Abstract
A sequence-based analysis of seven housekeeping and virulence-related genes shows that the species Vibrio vulnificus is subdivided into three phylogenetic lineages that do not correspond with the biotypes and that biotype 2 is polyphyletic. These results support the reclassification of biotype 2 as a pathovar that would group the strains with pathogenic potential to develop vibriosis in fish.
Vibrio vulnificus is a pathogenic bacterial species that inhabits brackish waters in warm and tropical ecosystems (9, 11, 24). The species is highly heterogeneous and has been subdivided into three biotypes. Biotype 1 is the most abundant; it is distributed worldwide and causes sporadic cases of human vibriosis. Biotype 2 is also distributed worldwide, and it is the only one that harbors the genetic information to infect both fish and humans. Finally, biotype 3 is geographically restricted to Israel, and it causes outbreaks of human vibriosis after fish handling (1, 4, 13, 25). Biotype 2 is further subdivided into serovars A, E, and I (7; unpublished results), serovar E being the one associated with human vibriosis (1).
The genes essential for fish vibriosis are located in a recently described virulence plasmid that can be transmitted between strains cointegrated with a conjugative plasmid (12) that is present in almost 90% of biotype 2 isolates (17). In contrast, the genetic basis of human infection is poorly understood, since the putative virulence factors identified so far are found in both clinical and environmental isolates of the three biotypes (24, 27).
Several studies based on multilocus sequence typing (MLST) of housekeeping genes (2, 3, 6) and on ribotyping (21) suggest that the species is subdivided into two main evolutionary lineages with apparently different human pathogenic potential; one includes a majority of the human clinical isolates of biotype 1 (clinical branch), and the other a majority of the environmental isolates of the same biotype (environmental branch). The few isolates of biotype 2 studied are in the environmental branch, while biotype 3 strains are in a variable position depending on the study (2, 3, 6).
Given this scenario, the aim of this work has been to analyze the evolutionary origins of biotype 2, starting from the hypothesis that horizontal transfer of the virulence plasmid together with recombination events could have played a major role in the emergence of this biotype. To this end, a sequence-based analysis of three virulence-associated (vvhA, wzz, and pilF) and four housekeeping (glp, mdh, pyrC, and pntA) genes (the last ones selected from the MLST scheme for V. vulnificus [3]) was applied to a collection of 115 isolates that included strains of the three biotypes from clinical (humans and fish) and environmental sources (Table 1). The primer pairs for the genes are listed in Table S1 in the supplemental material. The genetic variability (θ) at the locus and biotype level was examined by using DnaSP4.09 (20). pilF and wzz (genes involved in surface antigen biogenesis) showed the highest levels of genetic variability (Table 2). Regarding the biotypes, biotype 1 showed the highest genetic variability, while biotype 2 was highly homogeneous, and no genetic diversity was observed among the biotype 3 isolates (see Table S2 in the supplemental material).
TABLE 1.
