYVE on the intracellular vesicular structures (Figure 2, compare C with B). Likewise, a point mutation that interferes with PI(3)P binding (Gaullier et al., 1998) abolished the membrane localization of 2×FYVE when introduced into one (Figure 2D) or both (Figure 2E) of its constituent FYVE fingers. These results indicate that the 2×FYVE constructs were associated with vesicular membranes as a consequence of binding to PI(3)P.
Fig. 2. Confocal fluorescence microscopy of cells expressing 2×FYVE. BHK cells were transfected with 2×FYVE constructs, then permeabilized with saponin and fixed for confocal microscopy. (A) myc-2×FYVE (red) and GFP-PLC δ1 (green). (B and C) myc-2×FYVE (red) in the absence (B) and presence (C) of 100 nm wortmannin for 30 min. (D and E) myc-2×FYVE (red) with a single (D) or double (E) C215S mutation. (F) myc-2×FYVE (red) and endogenous mannose 6-phosphate receptor (green). (G and H) myc-2×FYVE (red) and endogenous EEA1 (green). (J) GFP-2×FYVE (green) and endogenous LBPA (red). (K and L) myc-2×FYVE (red) and endogenous EEA1 (green). In the merged images (A, F, I and J), yellow colour indicates co-localization between 2×FYVE and the organelle marker molecule. The arrow in (K) and (L) points to an expanded vacuolar structure that contains a high level of 2×FYVE and a low level of EEA1. Bars, 5 µm. Nuclei are indicated with ‘n’.
The localization of 2×FYVE was then compared with that of various organelle markers. myc- or GFP-tagged 2×FYVE co-localized extensively with endogenous EEA1 (Mu et al., 1995), a well-characterized marker of early endosomes (Figure 2G–I). 2×FYVE did not, however, co-localize significantly with any other markers of membranous compartments that were tested, including the Golgi marker, mannosidase II (not shown), the trans-Golgi network/late endosome marker, mannose-6-phosphate receptor (Figure 2F), the late endosome/lysosome marker, LAMP-1 (not shown), and the late endosome marker, lyso-bisphosphatidic acid (LBPA) (Kobayashi et al., 1998) (Figure 2J).
To investigate whether 2×FYVE could compete with endogenous FYVE finger proteins for PI(3)P binding, we examined the localization of EEA1 in cells that expressed a high level of 2×FYVE. While endogenous EEA1 co-localized with 2×FYVE at low expression (Figure 2K and L, right cell), EEA1 appeared to be partially displaced from membranes in the cells expressing 2×FYVE at a high level (Figure 2K and L, left cells). This was supported by quantitation of confocal images, which indicated that an ∼2.5-fold relative increase in 2×FYVE signal was associated with an ∼3-fold decrease in EEA1 signal (not shown). Moreover, the structures that were strongly positive for 2×FYVE were much larger than normal endosomes and had a vacuolar appearance (Figure 2L, inset). They could be labelled with internalized Alexa–transferrin (not shown), indicating that they were of endocytic origin. This effect is reminiscent of wortmannin treatment, which releases FYVE finger proteins from membranes and causes the vacuolation of endosomes (Patki et al., 1997; Fernandez-Borja et al., 1999; Komada and Soriano, 1999). While the size of the 2×FYVE-positive endosomes increased and the EEA1 labelling decreased as a function of the transfection time (not shown), we could also detect a nuclear labelling in the highly expressing cells (left cells in Figure 2L). The possible relevance of this labelling is discussed below.
The 2×FYVE probe can be used to detect PI(3)P in untransfected cells
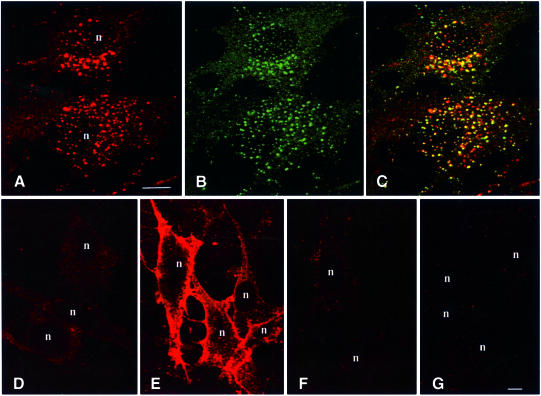
The finding that high level 2×FYVE expression led to alterations in endosome morphology underscores the potential caveats associated with the use of transfected phosphoinositide probes. We therefore investigated whether the 2×FYVE probe could be used to detect PI(3)P in untransfected cells. For this purpose, we permeabilized BHK cells by freeze–thawing prior to fixation, and we stained the cells with biotinylated GST–2×FYVE followed by Cy3–streptavidin. As shown in Figure 3A, a strong labelling of vesicular structures was detected, whereas unpermeabilized cells were not labelled (Figure 3D). The GST–2×FYVE labelling overlapped extensively with that of EEA1 (Figure 3B and C), indicating that this probe can be used for the direct detection of PI(3)P on endosomes. In agreement with this, the labelling was strongly reduced upon incubation of the cells with 100 nM wortmannin for 30 min prior to fixation (not shown). However, we could not rule out the possibility that 2×FYVE might only recognize PI(3)P in a certain cellular context, for instance in association with an endosomal protein. In order to investigate this, we studied the labelling of unpermeabilized cells that had been pre-incubated briefly with PI(3)P-containing liposomes, in order to incorporate PI(3)P into the outer leaflet of the plasma membrane. When these cells were stained with the 2×FYVE probe, a strong plasma membrane labelling was observed (Figure 3E). In contrast, no labelling was detected in cells incubated with buffer alone (Figure 3D), or with liposomes containing PI(3,4)P2 (Figure 3F) or PI(3,4,5)P3 (Figure 3G). This indicates that the 2×FYVE probe detects cell-associated PI(3)P as such, and not in conjunction with endosome-specific molecules. These data also confirm the high specificity of the 2×FYVE probe for PI(3)P.
