Abstract
The dairy industry adds starter bacterial cultures to heat-treated milk to control the fermentation process during the manufacture of many cheeses. These highly concentrated bacterial populations are susceptible to virulent phages that are ubiquitous in cheese factories. In this study, the dissemination of these phages by the airborne route and their presence on working surfaces were investigated in a cheese factory. Several surfaces were swabbed, and five air samplers (polytetrafluoroethylene filter, polycarbonate filter, BioSampler, Coriolis cyclone sampler, and NIOSH two-stage cyclone bioaerosol personal sampler) were tested. Samples were then analyzed for the presence of two Lactococcus lactis phage groups (936 and c2), and quantification was done by quantitative PCR (qPCR). Both lactococcal phage groups were found on most swabbed surfaces, while airborne phages were detected at concentrations of at least 103 genomes/m3 of air. The NIOSH sampler had the highest rate of air samples with detectable levels of lactococcal phages. This study demonstrates that virulent phages can circulate through the air and that they are ubiquitous in cheese manufacturing facilities.
A variety of food products, commodity chemicals, and biotechnology products are manufactured through large-scale bacterial fermentations (13). Because significant amounts of bacterial cells are cultivated in fermentation vats, most of these industries have experienced problems with phage contamination. Phage outbreaks represent an industrial concern because they can slow down the fermentation process and adversely impact product quality, resulting in extra costs and delays (13). The dairy industry has been working to control phage populations for decades (28). Cheesemakers can experience “slow vats” to various levels depending on, among others, the type of fermented milk product, the starter culture system, and the factory design. According to estimations, 0.1 to 10% of all milk fermentations are negatively affected by virulent phages (27).
The dairy industry relies on selected bacterial cultures for the production of an array of fermented milk products. For example, several strains of the low-GC Gram-positive bacterium Lactococcus lactis are extensively used for the transformation of milk into cheeses. The lactococcal starter culture population can reach levels of up to 109 CFU/ml of milk inside large industrial fermentation vats (24). This ecosystem is also ideal for the multiplication of virulent L. lactis phages, which are ubiquitous in dairy environments (4). If phage-sensitive host cells are dominant in the starter culture, a virulent phage inside one of these vats can initiate an infection, and its progeny will eventually lyse numerous starter cells and slow down the fermentation process, leading to low-quality fermented products (28). Significant progress in the control of phage populations within this food sector has been made, as several strategies are now available to keep these bacterial viruses at bay (for a review, see reference 13). For example, a typical cheese manufacturing plant uses several L. lactis strains on a rotation schedule to prevent the buildup of specific phages. Nonetheless, phage infection remains a considerable risk in cheese making because phage populations have evolved over time and can be disseminated through various routes. The source(s) of the phages and their dissemination routes must be identified in order to implement corrective actions to limit their propagation (13).
Virulent phages can originate from a variety of sources. New virulent phages can be introduced into a cheese manufacturing plant through raw milk, which can be contaminated at a low concentration (23). Moreover, some lactococcal phages can withstand pasteurization (1, 21). Novel virulent phages can also emerge within a factory through mutations and recombination events when wild-type phages infect sensitive hosts (18, 35). Reservoirs of phages include the materials and equipments used in the manufacturing process as well as the fermentation products and by-products (13). Milk with little or no contamination can also be put at risk by the addition of phage-contaminated ingredients, such as whey protein concentrates (13, 15). Although rarely documented, phage contamination can also occur through aerosolization. Concentrations of up to 108 PFU/m3 of air have been detected in some areas, mainly downstream of the fermentation process in a German cheese manufacturing plant (30-32).
Phage aerosolization can occur during air displacements or movements around contaminated surfaces or fluids. It can also occur by liquid splashes, which can aerosolize phages (40). Aerosol contamination represents a major microbiological challenge, especially when the aerosol sources and contents are not known. Studies of the airborne distribution of phages could help identify the sources and determine the impact of aerosols on failed milk fermentations.