Characteristics of the V. vulnificus isolates used in this study
| Strain | Country and yr of isolation | Source | Biotypea and serovar | STb | No. of allelic sequencesb of: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| glp | pntA | pyrC | pilF | wzz | vvhA | mdh | |||||
| YJ106 | Taiwan | Human blood | BT1/3 | 1 | 1 | 1 | 30 | 18 | 1 | 21 | 5 |
| CECT 4608 | Spain, 1990 | Eel farm water | BT1/3 | 2 | 1 | 2 | 12 | 11 | 11 | 2 | 13 |
| BT3-1033 | Israel, 1996 | Diseased human | BT1/3 | 3 | 2 | 5 | 2 | 10 | 13 | 5 | 10 |
| BT3-11028 | Israel, 1996 | Diseased human | BT1/3 | 3 | 2 | 5 | 2 | 10 | 13 | 5 | 10 |
| BT3-12 | Israel, 1996 | Diseased human | BT1/3 | 3 | 2 | 5 | 2 | 10 | 13 | 5 | 10 |
| BT3-162 | Israel, 1997 | Diseased human | BT1/3 | 3 | 2 | 5 | 2 | 10 | 13 | 5 | 10 |
| BT3-32 | Israel | Diseased human | BT1/3 | 3 | 2 | 5 | 2 | 10 | 13 | 5 | 10 |
| BT3-97 | Israel, 1997 | Diseased human | BT1/3 | 3 | 2 | 5 | 2 | 10 | 13 | 5 | 10 |
| A13 | Spain, 2002 | Diseased eel | BT2 non-serovar E (serovar A) | 4 | 3 | 7 | 3 | 1 | 2 | 7 | 6 |
| CECT 5198 | Spain, 1999 | Diseased eel | BT2 non-serovar E (serovar A) | 5 | 3 | 7 | 3 | 1 | 2 | 7 | 8 |
| CECT 7029 | Denmark, 2004 | Diseased eel | BT2 non-serovar E (serovar A) | 5 | 3 | 7 | 3 | 1 | 2 | 7 | 8 |
| CECT 7030 | Denmark, 2004 | Diseased eel | BT2 non-serovar E (serovar A) | 5 | 3 | 7 | 3 | 1 | 2 | 7 | 8 |
| A10 | Spain, 2002 | Diseased eel | BT2 non-serovar E (serovar A) | 5 | 3 | 7 | 3 | 1 | 2 | 7 | 8 |
| CECT 5343 | Spain, 2000 | Diseased eel | BT2 non-serovar E (serovar A) | 6 | 3 | 7 | 3 | 1 | 2 | 7 | 28 |
| A11 | Spain, 2002 | Diseased eel | BT2 non-serovar E (serovar A) | 7 | 3 | 7 | 3 | 1 | 2 | 7 | 32 |
| CECT 5689 | Spain, 2002 | Diseased eel | BT2 non-serovar E (serovar A) | 8 | 3 | 7 | 17 | 1 | 2 | 7 | 29 |
| CECT 5768 | Spain, 2001 | Diseased eel | BT2 non-serovar E (serovar A) | 9 | 3 | 7 | 17 | 1 | 2 | 7 | 30 |
| CECT 5769 | Spain, 2002 | Diseased eel | BT2 non-serovar E (serovar A) | 10 | 3 | 7 | 17 | 1 | 2 | 7 | 31 |
| L49 | Japan | Brackish water | BT1/3 | 11 | 3 | 7 | 26 | 1 | 2 | 7 | 6 |
| PD-2-66 | Spain, 2003 | Eel tank water | BT1/3 | 12 | 3 | 8 | 26 | 25 | 28 | 15 | 8 |
| JE | USA | Oyster | BT1/3 | 13 | 3 | 15 | 13 | 2 | 24 | 1 | 25 |
| CECT 5166 | USA | Human blood | BT1/3 | 14 | 3 | 18 | 15 | 14 | 16 | 2 | 14 |
| CECT 4867 | Unknown | Diseased eel | BT1/3 | 15 | 4 | 22 | 31 | 26 | 3 | 15 | 33 |
| 534 | Sweden | Diseased eel | BT1/3 | 16 | 5 | 8 | 4 | 2 | 4 | 12 | 8 |
| CECT 4869 | Belgium, 1990 | Diseased eel | BT1/3 | 17 | 5 | 8 | 13 | 2 | 14 | 19 | 1 |
| A2 | Spain, 2000 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| An4 | Spain, 2000 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| An5 | Spain, 2000 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| An6 | Spain, 2000 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| An7 | Spain, 2000 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| PD-1 | Spain, 2001 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| PD-3 | Spain, 2001 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| PD-5 | Spain, 2001 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| V1 | Spain, 2001 | Eel tank water | BT1/3 | 18 | 6 | 11 | 4 | 2 | 11 | 2 | 7 |
| PD-12 | Spain, 2001 | Eel tank water | BT1/3 | 19 | 6 | 11 | 4 | 23 | 11 | 2 | 7 |