Fig. 3. Confocal fluorescence microscopy of untransfected BHK cells with GST–2×FYVE as a probe. BHK cells on coverslips were either permeabilized by freeze–thawing (A–C) or left unpermeabilized (D–G). The unpermeabilized cells were incubated for 10 min at 37°C in the presence of buffer alone (D) or liposomes containing 10% PI(3)P (E), PI(3,4)P2 (F) or PI(3,4,5)P3 (G). The cells were then washed extensively prior to fixation. The coverslips were stained with biotinylated GST–2×FYVE (A and D–G) or anti-EEA1 (B) followed by Cy3–streptavidin or FITC–anti-IgG, respectively. Yellow colour in the merged image (C) indicates co-localization. Bar, 5 µm. Nuclei are indicated with ‘n’.
Electron microscopy with 2×FYVE reveals PI(3)P on early endosomes and intralumenal vesicles
To investigate the distribution of PI(3)P at the ultrastructural level, we sought to develop a post-embedding, in situ labelling approach to localize this lipid. This approach avoids cell permeabilization and other possible artefacts associated with pre-embedding labelling and is the method of choice to localize both proteins and lipids. Importantly, it also allows quantitative estimation of the distribution of a protein or lipid assuming similar labelling efficiencies in different compartments. However, to date, lipid localization has been limited by the availability of specific lipid probes and the difficulties in lipid retention. Ultrathin frozen sections (∼60 nm) were compared with freeze-substituted low temperature- embedded lowicryl sections of paraformaldehyde (PFA) or PFA/glutaraldehyde-fixed BHK cells or primary human fibroblasts. Sections were incubated with the GST–2×FYVE probe, washed and fixed. GST–2×FYVE was detected with anti-GST antibodies followed by protein A–gold. The GST–2×FYVE probe labelled discrete endocytic membranes in both frozen (Figures 4 and 5) and lowicryl sections (results not shown). As the frozen sections gave a more specific labelling, these were used in all further studies. To test the specificity of the labelling, sections were labelled in parallel with the same concentrations of the specific probe, GST–2×FYVE, and with the following control probes: GST alone and GST–2×FYVEC215S. The control probes gave negligible labelling (results not shown). Labelling of ultrathin sections of frozen pellets of liposomes containing PI(3)P or PI(3,4,5)P3 also showed the specificity of the labelling for PI(3)P (data not shown).
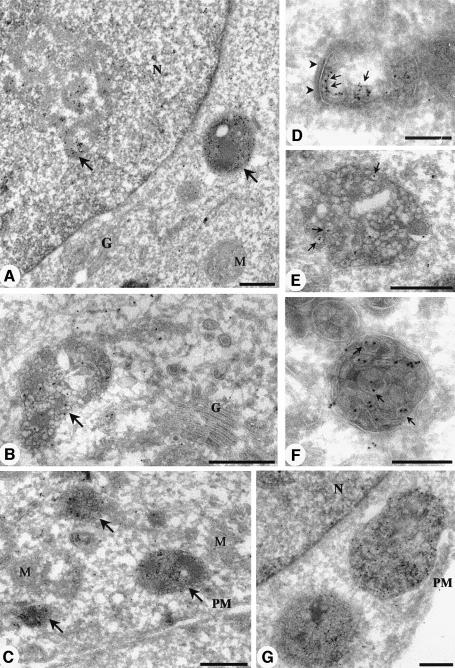
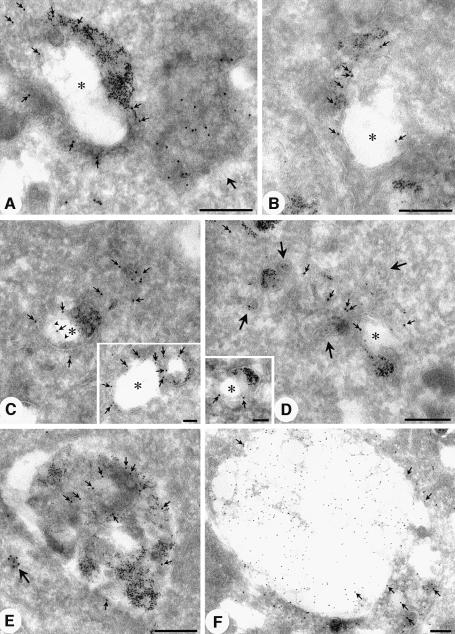
Fig. 4. Electron microscopic localization of PI(3)P using GST–2×FYVE as a probe. Ultrathin frozen sections of PFA/glutaraldehyde-fixed BHK cells were thawed and incubated with GST–2×FYVE. After washing and fixation, sections were incubated with antibodies to GST and then 10 nm protein A–gold. (A) A low-power overview demonstrating the specificity of the FYVE labelling. Specific labelling is associated with a multivesicular endosome and with a discrete region of the nucleus (arrows). Low or negligible labelling is associated with the Golgi complex (G), the nuclear envelope surrounding the nucleus (N) and mitochondria (M). (D–G) Higher magnification micrographs showing the specific labelling of early endosomes (D; recognizable by morphology and clear cytoplasmic coat, arrowheads), and multivesicular endosomes (B and C, arrows; and E–G). Note that there is negligible labelling of the Golgi complex (G) and of the plasma membrane (PM). The multivesicular endosomes show labelling concentrated on the internal membranes (arrows in E and F) with low labelling of the limiting membrane. Bars (A), (C) and (G), 500 nm; (B), (D), (E) and (F), 200 nm.
