Abstract
Nontyphoid salmonellosis caused by Salmonella enterica is the most common bacterial food-borne illness in humans, and fresh produce, including tomatoes, is a common vehicle. Accumulating data indicate that human enteric pathogenic bacteria, including S. enterica, interact actively with plants. Tomato plants were inoculated with S. enterica to evaluate plausible contamination routes and to determine if the tomato cultivar affects S. enterica colonization. S. enterica population levels on tomato leaves were cultivar dependent. S. enterica levels on Solanum pimpinellifolium (West Virginia 700 [WVa700]) were lower than on S. lycopersicum cultivars. S. enterica preferentially colonized type 1 trichomes and rarely interacted with stomata, unlike what has been reported for cut lettuce leaves. Early S. enterica leaf colonization led to contamination of all fruit, with levels as high as 105 CFU per fruit. Reduced bacterial speck lesion formation correlated with reduced S. enterica populations in the phyllosphere. Tomato pedicels and calyxes also harbored large S. enterica populations following inoculation via contaminated water postharvest. WVa700 green fruit harbored significantly smaller S. enterica populations than did red fruit or S. lycopersicum fruit. We found that plants irrigated with contaminated water had larger S. enterica populations than plants grown from seeds planted in infested soil. However, both routes of contamination resulted in detectable S. enterica populations in the phyllosphere. Phyllosphere S. enterica populations pose a risk of fruit contamination and subsequent human disease. Restricting S. enterica phyllosphere populations may result in reduced fruit contamination. We have identified WVa700 as a tomato cultivar that can restrict S. enterica survival in the phyllosphere.
Nontyphoid salmonellosis, a diarrheal disease caused by Salmonella enterica, is the most common bacterial food-borne illness in the United States. Consumption of contaminated produce, not animal products, has been the most repeated cause of salmonellosis in recent years (7). S. enterica colonization of tomato plants has caused several multistate and international outbreaks, each involving hundreds of cases (5, 9, 10, 33). Yearly salmonellosis outbreaks (2002 to 2008) have been caused by consumption of contaminated fresh tomatoes produced domestically (CDC, U.S. Food and Drug Administration [FDA]). Each reported clinical case represents only a fraction of the actual number of ill people; thus, hundreds reported equals thousands sick (21). The biology of this plant-microbe interaction is poorly understood, yet epidemiological evidence supports the idea that most outbreaks are due to preharvest contamination in the field.
Survival of bacterial populations on plants can be governed by the plants themselves. Plants have an immune system that can detect generic conserved components of most microorganisms (16). These conserved components have been termed microbe-associated molecular patterns (MAMPs). MAMPs are defense elicitors that are conserved and evolutionarily stable features of the microbes that cannot be sacrificed or even altered slightly without impairing viability (4). Many bacteria that are nonpathogenic to plants, such as salmonellae, express defense-eliciting molecules, such as flagellin, that trigger recognition when they arrive on leaves. Plant basal defense likely plays a role in limiting the growth of S. enterica in the phyllosphere. One basal-defense-activated response is stomatal closure, which limits the growth of bacteria by stranding them on the harsh leaf surface (12, 23).
The plant itself has been mostly ignored in the study of human pathogens in association with plants. However, host plants may play a role in determining the potential for human disease. Examination of domestic nontyphoid salmonellosis outbreaks associated with agricultural row crops reveals that the majority of these outbreaks are associated with either tomato or cantaloupe consumption. Only one outbreak has been reported in association with lettuce (FDA). In contrast, 25 Shiga toxin Escherichia coli outbreaks have been traced to lettuce (FDA). Previously, we examined the ability of S. enterica to colonize and survive on a variety of significant agricultural crop plants, including those that have previously been implicated in salmonellosis outbreaks (e.g., cilantro and tomato) and those not implicated (e.g., broccoli and radish) (1). We discovered that plants from seeds sown in contaminated soil became colonized with S. enterica, regardless of the plant type. However, among tomato cultivars, contamination rates varied. This differential level of S. enterica colonization among tomato cultivars suggests that plant genes may affect S. enterica survival on these plants. We hypothesize that plants which can limit or reduce S. enterica populations are less likely to be associated with salmonellosis outbreaks.
To test this hypothesis, we examined whether the S. enterica colonization differences we previously discovered among tomato cultivars are dependent upon the bacterium's arrival route by examining exposure via soil and water. In addition, we expanded our study of S. enterica population establishment to include two well-studied parental tomato lines, Solanum pimpinellifolium West Virginia 700 (WVa700) and S. lycopersicum Hawaii 7996 (H7996) (6, 27, 32). Finally, we identified preferred S. enterica colonization sites on tomato leaves and investigated the capability of S. enterica tomato phyllosphere populations to contaminate fruit.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The strains used in this study are listed in Table 1. Bacteria were plated on LB medium (Difco, Becton Dickinson, Franklin Lakes, NJ) from glycerol stocks. Plating onto Salmonella-Shigella agar (SS), a Salmonella semiselective indicator medium, was used to determine S. enterica populations (Difco). Pseudomonas syringae pv. tomato was grown on King's B medium (18). Plasmid pKT-kan, in which a 131-bp nptII promoter fragment from Tn5 was fused to the green fluorescent protein gene of plasmid pPROBE-KT, is a stable, broad-host-range vector that confers kanamycin resistance and constitutive green fluorescent protein expression (24). Plasmid pKT-kan was transformed into all of the S. enterica strains listed in Table 1. We used kanamycin at 50 μg/ml for S. enterica and rifampin at 25 μg/ml for P. syringae pv. tomato. Bacterial cultures were grown from −80°C glycerol stocks at 28°C (P. syringae pv. tomato) for 30 h or at 37°C (S. enterica) for 24 h. Bacterial cultures were suspended in sterile water to an optical density of 0.2 (S. enterica) or 0.4 (P. syringae pv. tomato), which approximates 108 CFU/ml of each strain. S. enterica strains were always inoculated as an eight-strain cocktail.