| CECT 4606 | Spain, 1990 | Healthy eel | BT1/3 | 20 | 6 | 11 | 4 | 4 | 3 | 2 | 8 |
| MLT 362 | USA | Environmental | BT1/3 | 21 | 7 | 4 | 21 | 8 | 25 | 16 | 33 |
| VV 1003 | USA | Environmental | BT1/3 | 22 | 8 | 23 | 28 | 27 | 29 | 14 | 24 |
| MLT 364 | USA | Environmental | BT1/3 | 23 | 9 | 24 | 24 | 8 | 9 | 16 | 8 |
| MLT 406 | USA | Environmental | BT1/3 | 24 | 10 | 19 | 26 | 2 | 3 | 2 | 1 |
| MLT404 | USA | Environmental | BT1/3 | 25 | 11 | 20 | 25 | 21 | 26 | 7 | 33 |
| ATCC 33816 | USA | Human blood | BT1/3 | 26 | 12 | 9 | 12 | 16 | 12 | 7 | 4 |
| CG100 | Taiwan, 1993 | Oyster | BT1/3 | 27 | 12 | 9 | 12 | 15 | 20 | 9 | 2 |
| V2 | Unkown | Environmental | BT1/3 | 28 | 12 | 9 | 12 | 16 | 12 | 13 | 4 |
| CG110 | Taiwan, 1993 | Seawater | BT1/3 | 29 | 12 | 9 | 19 | 15 | 12 | 4 | D3 |
| VV 352 | USA | Environmental | BT1/3 | 30 | 12 | 9 | 29 | 16 | 12 | 17 | 19 |
| CG111 | Taiwan, 1993 | Seawater | BT1/3 | 31 | 12 | 14 | 20 | 15 | 12 | 4 | 2 |
| CG118 | Taiwan, 1993 | Seawater | BT1/3 | 32 | 12 | 14 | 20 | 15 | 12 | 4 | 4 |
| CG106 | Taiwan, 1993 | Oyster | BT1/3 | 33 | 12 | 14 | 21 | 17 | 21 | 18 | 3 |
| CECT 5167 | Japan | Human blood | BT1/3 | 34 | 13 | 14 | 16 | 15 | 17 | 4 | 12 |
| VV 425 | USA | Environmental | BT1/3 | 35 | 13 | 21 | 4 | 18 | 31 | 5 | 27 |
| CECT 5164 | USA | Human blood | BT1/3 | 36 | 14 | 3 | 12 | 13 | 15 | 20 | 20 |
| CECT 5169 | USA | Human blood | BT1/3 | 37 | 15 | 7 | 4 | 17 | 19 | 11 | 22 |
| CS9133 | South Korea | Human blood | BT1/3 | 38 | 15 | 14 | 22 | 18 | 22 | 10 | 22 |
| KH03 | Japan, 2003 | Human blood | BT1/3 | 39 | 16 | 14 | 12 | 19 | 18 | 4 | 23 |
| E4 | USA | Seafood | BT1/3 | 40 | 16 | 14 | 21 | 19 | 18 | 4 | 26 |
| V4 | Australia | Human blood | BT1/3 | 41 | 17 | 14 | 27 | 18 | 30 | 22 | 23 |
| 94-9-118 | Denmark, 1994 | Expectoration from lungs | BT1/3 | 42 | 18 | 22 | 5 | 2 | 5 | 1 | 8 |
| G83 | South Korea | Fish | BT1/3 | 43 | 19 | 16 | 23 | 20 | 23 | 8 | 8 |
| CECT 5168 | USA | Human blood | BT1/3 | 44 | 20 | 7 | 4 | 16 | 18 | 4 | 21 |
| YN03 | Japan, 2003 | Human blood | BT1/3 | 45 | 21 | 13 | 9 | 12 | 32 | 7 | 7 |
| 94385 | Spain, 2001 | Leg wound | BT1/3 | 46 | 22 | 14 | 8 | 6 | 8 | 8 | 7 |
| 536 | Sweden | Diseased eel | BT2 non-serovar E | 47 | 23 | 10 | 3 | 3 | 2 | 7 | 8 |
| 535 | Sweden | Diseased eel | BT2 non-serovar E | 48 | 23 | 10 | 3 | 3 | 2 | 7 | 15 |
| CECT 5165 | USA | Seawater | BT1/3 | 49 | 23 | 21 | 14 | 14 | 3 | 2 | 8 |
| Riu-1 | Spain, 2003 | Seawater | BT1/3 | 50 | 24 | 16 | 8 | 20 | 7 | 8 | 7 |
| 94-9-130 | Denmark, 1994 | Water | BT1/3 | 51 | 25 | 8 | 7 | 5 | 7 | 1 | 8 |
| Riu-3 | Spain, 2003 | Seawater | BT1/3 | 52 | 25 | 12 | 3 | 24 | 12 | 1 | 8 |
| PD-2-58 | Spain, 2003 | Eel tank water | BT2 non-serovar E | 53 | 25 | 12 | 3 | 24 | 3 | 1 | 8 |
| PD-2-52 | Spain, 2003 | Eel tank water | BT2 non-serovar E | 54 | 25 | 12 | 3 | 24 | 27 | 1 | 8 |
| 94-9-119 | Denmark, 1994 | Wound infection | BT1/3 | 55 | 26 | 17 | 6 | 2 | 6 | 1 | 8 |
| A14 | Spain, 2002 | Diseased eel | BT2 non-serovar E | 56 | 27 | 7 | 3 | 1 | 2 | 7 | 8 |
| 95-8-161 | Denmark, 1995 | Diseased eel | BT2 non-serovar E (serovar I) | 57 | 28 | 7 | 9 | 7 | 2 | 7 | 16 |
| 95-8-162 | Denmark, 1995 | Diseased eel | BT2 non-serovar E (serovar I) | 58 | 28 | 7 | 9 | 7 | 2 | 7 | 17 |
| 95-8-6 | Denmark, 1995 | Diseased eel | BT2 non-serovar E (serovar I) | 59 | 28 | 7 | 10 | 7 | 2 | 7 | 8 |