Fig. 5. Early and late endosomal localization of PI(3)P. Sections were labelled for free PI(3)P using the 2×FYVE probe as described in the legend to Figure 4, or with antibodies to EEA1 (C, inset). BHK cells in (A–E) were incubated with 5 nm BSA–gold for 30 min at 37°C prior to fixation (Griffiths et al., 1989). (A–D) Representative examples of early endosomes labelled with the 2×FYVE probe (large gold; indicated by small arrows in all panels) and internalized 5 nm BSA–gold. Typical tubular vesicular endosomes and ring-shaped early endosomes with an electron-lucent central region (asterisk) surrounded by a double membraned cisterna are evident. Labelling is associated with the multivesicular region (B and C), the cytoplasmic face of the cisternal region (C and D, inset) and with the inner cisternal membrane (see arrowheads indicating inner membrane in C). Labelling is also associated with BSA–gold-labelled tubules (see D; large arrows indicate peripheral BSA–gold-labelled tubules). Note the multivesicular endosome (putative endosome carrier vesicle) containing no internalized gold which is heavily labelled with the 2×FYVE probe (A, large arrow). EEA1 labelling (C, inset, arrows) is associated predominantly with the cytoplasmic face of the BSA–gold-labelled early endosome. (E and F) Late endosomes. (E) A BSA–gold-labelled late endosome showing labelling throughout the multivesicular structure. A heavily labelled small vesicle is also evident (large arrow). (F) Sections of control BHK cells were labelled with specific antibodies to LBPA (10 nm gold) and with the 2×FYVE probe (15 nm gold). Labelling for LBPA is concentrated in the electron-lucent region of the large late endosome. In contrast, the 2×FYVE labelling (arrows) is concentrated in regions showing lower LBPA labelling and more obvious membrane profiles. Bars, main panels, 200 nm; insets, 100 nm.
In view of the high specificity of the probe, we went on to examine the subcellular distribution of PI(3)P. The plasma membrane (Figure 4C and G), Golgi complex (Figure 4A and B), nuclear envelope (Figure 4A and G), mitochondria and other internal membranes showed low labelling for PI(3)P (Table I). In contrast, early endosomes, recognized by their tubulovesicular morphology (Figure 4D), labelling for EEA1 (Figure 5C, inset) and internalized markers (Figure 5A–D), showed specific labelling with the 2×FYVE probe (∼11-fold enrichment per volume or 5-fold enrichment per membrane area as compared with the mitochondria; see Table I). To assess the distribution of 2×FYVE labelling with respect to distinct membranes of the early endosomal system (tubules, ring-shaped cisternae and multivesicular regions; see Griffiths et al., 1989), bovine serum albumin (BSA)–gold was internalized for 10 (not shown) or 30 min at 37°C. The latter was necessary to fill the early endosomal system but also caused labelling of late endosomes which could be distinguished morphologically. The 2×FYVE probe labelled the cytoplasmic face of the early endosome (∼50% of early endosomal gold labelling), the ‘inner’ membrane of the ring-shaped cisterna (see arrowheads in Figure 5C as well as Figure 4D) and the multivesicular region of the early endosome (Figures 4D and 5C). In contrast, EEA1 labelling was only observed on the cytoplasmic face of the early endosome (e.g. Figure 5C; and see Wilson et al., 2000).
Table I. Quantitative analysis of 2×FYVE gold labelling on ultrathin frozen sections of BHK cells.
| Feature | % gold associated with feature (± SEM) | Relative labelling per volumea | Relative labelling per membrane areab |
|---|---|---|---|
| Nucleus | 65 (± 8) | 2.5 | – |
| Nuclear envelope | 0.9 (± 0.3) | – | 0.52 |
| Mitochondria | 2.9 (± 0.2) | 1.0 | 1.0 |
| Early endosomesc | |||
| total | 5.9 (± 4.1) | 11.4 | 4.7 |
| limiting membrane | 3.2 (± 2.1) | ||
| internal membranes | 2.7 (± 2.0) | ||
| Multivesicular endosomesd | |||
| total | 22 (± 8) | 15.3 | 3.6 |
| limiting membrane | 1.7 (± 1.9) | ||
| internal membranes | 20 (± 6) | ||
| Golgi complex | 0.8 (± 0.3) | – | 0.55 |
| Plasma membrane | 2.5 (± 0.9) | – | 1.1 |
The gold distribution on 40 complete cell profiles was quantitated.
aGold labelling related to volume of compartments defined by point counting. Gold/volume related to labelling density in mitochondria.
bGold labelling related to total membrane (limiting and internal membranes) associated with compartments defined by intersection counting. Gold/membrane area related to labelling density of mitochondrial membranes.
The most striking labelling was, however, associated with spherical multivesicular bodies (MVBs) of 400–500 nm diameter (Figures 4 and 5A). Quantitation of gold labelling per membrane area within these MVBs was somewhat hindered by the dense packing of membranes within them (see Figure 4), but a comparison of labelling density in the MVBs as compared with the mitochondria showed a 15-fold enrichment per volume or 4-fold enrichment per membrane area as compared with the mitochondria (see Table I). The quantitative analysis suggests that, within the limits of the quantitation, the density of PI(3)P within the membranes of the early endosomes and MVBs is similar. The MVBs with the highest 2×FYVE probe labelling (e.g. see Figures 4A–C and E–G, and 5A) showed low labelling for late endosomal markers as compared with multivesicular late endosomes (Figure 5F), and they could be classified morphologically as endosomal carrier vesicles (ECVs), an intermediate between the early and late endosomes (Gruenberg and Maxfield, 1995). Remarkably, ∼90% of the labelling was associated with the internal membranes of the MVBs (Table I). This is consistent with a recruitment of cytoplasmically orientated PI(3)P into the interior of the ECVs (Figure 4). Interestingly, late endosomes, which show high labelling for lysosomal glycoproteins (not shown) and LBPA (Figure 5F), generally showed a lower PI(3)P labelling (Figure 5E and F) than putative ECVs. The PI(3)P labelling appeared in distinct regions low in LBPA, in which the internal membranes were well defined (Figure 5F). This suggests that two distinct classes of internal membranes may exist in the late endosomes, with the LBPA-containing internal vesicles being relatively resistant to lysosomal degradation (Kobayashi et al., 1998).