Viruses can be found on aerosol particles of various sizes, from the submicrometer range to tens of micrometers in aerodynamic diameter. Several types of samplers are available for characterization of the viral load in the air (for a review, see reference 40). However, no standard procedure for the sampling of airborne viruses exists at present. The efficacy of these samplers is often assessed on the basis of the recovery of infectious viral particles. Many of them have destructive effects on the virus structure, leading to false-negative results in viral culture assays (40, 41). Plaque assays are cumbersome because they require the use of several bacterial hosts, as the phage host range differs from one isolate to another (12, 25). Plaque assays may not always be feasible because pure culture may not be available if commercially defined-strain or mixed-strain bacterial cultures are used to make the fermented milk products. Consequently, it has been argued that analytical methods that are independent of viral infectivity, such as the detection of phage genomes by quantitative PCR (qPCR), are more suitable for the analysis of dairy samples (3, 9, 10, 12, 17, 33, 42) and air samples (41).
The aim of this study was to compare the efficiencies of five aerosol samplers in recovering airborne lactococcal bacteriophages in a cheese manufacturing plant and to assess subsequent detection of the phages by qPCR. The presence of these phages on various working surfaces was also investigated. Although L. lactis phages are currently classified into 10 genetically distinct groups (11), only the 2 most common groups, namely, 936 and c2, were targeted in this study. Members of these phage groups have been isolated worldwide and belong to the Siphoviridae family, each having a double-stranded DNA genome and a long, noncontractile tail (7, 8, 14, 16, 20-22, 25, 26, 34-36, 38, 39).
MATERIALS AND METHODS
Sampling site.
A factory producing cheddar cheese was chosen for aerosol sampling. This cheese factory uses commercial mesophilic starter cultures containing L. lactis strains, which are rotated on a daily basis. A typical aerosol sampling session started at the beginning of cheese production in the morning and lasted for approximately 12 h. Multiple fermentation vats were filled with pasteurized milk daily. The aerosol sampling was performed in the filling section at the end of the production line near a cheese whey source. Since the air from the initial sections of the cheese production line was moved toward this section, it was likely to contain the largest quantity of airborne lactococcal phages. Due to space restrictions within the factory, the sampling site was conducted in two zones separated by approximately 5 m.
Air sampling.
Air sampling in the cheese factory was done using the following five types of samplers (Table 1): 0.4-μm polycarbonate (PC) filters (SKC Inc., Eighty Four, PA), 0.3-μm polytetrafluoroethylene (PTFE) membrane filters (SKC Inc.), BioSamplers (SKC Inc.), a Coriolis cyclone sampler (Bertin Technologies, Montigny-le-Bretonneux, France), and a NIOSH two-stage bioaerosol cyclone (BC 251) personal sampler (5).
TABLE 1.
Sampler descriptions
| Sampling method | Protocol | Sampling rate used (liters/min) | Documented physical collection efficiency | Reference/source |
|---|---|---|---|---|
| PC filters (0.4 μm pore size) | Filtration | 2 | 0.035- to 1-μm particles: 78 to 99.99% (data obtained with similar filters from different manufacturers) | 19 |
| PTFE membrane filters (0.3 μm pore size) | Filtration | 2 | 10- to 900-nm particles: >96% | 6 |
| BioSampler | Liquid impaction | 12.5 | >1.0-μm particles: close to 100% | 37 |
| 0.5-μm particles: 90% | ||||
| Coriolis cyclone sampler | Liquid impaction | 300 | 0.8-μm particles: 62% | 2 |
| 1.6-μm particles: 70% | ||||
| 2.4-μm particles: 80% | ||||
| 4.4-μm particles: 100% | ||||
| 16-μm particles: 109% | ||||
| NIOSH two-stage bioaerosol cyclone (BC 251) personal samplers | Dry surface impaction (stages 1 and 2) and filtration (stage 3) | 10 | First stage 50% cutoff: 2.1-μm particles | Product data sheet |
| Second stage 50% cutoff: 0.41-μm particles |
PC and PTFE filters were mounted on cellulose pads and housed in 37-mm clear styrene three-piece cassettes (SureSeal; SKC Inc.) and were plugged into pumps (Gilian GilAir-5; Sensidyne, LP, Clearwater, FL) calibrated at a flow rate of 2 liters/min. Six filters of each type were evenly distributed in the two sampling areas and used for a period of 710 min. Thus, a total of 1,420 liters of air was sampled per filter. The closed-faced filters were positioned with the inlets facing the ground.