TABLE 1.
Bacterial strains and tomato cultivars used in this study
| Bacterial strain or tomato cultivar | Strain or cultivar characteristic(s) | Source or referencea |
|---|---|---|
| Salmonella enterica serovars | ||
| Baildon | 99A 23 | CSHL-Berkeley |
| Cubana | 98A 9878 | 25 |
| Enteritidis | 05x-02123 | CSHL-Berkeley |
| Havana | 98A 4399 | CSHL-Berkeley |
| Mbandaka | 99A 1670 | CSHL-Berkeley |
| Newport | 96 E01153c-TX | CSHL-Berkeley |
| Poona | 00A 3563 | CSHL-Berkeley |
| Schwarzengrund | 96 E01152c-TX | CSHL-Berkeley |
| Pseudomonas syringae pv. tomato | SM78-1Sm | 36 |
| Solanum pimpinellifolium WVa700 | D. Maxwell (32) | |
| Solanum lycopersicum cultivars or accession numbers | ||
| H7996 | D. Maxwell (32) | |
| Yellow Pear | BCHSC | |
| Nyagous | BCHSC | |
| LA2838A | TGRC | |
| LA3172 | LA2838A mutant, h | TGRC |
| LA3556 | LA2838A mutant, hl | TGRC |
| LA0337 | TGRC | |
| LA1049 | LA0337 mutant, af | TGRC |
| Micro-Tom | 29 | |
| Money Maker |
CSHL, California State Health Laboratory; BCHSC, Baker Creek Heirloom Seed Co., Mansfield, MO; TGRC, C. M. Rick Tomato Genetics Resource Center, Davis, CA.
S. enterica colonization of tomato cultivars via soil.
S. pimpinellifolium cultivar WVa700 and S. lycopersicum cultivars H7996, Yellow Pear, and Nyagous were used in these studies (details in Table 1). Soil was contaminated and tomato plants were directly seeded as described previously (1). Briefly, surface-sterilized tomato seeds were planted in potting mix (Metro Mix; Sun Gro Horticulture, Bellevue, WA) that had been soaked to field capacity the previous day with an S. enterica cocktail at 108 CFU/ml (or 1010 CFU/ml as indicated as a higher level). Four seeds were planted per 9-cm pot and then watered with 40 ml distilled water every other day by automated drip hoses to each pot (∼118 g). Each hose was directed away from the plant, and the water flow was kept low to avoid splashing of soil onto aboveground plant parts. Eight pots were used per cultivar, and one pot whose soil had been soaked with water served as a control. All pots were randomized throughout the growth chamber, and subsequent analysis was performed on the data as a completely randomized design. When plants were collected, they were severed directly below the cotyledons, with extreme care being taken to not touch the soil, and placed into a microcentrifuge tube containing 500 μl phosphate-buffered saline (PBS). The phyllosphere was ground, and 100 μl of the homogenate was plated directly onto SS with kanamycin (SS-kan) for population enumeration.
Ten plants per cultivar were harvested on days 10, 12, and 14 after planting, along with one control plant. In order to verify that the S. enterica soil populations were stable across pots and across multiple days, soil samples were taken the same day as plant samples. An aliquot of a sample was also enriched in LB-kan to determine if S. enterica was present but below the detection limit for direct plating of homogenates onto growth medium. A loop was used to streak the enrichment sample onto SS-kan plates to verify the presence or absence of S. enterica. S. enterica was identified on the semiselective SS plates by its characteristic black colony morphology.
S. enterica colonization of tomato cultivars via water.
The same tomato cultivars were used in both the soil and water inoculation experiments. Four seeds were sown per pot in Metro Mix medium and allowed to germinate for 12 days in a climate-controlled growth chamber. Eight pots were sown for each cultivar, with one used as a control. Plants were inoculated by dipping each pot with four seedlings into either water, as a control, or an S. enterica cocktail suspension (each strain at ∼108 CFU/ml) for 30 s. Plants were allowed to dry and placed back into the climate-controlled growth chamber. One plant was sampled per pot by the methods described above for the soil soak experiment for each cultivar at 2, 4, 6, and 8 days after inoculation. Dilutions were made for each plant and plated for population enumeration. Weight was used as a normalization factor to determine the number of CFU/g of tissue.
Four-week-old plants of WVa700 and H7996 were also used to test the ability of S. enterica to colonize each tomato cultivar. After 4 weeks of growth, the plants were dip inoculated with an S. enterica cocktail as described above. One plant per cultivar was dipped in sterile water as a control. Plants were allowed to air dry before being placed into the climate-controlled growth chamber. Plants were watered each day with 130 ml of distilled water by an automated drip tube placed in each pot directed away from the plant to avoid splashing. Two leaf discs (5 mm) were excised from individual leaflets from two plants per cultivar 7 and 14 days after inoculation. Leaf discs from the same plant were placed into a tube with 500 μl water. Leaf discs were homogenized, and aliquots were serially diluted and plated onto SS-kan for population enumeration.
Tomato cultivar susceptibility to P. syringae pv. tomato.