| 95-8-7 | Denmark, 1995 | Diseased eel | BT2 non-serovar E (serovar I) | 60 | 28 | 7 | 11 | 8 | 2 | 7 | 8 |
| CIP 81.90 | France | Human blood | BT2 serovar E | 61 | 29 | 7 | 4 | 4 | 3 | 3 | 8 |
| CECT 4862 | Japan, 1979 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| 90-2-11 | Denmark, 1990 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| 94-8-112 | Denmark, 1994 | Wound infection | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| 94-9-123 | Denmark, 1994 | Seawater | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CCUG 38521 | Sweden, 1997 | Wound infection | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4174 | Japan, 1979 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4601 | Spain, 1989 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4602 | Spain, 1990 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4603 | Spain, 1990 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4605 | Spain, 1990 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4607 | Spain, 1992 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4863 | USA | Leg wound infection | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4864 | Spain, 1994 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4865 | Taiwan | Diseased shrimp | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4866 | Australia | Human blood | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4870 | Sweden, 1991 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4917 | Spain, 1997 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 897 | Japan, 1979 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 898 | Japan, 1979 | Diseased eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| PD-2-47 | Spain, 2003 | Eel tank water | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| PD-2-55 | Spain, 2003 | Eel tank water | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| PD-2-56 | Spain, 2003 | Eel tank water | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| Riu-2 | Spain, 2003 | Seawater | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| UE516 | Taiwan | Diseased Japanese eel | BT2 serovar E | 62 | 29 | 8 | 4 | 4 | 3 | 3 | 8 |
| CECT 4868 | Norway, 1990 | Diseased eel | BT2 serovar E | 63 | 29 | 8 | 4 | 4 | 3 | 3 | 10 |
| CECT 4999 | Spain, 1999 | Diseased eel | BT2 serovar E | 64 | 29 | 8 | 4 | 4 | 3 | 3 | 16 |
| C1 | Spain, 2003 | Healthy eel | BT2 serovar E | 65 | 29 | 8 | 4 | 4 | 3 | 3 | 17 |
| CECT 4604 | Spain, 1990 | Diseased eel | BT2 serovar E | 66 | 29 | 8 | 4 | 4 | 3 | 3 | 18 |
| CECT 4998 | Spain, 1997 | Diseased eel | BT2 serovar E | 66 | 29 | 8 | 4 | 4 | 3 | 3 | 18 |
| CECT 5139 | Spain, 1998 | Diseased eel | BT2 serovar E | 66 | 29 | 8 | 4 | 4 | 3 | 3 | 18 |
| PD-2-50 | Spain, 2003 | Eel tank water | BT2 serovar E | 67 | 29 | 8 | 4 | 4 | 3 | 3 | 32 |
| PD-2-51 | Spain, 2003 | Eel tank water | BT2 serovar E | 67 | 29 | 8 | 4 | 4 | 3 | 3 | 32 |
| CECT 5762 | Spain, 2002 | Healthy eel | BT2 serovar E | 68 | 29 | 8 | 18 | 4 | 3 | 3 | 17 |
| CECT 5763 | Spain, 2002 | Eel tank water | BT2 serovar E | 69 | 29 | 8 | 18 | 4 | 3 | 3 | 8 |
| CECT 529T | USA | Human blood | BT1/3 | 70 | 29 | 18 | 15 | 14 | 16 | 2 | 14 |
| 960717-1/2F | Denmark, 1996 | Diseased eel | BT2 non-serovar E | 71 | 30 | 6 | 4 | 9 | 6 | 1 | 11 |
| 960426-1/4C | Denmark, 1996 | Diseased eel | BT2 non-serovar E | 72 | 30 | 6 | 4 | 9 | 10 | 6 | 11 |