BioSamplers were placed in Styrofoam boxes to prevent any broken glass from being scattered in the factory in the event of equipment breakage. The boxes were positioned on tables, and the samplers were plugged into pumps (Vac-U-Go sampling pump; SKC Inc.). Fifteen milliliters of sterile water with 0.01% Tween 20 was used as a sampling liquid in the BioSamplers. Twenty-minute samples were taken with the critical orifices, providing a sampling rate of 12.5 liters/min. Thus, a total of 250 liters of air was recovered per BioSampler sample.
Ten milliliters of sterile water with 0.01% Tween 20 was used in the sampling cones of the Coriolis sampler, which was run for a period of 10 min at 300 liters/min. The evaporated liquid was replaced to provide a final volume of 10 ml after the first sample was taken. The same sampling cone was used three times, and the remaining liquid volume was measured after the last sample was taken. Therefore, this sampler concentrated 9,000 liters of air in each sample in 30 min. The procedure was repeated with five sampling cones at one sampling site.
Four NIOSH samplers were distributed on two sites to provide 350-min sampling periods at 10 liters/min using calibrated AirCon-2 pumps (Gilian). Each NIOSH sampler was assembled using a 15-ml (stage 1) plastic tube, a 1.5-ml (stage 2) plastic tube, and a 0.4-μm PC capillary pore filter (SKC Inc.). Thus, a total of 3,500 liters of air was sampled per assay. The Coriolis sampler and the airborne particle counter (APC) (Met One model 3313-3-1; Pacific Scientific Instruments, Grants Pass, OR) were placed 1.3 m and 2 m above ground level, respectively, due to space restrictions. All the other samplers were placed approximately 1 m above ground level. The APC sampled the air at 28.3 liters/min, and airborne particles were analyzed in channels of 0.3, 0.5, 1.0, 5.0, and 10.0 μm.
The limit of detection for each sampler (see Table 3) was determined by taking into consideration the volume of air sampled, the dilution factor of the liquid suspension, and the limits of detection of the qPCRs.
Surface sampling.
Several surfaces in the cheese factory were swabbed with polyester fiber-tipped plastic applicators (Fisher Scientific Company, Ottawa, Ontario, Canada). The tips were first wet in sterile water with 0.01% Tween 20, cut after swabbing, and kept in 1 ml of the same liquid. Whenever possible, the areas swabbed were approximately 100 cm2. However, the exact size of the areas swabbed is not known in some cases, owing to the limited or uneven surfaces. Therefore, these results are considered qualitative.
Sample treatment and analysis.
All samples were kept on ice after on site sampling and then stored at 4°C until analyses, which were performed within 48 h. PTFE and PC filters were eluted by placing 5 ml of sterile water with 0.01% Tween 20 directly into the three-piece cassettes and by shaking the filters on a vortexer for 20 min. The volumes of liquid in the BioSamplers and the Coriolis samplers were measured, and the liquid was stored at 4°C. For the NIOSH samplers, 250 μl of sterile water with 0.01% Tween 20 was placed in each tube. The 1.5-ml tubes were pulse shaken on a vortexer until the pellet was no longer visible. The 15-ml tubes were processed in the same way; however, the tubes were also shaken upside down to clean the upper part of the tube. No DNA extraction was necessary for qPCR analysis. Every sample was analyzed by qPCR in triplicate at a minimum.
qPCR primer and standard curve design.