The same tomato cultivars were used in these experiments as in the water inoculation experiments, with the addition of S. lycopersicum cultivar Money Maker. Three-week-old tomato plants were dip inoculated in a 500-ml suspension of P. syringae pv. tomato at 108 CFU/ml for approximately 30 s. Plants were allowed to air dry (∼1 h) and placed into the climate-controlled growth chamber. Disease severity was determined by counting the lesions on two compound leaves of the same age and by counting all of the lesions per plant. Subsequent assays examined P. syringae pv. tomato populations following dip inoculation with a small population, 105 CFU/ml. P. syringae pv. tomato populations were enumerated by leaf disc homogenization as described above for S. enterica leaf population enumeration.
S. enterica contamination of fruit from leaves.
S. lycopersicum cultivar Mico-Tom was dip inoculated at the three-leaf stage as described above. At 1 and 7 days postinoculation (dpi), one leaf per plant was sampled as described above for S. enterica leaf population enumeration. Plants were watered each day by an automated drip tube at a low rate and placed in each pot directed away from the plant to avoid splashing. When fruit reached full maturity at the red stage, it was carefully removed from the plant with a sterile razor blade without making contact with the leaf surface. Fruits were weighed and placed in a Nasco Whirl-Pak sample bag (Nasco, Fort Atkinson, WI), and an equivalent amount of water was added. The fruit was then massaged by hand for 1 min, making sure not to break the surface of the tomato skin. Serial dilutions of samples were plated on SS-kan for S. enterica population enumeration. A 100-μl aliquot was enriched as described above to identify fruit that was contaminated with S. enterica whose populations were below enumeration levels.
Direct fruit inoculation with S. enterica.
Both ripe red and green tomatoes of cultivars WVa700 and H7996 were harvested on the same day. Tomatoes were stored at room temperature overnight. A 500-ml suspension of eight S. enterica strains at 108 CFU/ml was used for inoculation. Whole fruits, with the pedicel and calyx still attached, were placed in a beaker with the S. enterica solution and vigorously stirred for 2 min with a stir bar to ensure that the entire sample was submerged. Fruits were then removed from the solution and placed in a biological safety cabinet to dry completely in moving air for 1.25 h. All fruits were then placed into plastic boxes along with a beaker of water in order to keep the humidity stable. The boxes were stored at 18°C for 1 h, 2 days, or 7 days.
Fruits, pedicels, and calyxes were either tested intact or separated into fruit alone and pedicel and calyx for their S. enterica population sizes. The pedicel and calyx were placed in a microcentrifuge tube and weighed. The weight was multiplied by 5, and an equivalent amount of PBS was then added to each microcentrifuge tube. Tubes containing the pedicel and calyx were vortexed for 30 s, samples were serially diluted, and aliquots were plated on SS-kan for S. enterica population enumeration. The surface area of each fruit was determined by measuring the circumference. The circumference of each fruit was measured at the point of largest girth. The fruit and water (milliliters) equivalent to the fruit's surface area were added to a Whirl-Pak bag. Fruits were treated as described above. When the pedicel and calyx were removed, all fruit were partially broken open during sampling to standardize the method since the red WVa700 fruits, but not any of the green or H7996 red fruits, tended to rupture when the pedicel and calyx were removed.
Stomata and trichomes.
S. lycopersicum strains with accession no. LA2838A, LA3172, LA3556, LA0337, and LA1049 (details in Table 1) were examined for S. enterica leaf population survival. Ten-day-old plants with at least two true leaves were dip inoculated in an S. enterica cocktail as described above. S. enterica populations were enumerated on SS-kan at 2, 7, and 14 dpi. Ten-millimeter leaf discs were excised from fully expanded leaves chosen randomly. Two leaves per plant and four plants were sampled at each date. Leaf discs were placed in 500 μl of sterile water and homogenized. The homogenate was serially diluted and plated on SS-kan for S. enterica population enumeration.
Statistics.
The initial study of S. enterica contamination of tomato cultivars via soil (1) indicated that, on average, the different heirloom cultivars were contaminated at a rate of 29.6%. If the two cultivars with a significantly lower level of contamination, Yellow Pear and Nyagous, are removed, then the remaining cultivars are contaminated, on average, at a rate of 35%. These two rates (29.6 and 35%) give us a baseline probability of the contamination of tomato plants from an inoculated soil source and were used in our binomial distribution calculations. S. enterica population data were log transformed prior to mean calculations. All means were tested for significance using Duncan's multiple-range test provided by SAS (v 9.1; SAS, Inc., Cary, NC).
Microscopy.
S. lycopersicum cultivar Money Maker was dip inoculated as described above. At 5 dpi, leaves were examined microscopically with an Olympus BX-60 epifluorescence microscope (Opelco, Dulles, VA) equipped with separate excitation filters to identify preferred colonization sites on tomato leaves. Centimeter squares were excised from two leaflets of three plants. Leaf tissue was mounted on microscopic slides and examined for green fluorescence from bacteria. Fluorescent cell aggregates were photographed with an Olympus digital camera (Optronics, Goleta, CA) using Image-Pro Plus capture software. The trichome type was identified by visual comparison to previous descriptions (20).
S. lycopersicum cultivars WVa700 and H7996 were examined for stoma and type I glandular trichome density by light microscopy with an Axioscope A1 (Zeiss, Feldbach, Switzerland). Images were captured with an AxioCam MRc digital camera (Zeiss) and using AxioVision release 4.7 (Zeiss) capture software. Fully emerged leaves of 2-week-old plants were examined for stoma and trichome enumeration. One leaflet was chosen from five plants of each cultivar. Leaflets were oriented by the leaflet apex positioned 90° to the left of the microscope. Stomata were counted from the abaxial leaf surface. Five microscopic fields of view (×30 magnifications) were randomly chosen to exclude the midvein, but the field was filled with leaf tissue. Depth of view was manipulated with the fine focus control only to confirm stoma presence by visual confirmation of both guard cells and stomatal apertures for each individual. Type I trichomes were visually recognized by comparison with those previously reported (17). Whole, not broken, trichomes were counted along the leaf margin of ∼11 mm.