In addition to the cytoplasmic labelling with 2×FYVE, we noted a high labelling within the nucleolus of both BHK cells and human fibroblasts (Figure 4A, Table I). Like the cytoplasmic labelling, this labelling was absent in sections incubated with GST alone or the GST–2×FYVEC215S probe (results not shown), and the nuclear labelling was highly specific for the dense fibrillar component, rather than being dispersed throughout the entire nucleus. This observation is consistent with the fact that nuclei contain PI 3-kinase activity (Lu et al., 1998) and raises the possibility that, like PI(4,5)P2 (Boronenkov et al., 1998), PI(3)P may play a role in nuclear processes. As found previously for PI(4,5)P2, we found no association of PI(3)P labelling with the nuclear envelope. The generation, metabolism and function of the nuclear pool of phosphoinositides obviously will warrant further investigation (Maraldi et al., 1999).
Localization of PI(3)P in wild-type and vps mutant yeast
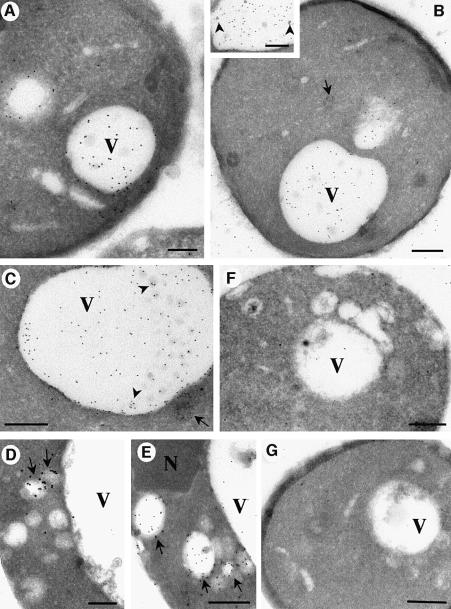
The involvement of PI(3)P in membrane trafficking was established when the product of a gene required for vacuolar protein sorting in Saccharomyces cerevisae, VPS34, was identified as a PI(3)P-producing PI3K (Schu et al., 1993). To investigate if PI(3)P trafficking is conserved between yeast and mammals, we labelled frozen sections of wild-type S.cerevisiae with the 2×FYVE probe and examined them by electron microscopy. High specific labelling was evident within the vacuole, in some cases in association with internal vesicles (Figure 6B, inset, and C). In addition, labelling was associated with cytoplasmic vesicles, possibly representing endosomal intermediates (Figure 6C). In contrast to mammalian cells, no significant labelling was observed within the yeast nuclei, and other organelles (e.g. mitochondria or the nuclear envelope) also showed negligible labelling. This shows that PI(3)P in yeast, like in mammalian cells, is internalized into intralumenal vesicles, as proposed by Wurmser and Emr (1998).
Fig. 6. Electron microscopic localization of PI(3)P in wild-type and mutant yeast cells. Saccharomyces cerevisae strains were fixed and processed for frozen sectioning. The Figure shows representative examples of wild-type (WT) cells (A–C), vps27Δ (D and E), vps34 (F) and vps15 (G). Sections were thawed and incubated with GST–2×FYVE. After washing and fixation, sections were incubated with antibodies to GST and then 10 nm protein A–gold. (A and B) Overviews demonstrating the specificity of the 2×FYVE labelling. Specific labelling is associated with the vacuole (V). Low or negligible labelling is associated with the plasma membrane and other organelles. The inset and (C) show higher magnification views of the vacuole in which some labelling is seen associated with small internal vesicles (arrowheads). Labelling is also associated with cytoplasmic vesicles, some of which appear multivesicular (C, arrow). In contrast to the WT yeast, the vps34 and vps15 strains show very low labelling with the 2×FYVE probe (F and G), showing the specificity of the probe. As compared with wild-type strains, vps27Δ cells show a marked accumulation of labelling in cytoplasmic vesicles (D and E, arrows) and on average a decreased vacuolar labelling (see Table I for quantitation). Note that there is some labelling between the cell profiles of the wild-type cells (A and B) which was not observed in the vps34 and vps15 strains and therefore presumably arises from broken cells or diffusion of the lipid. N, nucleus. Bars (A) and (D), 200 nm; (B), (C) and (E–G), 500 nm.
To assess definitively the specificity of the on-section labelling of ultrathin sections in an in vivo context, we examined 2×FYVE labelling of cells deficient in PI3K activity [vps34, lacking the PI3K enzyme, and vps15, lacking the Vps15p protein kinase which is required for PI3K activation (Stack et al., 1995)]. As compared with the wild-type strain, total labelling was decreased dramatically in the vps34 mutant and vps15 mutant cells (Figure 6F and G, Table II). This confirms the specificity of the 2×FYVE probe.