Primers for quantitative PCRs (qPCRs) were designed using the Beacon Designer 4.0 program (PREMIER Biosoft International, Palo Alto, CA) for SYBR green protocols. The primer pair used for the detection of the lactococcal 936 phage group has already been published (41). The targeted region was within orf6 of phage P008, a gene coding for a highly conserved structural protein within the 936 group (22, 35). The reverse and forward primers used were 5′-CCAGCAGTAGGGCGAACAAAG-3′ (positions 5187 to 5167) and (5′-TGAGGGAGACGGAACAAACGG-3′ (positions 5035 to 5055), respectively. For the c2 group, we targeted the gene coding for the highly conserved major capsid protein (20). The reverse and forward primers used were 5′-GCATTAAAGCCAACTGATAGC-3′ (positions 9642 to 9662) and 5′-AGTAAGAGGGATAGCGAACC-3′ (positions 9432 to 9451), respectively.
The qPCR experiments were performed using a DNA Engine Opticon 2 apparatus (Bio-Rad Laboratories, Hercules, CA) in 96-well plates. The qPCR mixture consisted of 0.5 μM forward primer, 0.5 μM reverse primer, and a 1× final concentration of iQ SYBR green supermix (Bio-Rad Laboratories) in a final volume of 25 μl, with 5 μl of which being the template DNA or sample. The standard curve was built using 100 to 5 × 106 plasmid copies (see below) per reaction tube and was repeated in duplicate on each plate. The results obtained using three plates were pooled to determine the equation of the slope used to calculate the phage concentration. The protocol used for qPCR was 94°C for 3 min (hot start), followed by 35 cycles of (i) 94°C for 20 s, (ii) 60°C or 53°C for 30 s for 936 and c2, respectively, (iii) plate reading, and (iv) 72°C for 25 s. A melting curve was calculated with readings every 0.2°C from 50°C to 95°C, holding the temperature for 1 s. The cycle threshold (CT) for all plates was set at a fluorescence intensity of 0.011.
For both phage groups, plasmids containing the amplicon obtained with the primers were used as templates for the construction of standard curves. Plasmids were obtained using the TOPO cloning reaction (Invitrogen, Carlsbad, CA) and purified using the QIAprep Spin miniprep kit (Qiagen Inc., Mississauga, ON, Canada). The optical density at 260 nm of the purified plasmid was used to determine its concentration, and aliquots of known concentrations were prepared.
For all aerosol samplers, viral extraction efficiency for DNA quantification was verified by inoculating the samplers with a known concentration of 936-like phages. The phage suspension was inoculated into the liquid sampling media for the Coriolis sampler and BioSampler and was extracted after sampling HEPA-filtered air. The phage suspension was allowed to dry in the NIOSH sampler and filters prior to air sampling. Viral extractions were done as described for the environmental samples.
Specificity of the primers.
The primer pairs were tested on 23 different lactococcal phages of the 936 group and 7 different phages of the c2 group (Table 2), as well as 1 phage of the P335 group used as a negative control. All these phages had previously been isolated from cheese factories and kept at the Felix d'Hérelle Reference Center for Bacterial Viruses (www.phage.ulaval.ca). Lysates were diluted in sterile water and used directly as a template for qPCR. Both primer pairs were used on all lysates to verify the specificity of the primers toward different isolates of the same phage groups. Results were confirmed both by the melting curves and by migration on 2% agarose gels for 30 min at 100 V. Furthermore, the PCR amplicons obtained from 28 randomly selected positive air and surface samples were sequenced to ensure primer specificity.
TABLE 2.