RESULTS
S. enterica population levels on tomato leaves are cultivar dependent.
Plants were grown from seeds planted in S. enterica-infested soil and sampled for S. enterica. S. enterica populations were below the level of enumeration; thus, contamination incidence was determined from enriched plant samples. A total of 30 plants were sampled from H7996 and 11 were contaminated with S. enterica (37% contamination incidence [CI]). Only 3 WVa700 plants out of 30 were contaminated (10% CI). H7996 had a CI similar to that of all of the tomato cultivars tested by Barak et al. in 2008 (1) (P = 0.106), while WVa700 had a significantly lower level of S. enterica contamination (P = 0.008). This experiment was repeated using a higher initial S. enterica concentration, with similar results (H7996, P = 0.19; WVa700, P = 0.02).
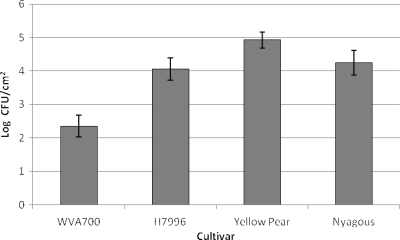
Plants were inoculated with contaminated water to simulate overhead sprinkler or pesticide application. The tomato cultivar had a significant effect on bacterial population levels (P < 0.001) across the three replicates of the contaminated-water experiment. Within each day, WVa700 always had significantly smaller S. enterica populations than the other three cultivars, similar to the data averaged across all 4 sampling days (Fig. 1). No significant difference was observed between the log S. enterica values for each cultivar across days for the first experimental replicate, meaning that the levels were relatively constant (P = 0.1072). However, for the remaining two replicates, the largest S. enterica population was detected 2 dpi and subsequently declined for the remainder of the sampling period (data not shown).
FIG. 1.
Mean S. enterica phyllosphere populations on tomato plants of different cultivars. Two-week-old tomato plants of S. pimpinellifolium cultivar WVa700 and S. lycopersicum cultivars H7996, Yellow Pear, and Nyagous were dip inoculated with an S. enterica cocktail. Plants were sampled at 2, 4, 6, and 8 dpi. Bacterial counts were log transformed, and populations were calculated as log CFU/cm2 across four sampling dates. The bars show the means across the 4 days, and the error bars represent the standard deviation of the mean.
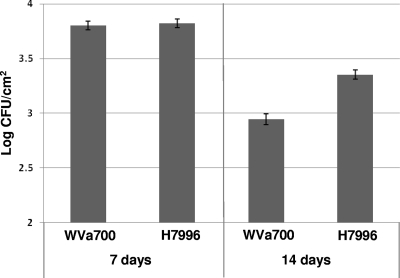
Plants were inoculated at 4 weeks old to investigate if plant age would affect differences among the S. enterica populations on the two tomato cultivars. Tomato cultivars had significantly different bacterial populations (P = 0.049) at 14 dpi but not at 7 dpi (Fig. 2). Incubation of plants at 95% humidity allowed S. enterica to maintain larger populations on both cultivars compared to plants incubated at 75% humidity (data not shown). At 24 h postinoculation, S. enterica populations on leaves following dip inoculation did not differ among cultivars or between young and old plants (2 versus 4 weeks).
FIG. 2.
Mean S. enterica phyllosphere populations of tomato plants. Four-week-old tomato plants of S. pimpinellifolium cultivar WVa700 and S. lycopersicum cultivar H7996 were dip inoculated with an S. enterica cocktail. Plants were sampled at 7 and 14 dpi. Bacterial counts were log transformed, and populations were calculated as log CFU/cm2. Error bars represent the standard deviation of the mean.
S. enterica preferentially colonizes trichomes.
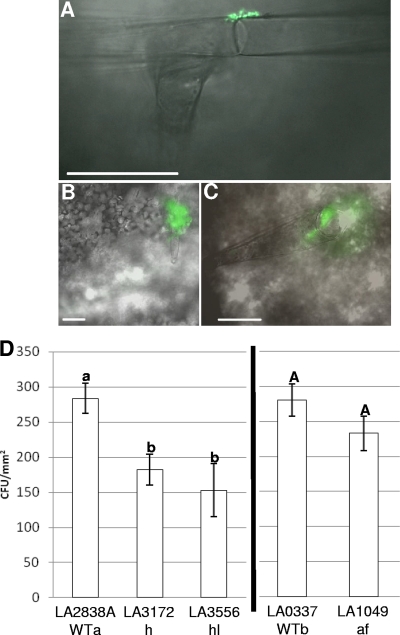
To identify preferred S. enterica colonization sites on tomato leaves, we dip inoculated tomato plants to simulate contaminated irrigation or pesticide water and examined them microscopically. We found S. enterica exclusively at the base of or on trichomes (Fig. 3 A to C). We identified type I glandular trichomes as the preferred tomato leaf colonization site by comparison of tomato trichomes described previously (20). To determine the role of trichomes in S. enterica leaf population survival, we examined populations among tomato trichome mutants. S. enterica populations were significantly smaller on LA3172 (h) and LA3556 (hl) than on the wild type (LA2838A). S. enterica population levels were the same on the wild type (LA0337) and LA1049 (af) (Fig. 3D). To determine whether physical traits such as trichomes play a role in the differential S. enterica colonization of WVa700 and H7996 observed, the density of trichomes was measured. WVa700 (6.0 ± 1.1) had significantly fewer type 1 glandular trichomes along the leaf margin than H7996 (9.5 ± 1.8; P = 0.0001), and this trend was also observed across the entire leaf (data not shown).