Table II. Quantitative analysis of 2×FYVE labelling for electron microscopy of yeast strains.
| Total FYVE labelling relative to wild-type yeasta |
Gold distribution; % relative to wild-type yeasta (% of total in each strainb) |
|||
|---|---|---|---|---|
| Strain | Total gold | Vacuole | Cyt Ves | Cyt other |
| Wild-type | 100% | 82.3 (82.3) | 9.2 (9.2) | 8.5 (8.5) |
| vps27Δ | 76.5% | 50.0 (65.4) | 18.7 (24.4) | 7.9 (10.3) |
| vps34 | 10.9% | 4.5 (41.3) | 2.1 (19.3) | 4.2 (38.5) |
| vps15 | 10.6% | 5.0 (47.2) | 1.6 (15.1) | 3.9 (36.8) |
The indicated strains of S.cerevisae were fixed in paraformaldehyde and then processed for frozen sectioning. Sections of pellets from each strain were labelled with 2×FYVE–GST followed by 10 nm protein A–gold. Quantitation was performed by randomly selecting cell profiles and determining the fraction of gold present in the vacuole, in cytoplasmic vesicles (Cyt Ves) (including multivesicular bodies and electron-lucent putative endosomes, see Figure 6) and in other areas of the cytoplasm (Cyt other).
aCounts were related to the total gold in the same number of wild-type cells (20 cell profiles for each strain).
bNumbers in parentheses show gold counts associated with each compartment represented as the fraction of the total gold associated with each strain.
To gain further insights into the nature of the PI(3)P-positive cytoplasmic vesicles and their putative endosomal origin, we also examined the distribution of PI(3)P in vps27Δ cells (Figure 6D and E). These cells with a classical class E vps defect accumulate pre-vacuolar endosomal intermediates due to a block in traffic to the vacuole (Raymond et al., 1992; Piper et al., 1995). Interestingly, these cells showed a significantly decreased vacuolar labelling and an increased labelling of cytoplasmic vesicles (Figure 6D and E, Table II). This is in agreement with the view that the PI(3)P-positive cytoplasmic vesicles represent endocytic intermediates. The 2×FYVE labelling of wild-type and vps mutant yeast cells establishes this probe as a highly versatile endocytic marker that can be used with diverse organisms, and highlights the conserved role of PI(3)P in endocytic traffic from yeast to mammals.
Discussion
Here we have used 2×FYVE as a specific probe for PI(3)P, and we show the first ultrastructural localization of a PI3K product in yeast and mammalian cells. We have rigorously tested the validity of 2×FYVE as a probe, and the following results show that it can be used for the specific detection of PI(3)P: (i) it binds with high affinity and specificity to PI(3)P-containing membranes, as detected biochemically and by electron microscopy; (ii) a point mutation that blocks binding to PI(3)P inhibits labelling of cells with 2×FYVE; (iii) treatment of mammalian cells with the PI3K inhibitor wortmannin abolishes 2×FYVE labelling; (iv) the plasma membranes of cells pre-incubated with PI(3)P, but not other PI3K products, are labelled by 2×FYVE; and (v) wild-type yeast cells, but not yeast cells that lack a functional PI3K, are labelled by 2×FYVE.
Our electron microscopic detection of PI(3)P illustrates the need for electron microscopy when determining the localization of a phosphoinositide: from the fluorescence microscopy of cells transfected with tagged 2×FYVE, we could conclude that PI(3)P is located to early endosomes, based on the high degree of co-localization with EEA1. However, electron microscopy was required in order to reveal the additional localization of PI(3)P to internal vesicles of MVBs. A transfected probe within the cytoplasm of the cells may not have accessibility to the inner membranes of the MVBs, particularly if the internal vesicles separate from the limiting membrane of the MVBs (Miller et al., 1986; Odorizzi et al., 1998). On the other hand, a transfected probe might compete efficiently with endogenous proteins for PI(3)P binding, whereas the GST–2×FYVE probe used for electron microscopy is likely to detect only unligated PI(3)P [i.e. PI(3)P which is not bound to FYVE finger proteins or other PI(3)P-binding proteins].
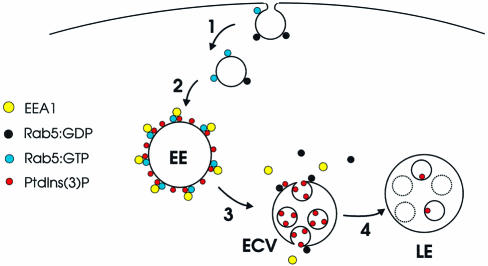
The detection of PI(3)P on early endosomes and on the intralumenal vesicles of multivesicular endosomes enables us to propose a model for a role for PI(3)P in endosomal dynamics (Figure 7). PI(3)P is generated on the early endosome or incoming endocytic vesicles via recruitment of the PI3K hvps34 by Rab5:GTP (Christoforidis et al., 1999). This facilitates the association of EEA1 with the limiting membrane of the early endosome, via its binding to Rab5:GTP and PI(3)P (Simonsen et al., 1998b). The inward budding of the limiting membrane of the vacuolar region of the early endosome removes the PI(3)P from the cytoplasmic face while generating the multivesicular region. This may physically prevent the binding of cytoplasmic proteins to PI(3)P. Consistent with this hypothesis, we detected PI(3)P on both the cytoplasmic face of the early endosome and internal vesicles, but have only observed EEA1 on the cytoplasmic face of the early endosome (Mu et al., 1995; Wilson et al., 2000; this study, Figure 5). Rab5 GTP hydrolysis may cause EEA1 to dissociate from the membrane (Simonsen et al., 1998b) (possibly facilitated by other FYVE proteins) and this may trigger the inward invagination. The relative depletion of PI(3)P from LBPA-positive late endosomes suggests that the internal vesicles containing PI(3)P may be degraded or that the PI(3)P is modified further, for example by kinases. This model is in agreement with findings from yeast, which show that vacuolar hydrolase and PI(3)P 5-kinase activities are involved in PI(3)P turnover (Gary et al., 1998; Wurmser and Emr, 1998). The observation that wortmannin blocks MVB formation (Fernandez-Borja et al., 1999) is also consistent with the above model and suggests that PI(3)P may be crucial to inward vesiculation. Moreover, the kinetics of wortmannin-induced EEA1 dissociation from early endosomes (Patki et al., 1997) are entirely consistent with the kinetics of endosome carrier vesicle formation (Gruenberg and Maxfield, 1995).