Primer specificity for c2 and 936 detection and quantification
| Phage isolate from cheese factories | Group | Result of indicated test |
|||
|---|---|---|---|---|---|
| 936 |
c2 |
||||
| qPCR | Gel | qPCR | Gel | ||
| DM1 | 936 | + | + | − | − |
| DM2 | 936 | + | + | − | − |
| DM3 | 936 | + | + | − | − |
| DM4 | 936 | + | + | − | − |
| DM5 | 936 | + | + | − | − |
| DM6 | 936 | + | + | − | − |
| DM7 | 936 | + | + | − | − |
| DM8 | 936 | + | + | − | − |
| DM9 | 936 | + | + | − | − |
| DM10 | 936 | + | + | − | − |
| DM11 | 936 | + | + | − | − |
| DM12 | 936 | + | + | − | − |
| DM13 | 936 | + | + | − | − |
| JG4 | 936 | + | + | − | − |
| JG9 | 936 | + | + | − | − |
| JG10 | 936 | + | + | − | − |
| GL1 | 936 | + | + | − | − |
| GL3 | 936 | + | + | − | − |
| GL7 | 936 | + | + | − | − |
| GL8 | 936 | + | + | − | − |
| GL9 | 936 | + | + | − | − |
| GL10 | 936 | + | + | − | − |
| GL11 | 936 | + | + | − | − |
| LS13 | c2 | − | − | + | + |
| CB17 | c2 | − | − | + | + |
| CB27 | c2 | − | − | + | + |
| GR3 | c2 | − | − | + | + |
| GR4 | c2 | − | − | + | + |
| GR6 | c2 | − | − | + | + |
| bIL67 | c2 | − | − | + | + |
| GR2 | P335 | − | − | − | − |
Statistical analysis.
For the air samples, data were expressed as medians and interquartile ranges. Statistical analyses were performed using the mean values. One-way analysis of variance (ANOVA) was used to analyze experimental factors associated with the comparison of samplers. For qPCR results, values were log transformed to stabilize the variances. Reported P values are based on these transformations. The univariate normality assumptions were verified with the Shapiro-Wilk test. Brown and Forsythe's variation of Levene's test was used to verify the homogeneity of variances. The Tukey multiple-comparison technique was applied post hoc to the ANOVA. The results with P values of ≤0.05 were considered significant. All analyses were conducted using the SAS statistical package, version 9.2 (SAS Institute Inc.).
RESULTS
Testing of primer pairs for specificity in qPCR assays.
To confirm that the selected primers were able to detect a diverse set of phages belonging to either the 936-like or the c2-like phages, we tested them in qPCR assays using the phages listed in Table 2. Both pairs of 21-nucleotide primers were specific to either the 936- or the c2-like phages (Table 2). No false-negative or false-positive results were obtained with the primers. According to the standard curves obtained, the limit of detection varied from 1 to 50 genome copies per reaction tube for the 936 qPCR protocol and from 5 to 50 genome copies per reaction tube for the c2 qPCR protocol. Moreover, all the sequenced qPCR amplicons were in accordance with the anticipated phage groups, as identified by the qPCR protocol.
Size distribution of airborne particles.
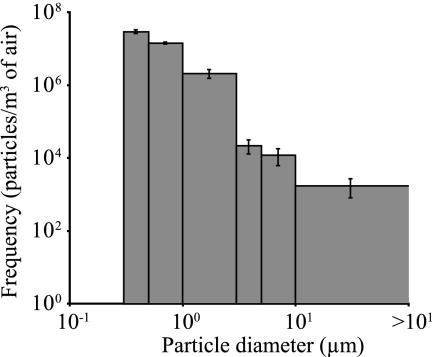
The size distribution of airborne particles in the cheese factory was monitored throughout the sampling period by collecting data with the APC for 5 min at a time, followed by 15-min waiting times. Mean counts of particles per cubic meter of air for each of the six channels of the APC are shown in Fig. 1. Airborne particle concentration and size distribution were stable throughout the study.
FIG. 1.
Mean size distribution of airborne particles in a cheese factory shown according to the frequency of the particle count in each channel of the APC. Error bars represent standard deviations.
Comparison of air samplers for the recovery of lactococcal phages.
Samples obtained from samplers spiked with known concentrations of 936-like phages were analyzed by qPCR. The concentrations detected were within the same order of magnitude as that of the theoretical concentrations (data not shown). Viral extractions from the samplers were satisfactory for proper comparison of sampler efficiencies. Results obtained from the qPCR analysis of the aerosol samples are summarized in Table 3. Interestingly, lactococcal phages belonging to both groups were detected with all air samplers, indicating that a significant number of phages were suspended in the air within the cheese manufacturing plant. However, sampler efficiency varied considerably.