FIG. 3.
Preferential S. enterica colonization of type 1 trichomes. (A to C) Micrographs of S. enterica(pKT-kan) colonizing the type 1 trichomes of S. lycopersicum cultivar Money Maker. The scale bars represent 100 μm. (D) Mean S. enterica populations on the wild type (LA2838A and LA0337) and mutants LA3172 (h), LA3556 (hl), and LA1049 (af). LA2838A is labeled WTa, as it is the genetic background of trichome mutants with accession numbers LA3172 and LA3556; LA0337 is labeled WTb, as it is the genetic background of the trichome mutant with accession number LA1049. Two-week-old S. lycopersicum tomato plants were dip inoculated with an S. enterica cocktail. Plants were sampled at 7 dpi. Populations were calculated as CFU/mm2. Error bars represent the standard deviation of the mean. Different letters represent significant differences between mean S. enterica population sizes.
Reduced bacterial speck lesion formation correlates with small S. enterica populations.
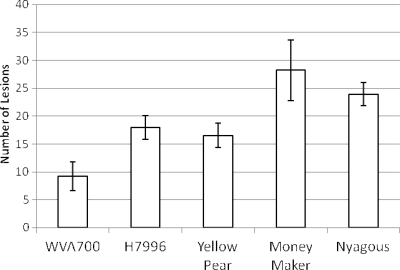
We examined the susceptibility of each tomato cultivar to bacterial speck of tomato. We inoculated each cultivar with P. syringae pv. tomato, the causal agent of bacterial speck of tomato, and visibly enumerated foliar lesions. All of the cultivars tested were susceptible to P. syringae pv. tomato, and lesions were visible on the cultivars tested (WVa700, H7996, Yellow Pear, Money Maker, and Nyagous). However, we observed a significant difference in the number of lesions among the cultivars (Fig. 4). Cultivars Yellow Pear and Money Maker had significantly more lesions (P < 0.001), while H7996 and Nyagous had fewer. Overall, WVa700 had the fewest lesions. This trend held for the total lesion number per plant (data not shown).
FIG. 4.
Mean bacterial speck lesions on two compound leaves of tomato plants. Three-week-old S. pimpinellifolium cultivar WVa700 and S. lycopersicum cultivar H7996, Yellow Pear, Money Maker, and Nyagous tomato plants were dip inoculated with a P. syringae pathovar tomato suspension. Bacterial speck lesions were enumerated at 5 to 7 dpi. Error bars represent the standard deviation of the mean.
To investigate a mechanism for the difference in bacterial speck lesion formation between WVa700 and H7996, we examined both P. syringae pv. tomato population survival and the physical traits of the leaves. P. syringae pv. tomato populations were larger on WVa700 (2.56 ± 0.26 CFU/cm2) than on H7996 (1.42 ± 0.32 CFU/cm2; P = 0.02) at 1 week after inoculation with 105 CFU/ml. To determine whether physical traits such as stomata play a role in the differential bacterial speck lesion formation observed between WVa700 and H7996, the density of stomata was measured. WVa700 (302 ± 91) had a significantly higher stoma density than H7996 (196 ± 37; P = 0.000001).
S. enterica leaf colonization leads to fruit contamination.
To determine if S. enterica in the phyllosphere can colonize fruit, we dip inoculated tomato plants (cultivar Micro-Tom) prior to flower development. Plants were left undisturbed throughout the experiment to avoid cross contamination of fruit by human intervention. Eleven fruits were collected from four plants; S. enterica was isolated from the surfaces of all 11 fruits. S. enterica populations per fruit ranged from below the enumeration limit (confirmed by enrichment) to 4.7 × 105 CFU.
Tomato pedicels and calyxes harbor large S. enterica populations.
WVa700 and H7996 fruits were dip inoculated to simulate deposition in an S. enterica-contaminated dump tank. First we investigated the whole fruit with the pedicel and calyx remaining attached during sampling (Table 2). There was no significant difference in S. enterica populations between red fruits of cultivar WVa700 and H7996 or green fruits at 2 dpi. Population levels were also not significantly different between either red fruits of WVa700 and H7996 (log10 3.77) or green fruits (log10 3.69). Also, no significant differences were observed after examining the interaction effect between cultivar and color on whole fruits with the pedicel and calyx attached.
TABLE 2.
S. enterica populations on tomato fruit
| Cultivar and fruit color | Mean log10 CFU ± SD |
||||
|---|---|---|---|---|---|
| 2 dpi |
Fruit |
||||
| Fruit, pedicel, calyx | Pedicel, calyx | Fruit | 7 dpi | 1 hpi | |
| H7996 | |||||
| Red | 3.8 ± 0.1 | 5.5 ± 0.1 | 3.7 ± 0.1 | 4.7 ± 0.8 | NTa |
| Green | 3.7 ± 0.1 | 5.6 ± 0.1 | 2.8 ± 0.1 | 3.1 ± 1.3 | NT |
| WVa700 | |||||
| Red | 3.7 ± 0.1 | 5.2 ± 0.2 | 3.7 ± 0.1 | 3.1 ± 1.6 | 2.2 ± 0.6 |
| Green | 3.7 ± 0.1 | 5.4 ± 0.1 | 2.9 ± 0.1 | 1.3 ± 1.4 | 1.2 ± 0.2 |
NT, not tested.