Fig. 7. Model for the role of PI(3)P in endocytic traffic. (1) The endocytic vesicle, which is devoid of EEA1 (Christoforidis et al., 1999), contains an activity that converts Rab5:GDP into Rab5:GTP (Horiuchi et al., 1995). (2) The vesicle fuses with the early endosome (EE) in a process regulated by EEA1 and other proteins (Simonsen et al., 1998b; McBride et al., 1999). The limiting membrane of the EE is rich in PI(3)P, caused by Rab5:GTP-mediated PI 3-kinase recruitment (Christoforidis et al., 1999), and in EEA1, which binds to the EE via interaction with both PI(3)P and Rab5:GTP (Simonsen et al., 1998b). (3) On the ECV, GTP hydrolysis by Rab5 is accompanied by the dissociation of EEA1 and Rab5:GDP, and PI(3)P is internalized into intralumenal vesicles. (4) The ECV fuses with (or matures into) a late endosome (LE), in which PI(3)P is metabolized.
The in situ detection of PI(3)P described here provides a new and powerful approach to detect a phosphoinositide and a unique tool for studies of endocytic traffic in diverse eukaryotic systems. While we have concentrated on fibroblasts and yeast cells, it will be interesting to determine the localization of PI(3)P in other cell types, such as neurons and antigen-presenting cells. Moreover, the successful detection of PI(3)P with 2×FYVE suggests that it may be possible to design similar probes for determining the intracellular localization of other phosphoinositides. This will greatly advance our understanding of lipid signalling.
Materials and methods
Plasmids and expression
All FYVE constructs were prepared using PCR. The FYVE finger used in this study consists of residues 147–223 of mouse Hrs (Komada and Kitamura, 1995). 2×FYVE consists of this domain in duplicate, with the linker QGQGS separating the two FYVE fingers. We also prepared a double FYVE finger of EEA1 (Mu et al., 1995), which consists of residues 1325–1411 in duplicate, separated by the linker GSGN. For transfections using the T7 RNA polymerase recombinant vaccinia system (Sutter et al., 1995), constructs tagged N-terminally with the myc epitope (Evan et al., 1985) or enhanced green fluorescent protein (EGFP; Clontech) were cloned behind the T7 promoter of pGEM-1 (Promega). Cells were transfected as described (Simonsen et al., 1998a) and analysed 6 h post transfection. For expression in Escherichia coli BL-21(DE3), constructs were cloned into the polylinker of pGEX-5X-3 (Pharmacia). Protein expression and purification were according to the manufacturer’s instructions.
Yeast strains
Yeast strains used in this study were cultured in rich media (1% yeast extract, 1% peptone, 2% dextrose) or standard minimal medium with appropriate supplements (Sherman et al., 1986). The strains used in this study were congenic to SF838-9D (wild-type) (Matα leu2-3,112 ura3-52 his4-519 ade6 gal2 pep4-3) (Rothman and Stevens, 1986) and are as follows: SF838-9Dα-mut.223 (vps34/vpl7-1) (Raymond et al., 1992) (Matα leu2-3,112 ura3-52 his4-519 ade6 gal2 pep4-3 vpl7-1) (Rothman and Stevens, 1986); SF838-9Dα-mut.2066 (vps15/vpl19-10) (Raymond et al., 1992) (Matα leu2-3,112 ura3-52 his4-519 ade6 gal2 pep4-3 vpl19-10) (Rothman and Stevens, 1986); and NBY123 (vps27Δ) (Matα leu2-3, 112 ura3-52 his4-519 ade6 gal2 pep4-3 vps27Δ::LEU2). NBY123 was constructed from SF838-9D using pKJH2 to disrupt VPS27 (Piper et al., 1995). Fifty OD600 units of cells in mid-log phase were harvested and fixed with 8% PFA in phosphate buffer, pH 7.35. Cells were processed for frozen sectioning and sections were labelled for electron microscopy with 2×FYVE–GST as described below.
Liposome-binding assay
Phosphoinositides (0.05–2%, Echelon) were incorporated into small unilamellar vesicles containing 63–65% phosphatidylcholine, 20% phosphatidylserine, 15% phosphatidylethanolamine and a trace amount of [3H]phosphatidylcholine, and the binding to GST fusion proteins immobilized on glutathione–Sepharose beads (Pharmacia) was assayed as described (Schiavo et al., 1996).
Surface plasmon resonance
Surface plasmon resonance was recorded at 25°C on a BIAcore X (BIAcore, Sweden). The liposomes used were the same as those used in the liposome-binding assay, except that they contained 2% PI(3)P and no radioactive tracer. Liposomes (0.35 mg/ml) were loaded onto a BIAcore L1 chip by three successive injections of 80 µl of liposomes at a flow rate of 5 µl/min. The reference cell was loaded with similar liposomes lacking PI(3)P. Sensorgrams were recorded upon the injections of 0.2–2 µg of protein at a flow rate of 20 µl/min. The lipid surface was regenerated using 10 mM NaOH. Analysis of association and dissociation kinetics was done with the BIAsimulation 3.0 software package supplied with the instrument. The fit to various kinetic models was evaluated on the basis of χ2 values and scatter of residuals.
Confocal fluorescence microscopy
Transfected cells on coverslips were permeabilized with 0.05% saponin or by freeze–thawing, then fixed with PFA and stained with the indicated antibodies followed by fluorescein isothiocyanate (FITC)- or lissamine rhodamine-labelled secondary antibodies (Jackson Immunoresearch), as described (Simonsen et al., 1998a). In some cases, the coverslips were stained with biotinylated GST–2×FYVE (20 µg/ml) followed by Cy3–streptavidin (2 µg/ml, Jackson Immunoresearch). The coverslips were examined with a Leica TCS NT confocal microscope equipped with a Kr/Ar laser and a PL Fluotar 100×/1.30 oil objective. The relative intensities of structures labelled by two antibodies were quantitated with the software provided with the instrument.