TABLE 3.
Airborne concentrations of 936 and c2 phages in a cheese factory
| Sampling method | Result for: |
|||||
|---|---|---|---|---|---|---|
| 936 |
c2 |
|||||
| Positive/total assaysa | Median concn (1st quartile/3rd quartile)b | LODc | Positive/ total assaysa | Median concn (1st quartile/3rd quartile)b | LODc | |
| NIOSH sampler (stage 1) | 12/12 (100) | 5,346 (2,494/6,010) | 10 | 10/12 (83) | 162 (124/208) | 50 |
| NIOSH sampler (stage 2) | 11/12 (92) | 280 (159/449) | 10 | 5/12 (42) | 223 (139/465) | 50 |
| Coriolis sampler | 43/52 (83) | 580 (249/2,905) | 62 | 32/52 (62) | 1,848 (792/31,077) | 318 |
| PC filter | 15/18 (83) | 2,399 (1,372/3,483) | 492 | 5/18 (28) | 5,377 (3,562/5,595) | 2,523 |
| PTFE filter | 14/18 (78) | 1,538 (1,271/2,093) | 492 | 5/18 (28) | 3,378 (3,232/3,917) | 2,523 |
| BioSampler | 1/18 (6) | 27,903 | 6,986 | 3/18 (17) | 51,750 (44,905/66,148) | 35,821 |
Number of positive qPCR assays out of the total number of assays (percentage of positive samples shown in parentheses).
Median concentration of genomes per m3 of air, taking only positive samples into account.
LOD, limit of detection of viral genomes per m3 of air by qPCR.
The first stage of the NIOSH sampler was more efficient at detecting both 936-like and c2-like phages (Table 3). When the results from the detection of both phage groups are combined, 22 out of 24 samples (92%) were positive in the first stage and 16 out of 24 (67%) were positive in the second stage. The differences in the concentrations of airborne phages found in the first and second stages of the NIOSH sampler were very significant, with 16-fold more phages being detected in the first stage than in the second stage (P < 0.0001) for 936-like phages. No difference was observed for c2-like phages. It should be noted that the NIOSH sampler had the lowest limit of detection.
A total of 75 out of 104 (72%) samples from the Coriolis sampler were positive for lactococcal phages when the results of the two phage groups are combined (Table 3). The concentrations of airborne c2-like phages detected were significantly higher with the Coriolis sampler than with stages 1 (23-fold; P < 0.0001) and 2 (17-fold; P = 0.002) of the NIOSH sampler. In contrast, detection of 936-like phages was significantly lower than that with stage 1 (5-fold; P < 0.0001) and significantly higher than that with stage 2 (3-fold; P = 0.002) of the NIOSH sampler.
The PC and PTFE filters showed equivalent phage recovery, giving almost the same number of positive samples. No significant difference between the two filter types in the concentration of airborne viruses recovered was found. However, statistical analyses based on mean values showed that PC filters allowed the detection of 3-fold more airborne 936 phages than the Coriolis sampler. Overall, the two filters were still less efficient than the NIOSH and Coriolis samplers (Table 3). They were particularly deficient in the recovery of c2-like phages.
The BioSampler showed a very high detection limit in terms of viral genomes per m3 of air (Table 3). Every phage genome detected in qPCR analysis of the BioSampler samples represented 104 phage genomes/m3 of air. Only 1 sample out of 18 allowed the detection of airborne 936-like phages, whereas 3 out of 18 qPCRs revealed c2-like phages. However, for positive samples, the BioSampler detected the highest concentrations of airborne viruses. This sampler allowed the detection of >100-fold-higher concentrations of airborne viruses than stages 1 and 2 of the NIOSH sampler and >10-fold-higher concentrations than the two filters and the Coriolis sampler.
Surfaces.