When the pedicel and calyx were removed from the fruits and analyzed alone, there was no significant difference between the population levels of S. enterica on the pedicels and calyxes from WVa700 or H7996 at 2 dpi. Fruit color also did not make a difference in S. enterica levels; the population levels were similar, at log10 5.34 and log10 5.55 for the pedicels and calyxes from red WVa700 and H7996 fruits and green fruits, respectively. No significant differences were found by examining the interaction between the cultivar and color effects on the pedicel and calyx S. enterica populations. However, S. enterica populations were significantly different between the pedicel and calyx and fruit overall and for the variables of cultivar and color.
WVa700 green fruits harbor the smallest S. enterica populations.
When just the fruits were analyzed without the pedicels and calyxes, color was significant at 2 dpi. Red fruits of WVa700 and H7996 combined had a higher level of S. enterica (log10 3.69), and green fruit had around half a log less (log10 2.91). We also examined S. enterica population survival at 7 dpi and found that when fruits alone were analyzed there was no significant difference between either cultivar or color. The population trend was smaller S. enterica populations on WVa700 red and green fruits (log10 2.16) than on H7996 (log10 3.94) and on green fruits of the combined cultivars (log10 1.80) than on red fruits (log10 3.64). However, significant differences were found by examining the interaction between the cultivar and color effects, with WVa700 green fruit having the smallest S. enterica population and H7996 red fruit having the largest (Table 2). To determine the role of initial attachment, WVa700 fruit was assayed 1 h postinoculation and green fruit had a significantly smaller S. enterica population than red fruit.
DISCUSSION
The plant itself has been mostly ignored in the study of human pathogens' association with plants. In this study, we found differential levels of S. enterica populations among tomato cultivars. Tomato plants were colonized by S. enterica following direct seeding in contaminated soil (1) or exposure to contaminated water (this study). Direct seeding in S. enterica-infested soil resulted in low plant colonization rates (10 to 40%), although potting mix may support little microbial competition, unlike an agricultural soil environment. These results suggest that although tomato plants directly seeded into S. enterica-infested soil may become colonized, the risk of the edible tomato fruit becoming contaminated in this manner is probably low. The risk of tomato fruit contamination is most likely greater following direct introduction of S. enterica into the phyllosphere or fruit via contaminated irrigation or pesticide water.
Leaves, pedicels, calyxes, and fruits were readily colonized by S. enterica from contaminated water. We found low variance among S. enterica levels in the tomato phyllosphere, although these samples were randomly chosen leaves that included dip-inoculated and recently emerged leaves. These results support the previously drawn conclusion that S. enterica can actively colonize the phyllosphere (8). WVa700 plants failed to be colonized to the same level as H7996. This difference in phyllosphere S. enterica populations is not an attachment difference, as similar populations were found among cultivars at 24 h postinoculation. This differential level of S. enterica contamination among tomato cultivars suggests that plant genetic traits can permit or inhibit colonization by enteric human pathogens. Since younger leaves have been described as more susceptible to plant pathogens (11), we examined whether S. enterica survival would differ between WVa700 and H7996 in older plants. S. enterica populations were smaller on WVa700 leaves, regardless of plant age at inoculation. In general, larger S. enterica populations were recovered from 4-week-old plants than from 2-week-old plants but S. enterica was unable to sustain substantial populations on the leaves of WVa700. The genes and mechanisms that keep some tomato cultivars comparatively free of S. enterica remain to be discovered.
Since tomato leaves are seldom eaten, we investigated whether fruit became colonized from S. enterica phyllosphere populations. Indeed, S. enterica was isolated from every fruit of plants with phyllosphere populations of S. enterica. These plants were kept in an undisturbed environment to minimize outside factors that could facilitate the transfer of S. enterica from leaf to fruit. In a real-world setting, where leaves often come in direct contact with fruit or bacterial dispersal mechanisms such as wind and water, S. enterica phyllosphere populations may easily lead to fruit contamination. In this study, large S. enterica populations were readily isolated from the surface of red fruit more than 1 month after plant exposure. Controlling S. enterica phyllosphere populations may lead to reduced fruit contamination. Our results support the conclusion that S. enterica phyllosphere populations are a risk factor for tomato fruit contamination and subsequent human disease.
How S. enterica phyllosphere populations contaminate tomato fruit is unknown. However, seed contamination has been reported following S. enterica root inoculation (8). Cooley et al. reported rapid S. enterica movement from the inoculation point along the Arabidopsis thaliana root exterior to the crown and slower progression in the phyllosphere to the plant apex and seed. The S. enterica movement was flagellum mediated. We hypothesize that tomato fruit contamination from S. enterica phyllosphere populations occurs in a similar manner. Future studies of S. enterica plant colonization mechanisms will examine the similarity of movement from the tomato phyllosphere to fruit as reported for A. thaliana from root to seed.
The specific plant traits that facilitate colonization by human enteric pathogens are largely unknown, but certain morphological traits appear to be important. Stomata serve as a gateway to the leaf interior for some foliar phytobacterial pathogens. Bacterial plant pathogens such as P. syringae pv. tomato and Xanthomonas campestris pv. campestris override stoma closure, a plant defense mechanism against infection, and invade the leaf interior through this opening (12, 23). Without access to the leaf apoplast, bacteria struggle to survive the restricted nutrients, UV radiation, and drought of the leaf surface (3, 26). S. enterica fails to grow significantly in the phyllosphere of agricultural plants, although it can survive for extended periods (14, 15). However, S. enterica grows in the tomato phyllosphere in the presence of Xanthomonas vesicatoria, the causal agent of bacterial spot of tomato, in the absence of plant disease (2). The mechanism that allows S. enterica growth has not been identified. S. enterica may access the substomatal chamber with X. vesicatoria during the plant pathogen's infection cycle. In the absence of plant pathogens, S. enterica colonizes the intercellular margins and stomata of cut lettuce leaves (19, 35). E. coli O157:H7 can survive bleach treatment of lettuce because bacterial cells are protected in cuticle cracks and on trichomes, possibly in aggregates (31). However, it is unclear whether this happens on living plants. We studied initial attachment and colonization sites of S. enterica on tomato leaves. To our surprise, S. enterica was rarely found near stomata. S. enterica overwhelmingly preferred to initiate colonization on trichomes of tomato leaves.