| N87 | Japan, 1987 | Human blood | BT1/3 | 73 | 31 | 4 | 1 | 22 | 33 | 4 | 9 |
BT, biotype.
Sequences from each locus were aligned using Vector NTI 9.0.0 software (Infomax) and were edited manually by visual inspection. Different allelic sequences within a locus were assigned arbitrary numbers. Each isolate was consequently given a 7-number sequence designated a sequence type (ST).
TABLE 2.
Genetic diversity parameters for the 4 housekeeping and 3 virulence-associated loci studied in 115 V. vulnificus isolates
| Locus | Chromosome | Sequence length | No. of haplotypes | Haplotype diversity | Nucleotide diversity (π) | Polymorphic sites (S) | θ (from S) | Pairwise nucleotide differences (k) |
|---|---|---|---|---|---|---|---|---|
| mdh | I | 489 | 32 | 0.82 | 8.5·10−3 | 23 | 0.01 | 4.2 |
| pilF | I | 480 | 27 | 0.87 | 3.3·10−2 | 57 | 0.025 | 15.72 |
| wzz | I | 460 | 33 | 0.84 | 2.7·10−2 | 61 | 0.027 | 12.65 |
| glp | I | 479 | 31 | 0.87 | 1.8·10−2 | 50 | 0.02 | 8.55 |
| pyrC | II | 423 | 31 | 0.79 | 1.6·10−2 | 39 | 0.02 | 6.99 |
| pntA | II | 396 | 24 | 0.85 | 1.5·10−2 | 33 | 0.018 | 5.95 |
| vvhA | II | 432 | 22 | 0.85 | 1.8·10−2 | 32 | 0.015 | 7.921 |
| Value for: | ||||||||
| All genes | 3,159 | 73 | 0.95 | 2·10−2 | 295 | 0.017 | 20.87 | |
| Chromosome I | 1,908 | 66 | 0.94 | 2.1·10−2 | 191 | 0.02 | 41.11 | |
| Chromosome II | 1,251 | 53 | 0.91 | 1.6·10−2 | 104 | 0.017 | 20.88 | |
| Housekeeping genes | 1,787 | 68 | 0.95 | 1.4·10−2 | 145 | 0.016 | 25.69 | |
| Virulence genes | 1,372 | 49 | 0.89 | 2.6·10−2 | 150 | 0.023 | 36.3 |
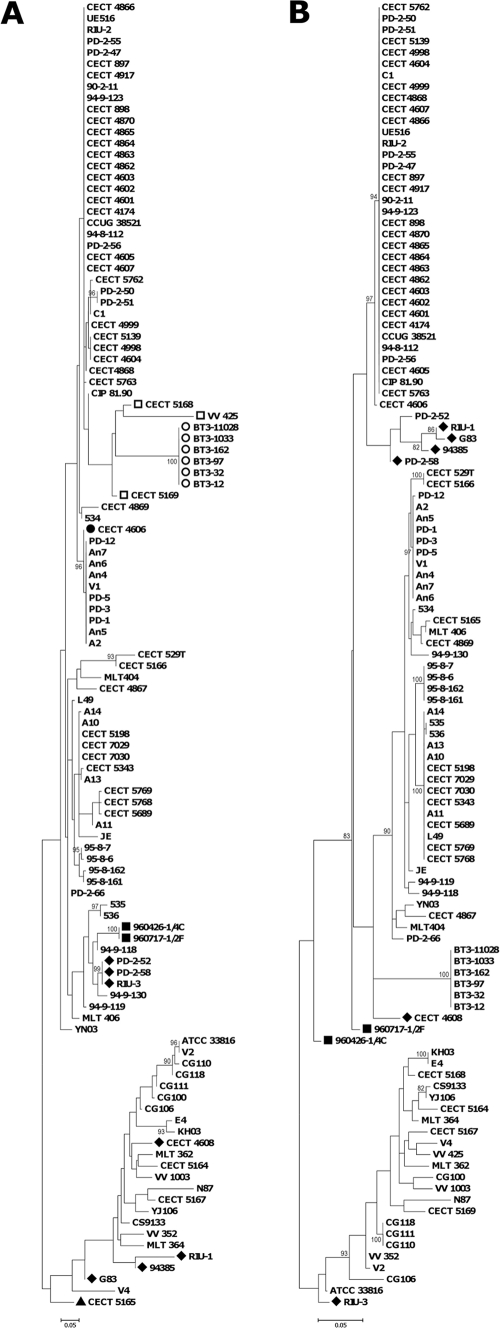
To analyze the phylogeny of the V. vulnificus collection, we constructed a maximum likelihood (ML) tree from the 3,159-bp concatenated sequence of the seven loci (Fig. 1) by using PHYML 2.4.4 (8). The most appropriate model for nucleotide substitution was assessed with Modeltest version 3.7 (16). The concatenated tree shows the isolates clustered into three main lineages (Fig. 1). Lineage I (LI) contained isolates of biotypes 1 and 2 from fish farms and isolates from diseased fish and humans infected through fish handling or water contact. This lineage is enriched in European isolates, probably because the fish-farming industry is especially developed in Europe, whose countries apply specific-pathogen-control programs. LII was formed by biotype 3 strains from Israel, and LIII included biotype 1 isolates mostly recovered from environmental samples or from human septicemic cases registered in the United States and Asia. The nucleotide diversity within each lineage was then examined, and it was found that LI and LIII have similar values (π in Table S2 in the supplemental material). The human isolates are genetically more diverse than those from environmental origins, and both are much more diverse than isolates from diseased animals. This result would suggest that multiple environmental clones have the ability to infect humans, which correlates with human cases being presented as sporadic infections worldwide, and that only a few clones are able to infect fish, although they are overrepresented by clone amplification after epizootics in fish farms. The exception would be the clone formed by biotype 3 isolates, the only ones capable of causing outbreaks of human vibriosis.
FIG. 1.