Various surfaces were swabbed in the factory to check for the presence of c2-like and 936-like phages. Using the swabbing method, the limit of detection for the 936-like phages was 1.4 phage genomes per cm2 and was 5-fold higher for the c2 group. Both phage groups were found on most of the surfaces swabbed (100 cm2), as evidenced by qPCR assays (Table 4). Concentrations of 936-like phages ranged from 3 genome copies/cm2 on the top of a closed fermentation tank to 9.1 × 103 genome copies/cm2 on the handle of a door leading to the office area of the cheese manufacturing plant, whereas c2 concentrations reached 3.1 × 103 genome copies/cm2 on the surface of cleaning material.
TABLE 4.
Estimated concentrations of 936-like and c2-like genomes per cm2 of a swabbed surface
| Surface description | Result for: |
|||
|---|---|---|---|---|
| 936 |
c2 |
|||
| No. of genomes/cm2 | Positive/total assaysa | No. of genomes/cm2 | Positive/total assaysa | |
| Door handle | 9,106 ± 789 | 3/3 | 869 ± 69 | 3/3 |
| Rough floor | 6,115 ± 785 | 3/3 | 1,113 ± 120 | 3/3 |
| Top of paper towel dispenser | 6,079 ± 800 | 3/3 | 458 ± 109 | 3/3 |
| Cleaning material | 1,854 ± 271 | 3/3 | 3,138 ± 449 | 3/3 |
| Smooth floor | 448 ± 145 | 15/15 | 564 ± 403 | 9/10 |
| Wall | 407 ± 169 | 3/3 | 945 ± 66 | 3/3 |
| Push for hand sanitizer | 266 ± 126 | 3/3 | 0 | 0/3 |
| Office table | 60 ± 28 | 3/3 | 83 ± 41 | 3/3 |
| Top of electrical panel | 53 | 1/4 | 0 | 0/5 |
| Top of stainless pipe | 30 ± 7 | 3/3 | 260 ± 59 | 3/3 |
| Stair ramp | 11 ± 10 | 4/6 | 67 ± 44 | 3/6 |
| Top of closed tank | 3 ± 1 | 3/3 | 12 ± 2 | 3/3 |
| Vertical panel | 2 | 1/3 | 9 | 1/3 |
| Vertical surface (electrical) | 0 | 0/3 | 43 ± 5 | 3/3 |
Number of positive qPCR assays out of the total number of assays.
DISCUSSION
The source(s) of phages as well as their dissemination routes should be identified in a cheese manufacturing plant in order to implement long-term corrective actions to limit phage propagation and improve overall product quality (13). Contamination sources and dissemination routes are not easy to identify, as several control points need to be checked. Because virulent dairy phages have a narrow host range (12, 25), it is not practicable to use culture assays to detect them in various dairy environments, including air samples. Additionally, phages can deteriorate and lose their infectivity during sampling and sample processing (40). Molecular biology methods independent of phage infectivity and of bacterial hosts can facilitate the analysis of the viral content of environmental samples (40). In this study, quantitative PCR using SYBR green fluorescence and primers specific to conserved regions was successfully performed to detect the two main lactococcal phage groups in various samples obtained from a cheese manufacturing plant, including air samples.
Only a few studies to date have shown that airborne phages can be detected in industrial cheese manufacturing plant settings (30-32). We also had some anecdotal evidences that ventilation breakdowns lead to increased phage contamination. Our analyses of air samples confirmed that lactococcal phages can be disseminated through the airborne route, since up to 2.7 × 104 and 6.6 × 104 lactococcal phage genomes per m3 of air were detected using the BioSampler (Table 3). Considering that it takes only a few infectious phages to infect phage-sensitive L. lactis cells and start the phage lytic cycle, cheese factories should possess adequate ventilation and control the airflow to minimize phage dissemination as much as possible.