Electron microscopy
BHK cells were fixed with 4% PFA/0.01 glutaraldehyde or 8% PFA only in 0.1 M phosphate buffer, pH 7.35, for 2 h at room temperature. Where appropriate, cells were incubated with BSA–gold for 10–30 min at 37°C to label the early endocytic compartments. They were then processed for frozen sectioning and sections retrieved using methylcellulose/sucrose as described previously (Liou et al., 1997). Sections were incubated with 5–10 µg/ml GST–2×FYVE (or the same concentration of control probe GST or GST–2×FYVEC215S) for 30 min at room temperature and then washed [4× 5 min with phosphate-buffered saline (PBS)] and fixed with 4% PFA in PBS. They were then washed and incubated with rabbit polyclonal antibodies to GST (AmRad) followed by protein A–gold (University of Utrecht, The Netherlands). Identical results were obtained with primary human fibroblasts fixed in 8% PFA for 2 h at room temperature. Some variability in labelling density (but not the pattern of labelling) was observed between different preparations of mammalian cells for unknown reasons. BHK cells fixed as above were processed for freeze substitution and embedding in HM23 resin as described (van Genderen et al., 1991). Yeast cells were fixed in 8% PFA, processed, sectioned and labelled as described above for frozen sections, with all strains processed and labelled together. Quantitation of labelling of BHK cells was performed by analysing cell profiles at random and determining the percentage of gold particles associated with specific compartments. This was related to the volume occupied by each compartment, estimated by point counting on the same sections. To allow the estimation of the volume of endosomal compartments, BHK cells that had internalized BSA–gold for 30 min were used for the quantitation. This incubation time was required to label fully the complex tubular/vesicular regions of the early endocytic compartments. The density of GST–2×FYVE gold labelling per membrane area was determined by relating gold particles to membrane area by intersection counting using a lattice grid overlaid on the images (Griffiths et al., 1989). Quantitation of yeast cell labelling was performed on 20 random cell profiles.
Acknowledgments
Acknowledgements
We thank Jean Gruenberg, Peter Schu, Sue-Goo Rhee and Naomi Kitamura for the gifts of anti-LBPA, anti-mannose 6-phosphate receptor, PLC-δ1 DNA and Hrs DNA, respectively, and Tom H.Stevens for yeast strains. We are particularly grateful to David James for comments on the manuscript. H.S. was supported by the Top Research Programme, the Research Council of Norway and by the Novo-Nordisk Foundation. R.G.P was supported by the Australian Research Council and the Human Frontier Science Program.
References
- Boronenkov I.V., Loijens,J.C., Umeda,M. and Anderson,R.A. (1998) Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell, 9, 3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G. and Emr,S.D. (1998) Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell, 2, 157–162. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska,M., Ashman,K., Wilm,M., Zhao,L., Yip,S.-C., Waterfield,M.D., Backer,J.M. and Zerial,M. (1999) Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nature Cell Biol., 1, 249–252. [DOI] [PubMed] [Google Scholar]
- Corvera S., D’Arrigo,A. and Stenmark,H. (1999) Phosphoinositides in membrane traffic. Curr. Opin. Cell Biol., 11, 460–465. [DOI] [PubMed] [Google Scholar]
- Evan G.I., Lewis,G.K., Ramsay,G. and Bishop,J.M. (1985) Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol. Cell. Biol., 5, 3610–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Borja M., Wubbolts,R., Calafat,J., Janssen,H., Divecha,N., Dusseljee,S. and Neefjes,J. (1999) Multivesicular body morphogenesis requires phosphatidylinositol 3-kinase activity. Curr. Biol., 14, 55–58. [DOI] [PubMed] [Google Scholar]
- Gary G.D., Wurmser,A.E., Bonangelino,C.J., Weisman,L.S. and Emr,S. (1998) Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol., 143, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier J.-M., Simonsen,A., D’Arrigo,A., Bremnes,B., Aasland,R. and Stenmark,H. (1998) FYVE fingers bind PtdIns(3)P. Nature, 394, 432–433. [DOI] [PubMed] [Google Scholar]
- Griffiths G., Back,R. and Marsh,M. (1989) A quantitative analysis of the endocytic pathway in baby hamster kidney cells. J. Cell Biol., 109, 2703–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J. and Maxfield,F.R. (1995) Membrane transport in the endocytic pathway. Curr. Opin. Cell Biol., 7, 552–563. [DOI] [PubMed] [Google Scholar]
- Hirose K., Kadowaki,S., Tanabe,M., Takeshima,H. and Iino,M. (1999) Spatiotemporal dynamics of inositol 1,4,5 trisphosphate that underlies complex Ca2+ mobilization patterns. Science, 284, 1527–1530. [DOI] [PubMed] [Google Scholar]
- Horiuchi H., Giner,A., Hoflack,B. and Zerial,M. (1995) A GDP/GTP exchange-stimulatory activity for the Rab5–RabGDI complex on clathrin-coated vesicles from bovine brain. J. Biol. Chem., 270, 11257–11262. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Stang,E., Fang,K.S., De Moerloose,P., Parton,R.G. and Gruenberg,J. (1998) A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature, 392, 193–197. [DOI] [PubMed] [Google Scholar]
- Komada M. and Kitamura,N. (1995) Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol. Cell. Biol., 15, 6213–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M. and Soriano,P. (1999) Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev., 13, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawe D.C., Patki,V., Heller-Harrison,R., Lambright,D. and Corvera,S. (2000) The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. J. Biol. Chem., 275, 3699–3705. [DOI] [PubMed] [Google Scholar]