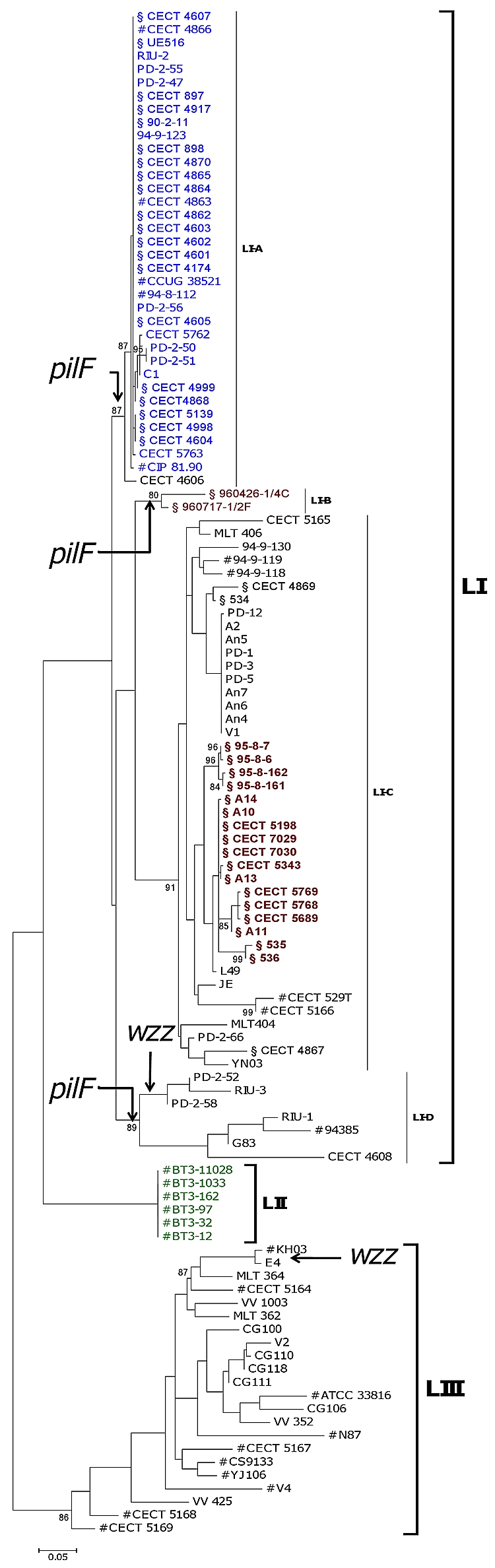
Maximum likelihood phylogenetic tree of 115 V. vulnificus isolates obtained from the alignment of 7 concatenated loci. Black, biotype 1 isolates; blue, biotype 2 serovar E isolates; red, biotype 2 non-serovar E isolates; green, biotype 3 isolates; #, human isolates; §, diseased fish isolates. Branches where recombination events involving the indicated loci might have occurred are indicated by arrows. The numbers at the nodes represent the percentage values given by bootstrap analysis of 1,000 replicates.
Furthermore, LI can be subdivided into four main groups, as follows: LI-A, grouping all biotype 2 serovar E isolates plus a Spanish biotype 1 isolated from a fish tank (CECT 4606); LI-B, formed by two atypical isolates of biotype 2 from Denmark; LI-C, clustering all non-serovar E biotype 2 isolates together with biotype 1 isolates from fish farms and humans; and lastly, LI-D, grouping biotype 1 isolates from the environment and humans (mainly from blood). Thus, biotype 2 is polyphyletic and appears to be divided into serovar-related subgroups, the isolates in each subgroup being more related to biotype 1 isolates from fish farms than to each other. This result is compatible with the hypothesis that biotype 2 emerged by acquisition of the virulence plasmid by V. vulnificus strains from fish farms.
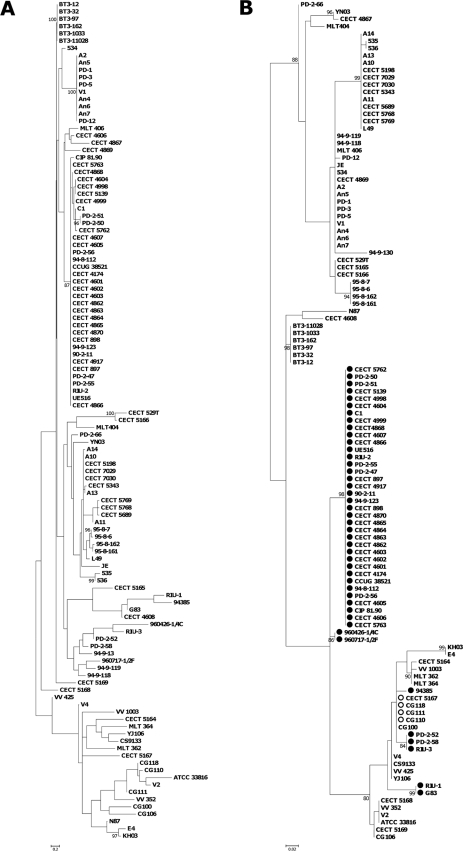
The groupings of strains changed when only virulence or housekeeping genes were considered (Fig. 2). In both cases, the position of biotype 3 changed, and this also occurred in the individual gene trees (Fig. S1 to S5 in the supplemental material). Previous studies of the phylogeny of V. vulnificus (based on different techniques, multilocus enzyme electrophoresis, MLST, or 16S rRNA or gene sequencing) divided the species into two main lineages, the clinical and the environmental lineages (5, 6, 10, 15, 19, 23, 26). In our study, the division in the two lineages was observed in the phylogenetic trees from virulence-related or from housekeeping genes but not in the concatenated phylogenetic tree. Thus, it can be concluded that combining four housekeeping and three virulence gene sequences in the analysis gives enough resolution to show biotype 3 as an independent lineage. Bisharat et al. (3) proposed that biotype 3 contains a mosaic genome that would have evolved by hybridization of genomes of representative strains of the other two lineages. Our results support this hypothesis, although the change of position in the trees affected not only biotype 3 isolates but also biotype 2 non-serovar E isolates and biotype 1 strains, mostly from fish-farming environments (Fig. 2; also see Fig. S1 to S5 in the supplemental material).