S. enterica initiates leaf colonization on glandular trichomes of tomato plants. Tomato plants have seven types of glandular trichomes (20). Glandular trichomes have developed on plants as a physical barrier to small herbivorous insects and chemical deterrents. Exudates from glandular trichomes can entrap or poison insects and act as a source of nutrients for bacteria (34). Some tomato glandular trichomes facilitate phytobacterial survival (30). Under dry conditions, P. syringae pv. tomato uses trichomes as a major habitat; however, whether the bacterium utilizes these trichomes for their ability to retain moisture or their ability to release nutrients remains unknown. Tomato mutants that lack one or more types of trichomes support relatively low levels of P. syringae pv. tomato during dry periods, whereas wild-type plants retain significantly larger populations. We found fewer S. enterica bacteria present 1 week after inoculation onto h and hl mutant cultivars, although similar numbers of S. enterica bacteria initially attached. The h mutant has few type I trichomes, the hl mutant has twisted and bent type I and VI trichomes, and the af mutant has few short trichomes (17). The leaf and stem of the hl mutant appear smooth due to the distorted type I and VI trichomes. Our data support the conclusion that type I glandular trichomes of tomato plants support S. enterica phyllosphere colonization. Plant-associated bacteria congregate at the base of trichomes to take advantage of an oasis that offers nutrients and moisture (3, 26). Leaf canopy architecture, which includes density of trichomes and their proclivity to retain leaf surface moisture, can contribute to quantitative resistance against phytopathogenic bacteria (28). Thus, we measured trichome density and found more type I glandular trichomes on WVa700 than on H7996. The heritable trait that limits S. enterica colonization in WVa700 remains to be determined.
We investigated additional WVa700 and H7996 traits to understand differences between these cultivars that might suggest mechanisms involved in the restriction of S. enterica colonization. We examined the development of bacterial speck disease, a well-studied foliar disease of tomato plants (22). Prior to infection, epiphytic populations of P. syringae multiply to greater than 105 CFU (13). Following this pathogen population buildup, P. syringae invades leaves and disease symptoms, lesions, appear. We observed fewer bacterial speck lesions on WVa700 leaves, than on H7996 leaves; however, a larger P. syringae pv. tomato population prior to symptom development was observed in WVa700. A tomato cultivar with fewer stomata may restrict P. syringae pv. tomato entry into the leaf apoplast, regardless of large populations on the leaf surface, and account for the appearance of fewer lesions. In contrast, WVa700 has a higher stoma density than H7996. Thus, points of entry are plentiful for P. syringae pv. tomato to invade WVa700. Our sampling method did not differentiate endophytic P. syringae pv. tomato populations from epiphytic populations. Although reduced bacterial speck lesion formation correlates with smaller S. enterica populations in the phyllosphere of WVa700, the mechanisms that control each event are most likely distinct.
S. enterica contamination of tomato fruit differs among cultivars. WVa700 green fruit had fewer S. enterica cells attach in our contaminated dump tank model experiment. This reduction in initial attachment was also apparent at 7 dpi in the survival of smaller S. enterica populations on WVa700 green fruit. Large-scale commercial fresh market tomatoes are harvested when the fruit is green; thus, specific genotypic traits of tomato plants, still unidentified, could reduce contaminated fruit in the food supply. Both the phyllosphere and fruits of WVa700 supported smaller S. enterica populations, suggesting that the pursuit of the heritable traits that restrict S. enterica survival may lead to a novel intervention strategy for the control of salmonellosis associated with tomatoes.
This study lays the groundwork for the discovery of heritable tomato traits that restrict S. enterica colonization of tomato plants and subsequent fruit contamination. Our results support the conclusion that contaminated water leads to larger S. enterica populations on tomato plants than does the contamination caused by infested soil. However, both routes of contamination result in detectable S. enterica populations in the phyllosphere. Phyllosphere S. enterica populations can lead to fruit contamination. Restricting S. enterica phyllosphere populations may result in reduced fruit contamination. The significant S. enterica populations harbored in the tomato pedicel and calyx of ripe tomatoes suggest that the pedicel and calyx may serve as a risk factor for cross contamination in food preparation areas, i.e., tomato fruit sold on the vine. We have identified WVa700 as a tomato cultivar that can restrict S. enterica survival in the phyllosphere. Screening of WVa700 × H7996 progeny is the next step toward the identification of heritable traits correlated with reduced S. enterica phyllosphere populations.
Acknowledgments
This work was partially supported by the Food Research Institute, University of Wisconsin—Madison.
The authors have no financial interest in cultivar WVa700.
We thank Russell Spear for fluorescent microscopic imaging; William Zaragoza for assistance with the P. syringae pv. tomato and initial attachment to fruit assays; and Eric Ewert, James Luo, and Steven Molinarolo for assistance with the trichome and stoma studies.
Footnotes
Published ahead of print on 12 November 2010.