- Leevers S.J., Vanhaesebroeck,B. and Waterfield,M.D. (1999) Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Curr. Opin. Cell Biol., 11, 219–225. [DOI] [PubMed] [Google Scholar]
- Levine T.P. and Munro,S. (1998) The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol., 8, 729–739. [DOI] [PubMed] [Google Scholar]
- Liou W., Geuze,H.J., Geelen,M.J. and Slot,J.W. (1997) The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol., 136, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P.J., Hsu,A.L., Wang,D.S., Yan,H.Y., Yin,H.L. and Chen,C.S. (1998) Phosphoinositide 3-kinase in rat liver nuclei. Biochemistry, 37, 5738–5745. [DOI] [PubMed] [Google Scholar]
- Mao Y., Nickitenko,A., Duan,X., Lloyd,T.E., Wu,M.N., Bellen,H. and Quiocho,F.A. (2000) Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell, 100, 447–456. [DOI] [PubMed] [Google Scholar]
- Maraldi N.M., Zini,N. and Manzoli,F.A. (1999) Topology of inositol lipid signal transduction in the nucleus. J. Cell Physiol., 181, 203–217. [DOI] [PubMed] [Google Scholar]
- McBride H.M., Rybin,V., Murphy,C., Giner,A., Teasdale,R. and Zerial,M. (1999) Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell, 98, 377–386. [DOI] [PubMed] [Google Scholar]
- Miller K., Beardmore,J., Kanety,H., Schlessinger,J. and Hopkins,C.R. (1986) Localization of the epidermal growth factor (EGF) receptor within the endosome of EGF-stimulated epidermoid carcinoma (A431) cells. J. Cell Biol., 102, 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu F.T. et al. (1995) EEA1, an early endosome-associated protein. EEA1 is a conserved α-helical peripheral membrane protein flanked by cysteine ‘fingers’ and contains a calmodulin-binding IQ motif. J. Biol. Chem., 270, 13503–13511. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst,M. and Emr,S.D. (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell, 95, 847–858. [DOI] [PubMed] [Google Scholar]
- Patki V., Virbasius,J., Lane,W.S., Toh,B.H., Shpetner,H.S. and Corvera,S. (1997) Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc. Natl Acad. Sci. USA, 94, 7326–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patki V., Lawe,D.C., Corvera,S., Virbasius,J.V. and Chawla,A. (1998) A functional PtdIns(3)P-binding motif. Nature, 394, 433–434. [DOI] [PubMed] [Google Scholar]
- Piper R.C., Cooper,A.A., Yang,H. and Stevens,T.H. (1995) VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol., 131, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh L.E. and Cantley,L.C. (1999) The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem., 274, 8347–8350. [DOI] [PubMed] [Google Scholar]
- Raymond C.K., Howald-Stevenson,I., Vater,C.A. and Stevens,T.H. (1992) Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E mutants. Mol. Biol. Cell, 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.H. and Stevens,T.H. (1986) Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell, 47, 1041–1051. [DOI] [PubMed] [Google Scholar]
- Schiavo G., Gu,Q.-M., Prestwich,G.D., Söllner,T.H. and Rothman,J.E. (1996) Calcium-dependent switching of the specificity of phosphoinositide binding to synaptotagmin. Proc. Natl Acad. Sci. USA, 93, 13327–13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu P.V., Takegawa,K., Fry,M.J., Stack,J.H., Waterfield,M.D. and Emr,S.D. (1993) Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science, 260, 88–91. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G.F. and Hicks,J. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Simonsen A., Bremnes,B., Rønning,E., Aasland,R. and Stenmark,H. (1998a) Syntaxin-16, a putative Golgi t-SNARE. Eur. J. Cell Biol., 75, 223–231. [DOI] [PubMed] [Google Scholar]
- Simonsen A. et al. (1998b) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature, 394, 494–498. [DOI] [PubMed] [Google Scholar]
- Stack J.H., DeWald,D.B., Takegawa,K. and Emr,S.D. (1995) Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol., 129, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer T.P., Ahn,S. and Meyer,T. (1998) Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol., 8, 343–346. [DOI] [PubMed] [Google Scholar]
- Stenmark H. and Aasland,R. (1999) FYVE-finger proteins—effectors of an inositol lipid. J. Cell Sci., 112, 4175–4183. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Aasland,R., Toh,B.H. and D’Arrigo,A. (1996) Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem., 271, 24048–24054. [DOI] [PubMed] [Google Scholar]
- Sutter G., Ohlmann,M. and Erfle,V. (1995) Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett., 371, 9–12. [DOI] [PubMed] [Google Scholar]
- van Genderen I.L., van Meer,G., Slot,J.W., Geuze,H.J. and Voorhout,W.F. (1991) Subcellular localization of Forssman glycolipid in epithelial MDCK cells by immuno-electron microscopy after freeze-substitution. J. Cell Biol., 115, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P. and Balla,T. (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labelled phosphoinositide pools. J. Cell Biol., 143, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P., Rother,K.I. and Balla,T. (1999) Phosphatidylinositol 3-kinase-dependent membrane association of the Bruton’s tyrosine kinase pleckstrin homology domain visualized in single living cells. J. Biol. Chem., 274, 10983–10989. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K., Oatey,P.B., Tavaré,J.M. and Cullen,P.J. (1998) Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr. Biol., 8, 463–466. [DOI] [PubMed] [Google Scholar]
- Wilson J.M., de Hoop,M., Zorzi,N., Toh,B.-H., Dotti,C.G. and Parton,R.G. (2000) EEA1, a tethering protein of the early sorting endosome, shows a polarized distribution in hippocampal neurons, epithelial cells and fibroblasts. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A.E. and Emr,S.D. (1998) Phosphoinositide signaling and turnover: Ptdlns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J., 17, 4930–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]