FIG. 2.
Maximum likelihood phylogenetic tree of 115 V. vulnificus isolates obtained from the alignment of 4 housekeeping (A) and 3 virulence-associated (B) concatenated loci. Symbols indicate isolates that cluster in LI-A (•), LI-B (▪), LI-C (▴), LI-D (⧫), LII (○), or LIII (□) in the concatenated tree. The numbers at the nodes represent the percentage values given by bootstrap analysis of 1,000 replicates.
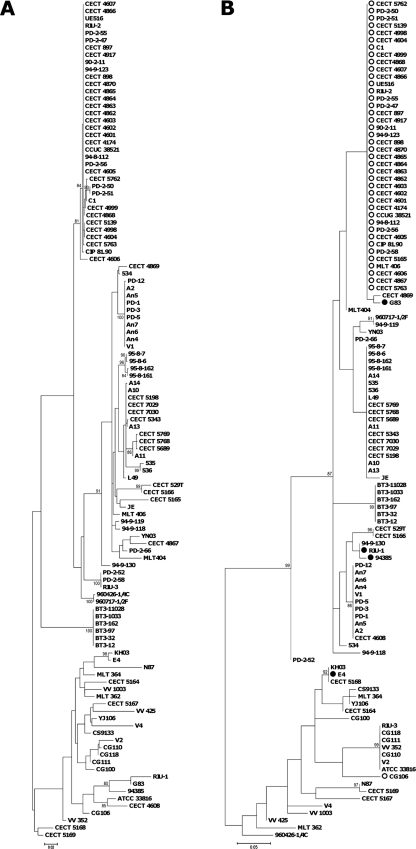
The Recombination Detection Program, version 3 (RDP3) (14) detected recombination in chromosome I by at least four of the implemented methods, involving two of the four loci (pilF and wzz). The putative recombinant events were mapped onto the phylogenetic tree of the seven concatenated loci (Fig. 1). To further corroborate the RDP3 results, two different ML trees were constructed for each recombinant locus. The tree obtained from the multiple sequence alignment of each locus was compared with the one obtained from the concatenated sequences of all the other loci by using TreePuzzle version 5.2 (22). In both cases, Shimodaira-Hasegawa (SH) and expected likelihood weight (ELW) tests revealed significant differences between the two topologies (Table 3). Figures 3 and 4 each show the phylogenetic tree of one of the loci involved in recombination (pilF and wzz) and the tree derived from the remaining aligned loci. In both cases, biotype 2 serovar E strains are involved in the recombination events, either as predicted parental strains, in wzz, or as daughter strains, in pilF. The other strains involved in recombination events are biotype 2 nontypeable strains and clinical and environmental biotype 1 isolates. This result suggests that the conditions of aquaculture settings (e.g., high nutrient loads and high host density) might favor the exchange of genetic material among strains of V. vulnificus, originating new variants in the V. vulnificus species.
TABLE 3.
Likelihood scores for the loci identified as involved in recombination eventsa
| Alignment | −lnL for ML tree of the: |
|
|---|---|---|
| Locus | Concatenated loci | |
| pilF | −3,136.12 | −4,148.14*/* |
| Concatenated loci without pilF | −11,095.79*/* | −9,753.16 |
| wzz | −3,125.20 | −4,001.95*/* |
| Concatenated loci without wzz | −11,302.35*/* | −9,503.07 |
For each recombinant locus, we considered two multiple alignments, the one from that locus and that obtained from the concatenated alignment of the other 6 loci. These two alignments were used to evaluate the likelihood (−lnL, negative natural log of the likelihood) of the ML trees obtained with each of them, and the tree obtained with each alignment is compared with that derived from the other by the Shimodaira-Hasegawa (SH) and expected likelihood weight (ELW) tests. Levels of significance for the SH/ELW tests: *, P < 0.001.
FIG. 3.
Maximum likelihood phylogenetic trees from 6 of the 7 concatenated loci, excluding region pilF (A) and the pilF locus (B). Isolates identified as recombinants are marked with a filled circle, while the putative parental isolates are marked with an open circle. The numbers at the nodes represent the percentage values given by bootstrap analysis of 1,000 replicates.
FIG. 4.
Maximum likelihood (ML) phylogenetic trees from 6 of the 7 concatenated loci, excluding region wzz (A) and the wzz locus (B). Isolates identified as recombinants are marked with a filled circle, while the putative parental isolates are marked with an open circle. The numbers at the nodes represent the percentage values given by bootstrap analysis of 1,000 replicates.