REFERENCES
- 1.Barak, J. D., A. Liang, and K.-E. Narm. 2008. Differential attachment and subsequent contamination of agricultural crops by Salmonella enterica. Appl. Environ. Microbiol. 74:5568-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barak, J. D., and A. S. Liang. 2008. Role of soil, crop debris, and a plant pathogen in Salmonella enterica contamination of tomato plants. PLoS One 3:e1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beattie, G. A., and S. E. Lindow. 1999. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology 89:353-359. [DOI] [PubMed] [Google Scholar]
- 4.Bent, A. F., and D. Mackey. 2007. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45:399-436. [DOI] [PubMed] [Google Scholar]
- 5.Bidol, S. A., et al. 2007. Multistate outbreaks of Salmonella infections associated with raw tomatoes eaten in restaurants—United States, 2005-2006. MMWR Morb. Mortal. Wkly. Rep. 56:909-911. [PubMed] [Google Scholar]
- 6.Carmeille, A., et al. 2006. Identification of QTLs for Ralstonia solanacearum race 3-phylotype II resistance in tomato. Theor. Appl. Genet. 113:110-121. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2009. Surveillance for foodborne disease outbreaks—United States, 2006. MMWR Morb. Mortal. Wkly. Rep. 58:609-615. [PubMed] [Google Scholar]
- 8.Cooley, M. B., W. G. Miller, and R. E. Mandrell. 2003. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 69:4915-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corby, R., et al. 2005. Outbreaks of Salmonella infections associated with eating Roma tomatoes—United States and Canada, 2004. MMWR Morb. Mortal. Wkly. Rep. 54:325-328. [PubMed] [Google Scholar]
- 10.Cummings, K., et al. 2001. A multistate outbreak of Salmonella enterica serotype Baildon associated with domestic raw tomatoes. Emerg. Infect. Dis. 7:1046-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doster, M. A., and W. C. Schnathorst. 1985. Effects of leaf maturity and cultivar resistance on development of the powdery mildew fungus on grapevines. Phytopathology 75:318-321. [Google Scholar]
- 12.Gudesblat, G. E., P. S. Torres, and A. A. Vojnov. 2009. Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiol. 149:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano, S. S., and C. D. Upper. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam, M., et al. 2004. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl. Environ. Microbiol. 70:2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam, M., et al. 2004. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog. Dis. 1:27-35. [DOI] [PubMed] [Google Scholar]
- 16.Jones, J. D. G., and J. L. Dangl. 2006. The plant immune system. Nature 444:323-329. [DOI] [PubMed] [Google Scholar]
- 17.Kang, J.-H., F. Shi, A. D. Jones, M. D. Marks, and G. A. Howe. 2010. Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J. Exp. Bot. 61:1053-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 19.Kroupitski, Y., et al. 2009. Internalization of Salmonella in leaves is induced by light and involves chemotaxis and penetration through open stomata. Appl. Environ. Microbiol. 75:6076-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luckwill, L. 1943. The genus Lycopersicon: historical, biological, and taxonomic survey of the wild and cultivated tomatoes. Aberdeen University Press, Aberdeen, Scotland.
- 21.Mead, P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melotto, M., W. Underwood, and S. Y. He. 2008. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46:101-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melotto, M., W. Underwood, J. Koczan, K. Nomura, and S. Y. He. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126:969-980. [DOI] [PubMed] [Google Scholar]
- 24.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 25.Mohle-Boetani, J. C., et al. 2001. Escherichia coli O157 and Salmonella infections associated with sprouts in California, 1996-1998. Ann. Intern. Med. 135:239-247. [DOI] [PubMed] [Google Scholar]
- 26.Monier, J. M., and S. E. Lindow. 2004. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 70:346-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau, P., P. Thoquet, J. Olivier, H. Laterrot, and N. Grimsley. 1998. Genetic mapping of Ph-2, a single locus controlling partial resistance to Phytophthora infestans in tomato. Mol. Plant Microbe Interact. 11:259-269. [Google Scholar]
- 28.Niks, R. E., and D. Rubiales. 2002. Potentially durable resistance mechanisms in plants to specialised fungal pathogens. Euphytica 124:201-216. [Google Scholar]
- 29.Reeves, A. F. 1977. Tomato trichomes and mutations affecting their development. Am. J. Bot. 64:186-189. [Google Scholar]
- 30.Schneider, R. W., and R. G. Grogan. 1977. Tomato leaf trichomes, a habitat for resident populations of Pseudomonas tomato. Phytopathology 67:898-902. [Google Scholar]
- 31.Takeuchi, K., and J. F. Frank. 2001. Quantitative determination of the role of lettuce leaf structures in protecting Escherichia coli O157:H7 from chlorine disinfection. J. Food Prot. 64:147-151. [DOI] [PubMed] [Google Scholar]
- 32.Thoquet, P., et al. 1996. Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii7996. Mol. Plant Microbe Interact. 9:826-836. [Google Scholar]
- 33.Toth, B., et al. 2002. Outbreak of Salmonella serotype Javiana infections—Orlando, Florida, June 2002. MMWR Morb. Mortal. Wkly. Rep. 51:683-684. [PubMed] [Google Scholar]
- 34.Wagner, G. J., E. Wang, and R. W. Shepherd. 2004. New approaches for studying and exploiting an old protuberance, the plant trichome. Ann. Bot. 93:3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warner, J. C., S. D. Rothwell, and C. W. Keevil. 2008. Use of episcopic differential interference contrast microscopy to identify bacterial biofilms on salad leaves and track colonization by Salmonella Thompson. Environ. Microbiol. 10:918-925. [DOI] [PubMed] [Google Scholar]
- 36.Willis, D. K., and T. G. Kinscherf. 2009. Population dynamics of Pseudomonas syringae pv. tomato strains on tomato cultivars Rio Grande and Rio Grande-Pto under field conditions. J. Phytopathol. 157:219-227. [Google Scholar]