Interestingly, the pilF ML tree splits the strains into two groups, one that clusters most clinical biotype 1 isolates from humans together with biotype 3 and biotype 2 serovar E isolates from human origins and another that groups serovar A and I isolates together with environmental biotype 1 isolates (Fig. 3B), suggesting that pilF could be used as a genetic marker to distinguish isolates potentially dangerous to humans. A PCR-based protocol to distinguish V. vulnificus isolates with pathogenic potential against humans based on the polymorphism in pilF has been designed and validated in our laboratory (18).
Conclusion.
The V. vulnificus species is subdivided into three different phylogenetic lineages which do not correspond to the current intraspecific biotype classification. LI and LII seem to have evolved in fish-farming-related environments where recombination or/and horizontal transfer phenomena would have favored the emergence of pathogenic clones for fish or humans, which would have been amplified after outbreaks of fish (biotype 2) or human (biotype 3) vibriosis. The polyphyletic origin of the so-called biotype 2 supports its reclassification within the species as a pathovar that would group the strains with pathogenic potential to infect and develop vibriosis in fish.
Supplementary Material
Acknowledgments
This work was financed by grants AGL2008-03977/ACU, BFU2008-03000, and Programa Consolider-Ingenio 2010 CSD2009-00006 from MICINN and by ACOMP/2009/240 from Conselleria d'Educació (Generalitat Valenciana).
We thank the SCSIE of the University of Valencia for technical support in determining the sequences.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amaro, C., and E. G. Biosca. 1996. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl. Environ. Microbiol. 62:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisharat, N., et al. 2007. The evolution of genetic structure in the marine pathogen, Vibrio vulnificus. Infect. Genet. Evol. 7:685-693. [DOI] [PubMed] [Google Scholar]
- 3.Bisharat, N., et al. 2005. Hybrid Vibrio vulnificus. Emerg. Infect. Dis. 11:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisharat, N., et al. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 5.Chatzidaki-Livanis, M., M. A. Hubbard, K. Gordon, V. J. Harwood, and A. C. Wright. 2006. Genetic distinctions among clinical and environmental strains of Vibrio vulnificus. Appl. Environ. Microbiol. 72:6136-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, A. L., J. D. Oliver, A. DePaola, E. J. Feil, and E. F. Boyd. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553-5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouz, B., and C. Amaro. 2003. Isolation of a new serovar of Vibrio vulnificus pathogenic for eels cultured in freshwater farms. Aquaculture 217:677-682. [Google Scholar]
- 8.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 9.Gulig, P. A., K. L. Bourdage, and A. M. Starks. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43(Spec. No.):118-131. [PubMed] [Google Scholar]
- 10.Gutacker, M., et al. 2003. Population genetics of Vibrio vulnificus: identification of two divisions and a distinct eel-pathogenic clone. Appl. Environ. Microbiol. 69:3203-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, M. K., and J. D. Oliver. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, C. T., et al. 2008. A common virulence plasmid in biotype 2 Vibrio vulnificus and its dissemination aided by a conjugal plasmid. J. Bacteriol. 190:1638-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marco-Noales, E., M. Milan, B. Fouz, E. Sanjuan, and C. Amaro. 2001. Transmission to eels, portals of entry, and putative reservoirs of Vibrio vulnificus serovar E (biotype 2). Appl. Environ. Microbiol. 67:4717-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson, W. B., R. N. Paranjype, A. DePaola, and M. S. Strom. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 17.Roig, F. J., and C. Amaro. 2009. Plasmid diversity in Vibrio vulnificus biotypes. Microbiology 155:489-497. [DOI] [PubMed] [Google Scholar]
- 18.Roig, F. J., E. Sanjuan, A. Llorens, and C. Amaro. 2010. pilF polymorphism-based PCR to distinguish Vibrio vulnificus strains potentially dangerous to public health. Appl. Environ. Microbiol. 76:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosche, T. M., Y. Yano, and J. D. Oliver. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381-389. [DOI] [PubMed] [Google Scholar]
- 20.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 21.Sanjuan, E., B. Fouz, J. D. Oliver, and C. Amaro. 2009. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 75:1604-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 23.Senoh, M., et al. 2005. The cytotoxin-hemolysin genes of human and eel pathogenic Vibrio vulnificus strains: comparison of nucleotide sequences and application to the genetic grouping. Microbiol. Immunol. 49:513-519. [DOI] [PubMed] [Google Scholar]
- 24.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microb. Infect. 2:177-188. [DOI] [PubMed] [Google Scholar]
- 25.Tison, D. L., M. Nishibuchi, J. D. Greenwood, and R. J. Seidler. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warner, J. M., and J. D. Oliver. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other vibrio species. Appl. Environ. Microbiol. 65:1141-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization and Food and Agriculture Organization of the United Nations. 2005. Risk assessment of Vibrio vulnificus in raw oysters. WHO Press, Geneva, Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.