Our study also clearly demonstrates that phage genomes can be found on various surfaces, including floors, walls, cleaning materials, pipes, door handles, and office tables. While it is not known if these viral nucleic acids are still part of infectious phages, it is safe to assume that they were at some point. These findings underscore the need to train workers regarding the importance of surfaces as sources of phage contaminations. They also suggest that the use of appropriate cleaning procedures and effective sanitizers needs to be carefully evaluated to reduce the risks of phage problems.
Although we successfully detected phages in aerosols using different air samplers, not all samplers showed the same level of efficiency. The air sampling devices used in earlier studies on airborne phages in cheese manufacturing plants (30, 32) were based on gelatin membrane filtration and on impaction on agar. Although the devices were able to detect phages, they have practical drawbacks. For example, gelatin membrane filtration can dry and break during prolonged sampling, or it can dissolve if liquid droplets are sampled. In contrast, impaction on agar relies on plaque assays, which is difficult to apply when a variety of bacterial strains are employed daily. Besides the concern about drying during prolonged sampling with the latter system, only large aerosol particles can be sampled. In fact, there is no standard and approved protocol to detect viral aerosols, let alone phages.
The NIOSH sampler gave the most reliable results in this study. It had the highest proportion of positive samples, low between-sample variability, and the lowest detection limit. The two stages of the NIOSH sampler were analyzed separately to take advantage of the aerodynamic size separation that took place with this sampler. At a sampling rate of 10 liters/min, the 50% cutoff is 2.1 μm for the first stage and 0.41 μm for the second stage, while the remainder of the aerosol is captured by the third stage. The second stage of this sampler detected the lowest airborne concentrations of 936-like phages, while the first stage detected the highest concentration. This suggests that most airborne 936-like phages where bound to larger particles. However, for the c2-like phages, there was no difference in the concentrations detected in the two stages of the sampler, indicating that these phages were present at similar concentrations on the smaller and the larger particles. It is not clear at present why this difference was observed. Both phage groups belong to the Siphoviridae family but have somewhat different morphology. Phages belonging to the 936 group have an isometric capsid that is approximately 60 nm in diameter and a long noncontractile tail ranging from 140 nm to 200 nm (morphotype B1), whereas c2-like phages have a prolate capsid (60 nm by 40 nm) and a 100-nm-long noncontractile tail (morphotype B2) (26).
Of all the samplers tested, the BioSampler allowed the highest recovery of airborne phage concentrations. However, given its very high detection limit and the few positive samples collected, this sampler was less suitable for the determination of the airborne phage concentrations.
The Coriolis sampler has the advantage of collecting a large volume of air in a very short time period. However, with its very high flow rate (300 liters/min), this sampler can draw in particles with greater inertia (greater aerodynamic size) than the filters (2 liters/min), the NIOSH sampler (10 liters/min), or the BioSampler (12.5 liters/min). Considering that the mass of a particle is proportional to the cubic value of the radius, a few large particles can drastically raise the concentration of airborne viruses detected. The presence of these larger aggregated particles in the liquid sample can also cause large differences in viral concentrations between aliquots (Table 3, c2-like phages). Our attempts to reduce these variations consisted of purifying the viral DNA with commercial kits and exposing the samples to sonication (data not shown). However, viral DNA purification lowered the concentrations of DNA and led to the underestimation of the viral load in the air sample, while sonication had no effect on the sample variability (data not shown). This inherent variability may be due to the variation of the concentrations of airborne viruses over the course of a day, reflected by the short sampling period of the Coriolis sampler. A sampler that slowly collects its sample, like the NIOSH sampler or PC and PTFE filters, likely provides a more representative evaluation of the airborne viral concentration over the course of a day.
In conclusion, various types of samplers were successfully used to collect airborne viruses, but the NIOSH sampler was the most efficient. Most samplers detected concentrations of at least 103 genomes/m3 of air for both the lactococcal 936 and c2 phage groups. The NIOSH sampler results indicate that a significant portion of the airborne phages was bound to small particles (<2.1 μm). Since these smaller particles can remain airborne for longer periods of time and are influenced by air movements, it is likely that they can be carried far away from their aerosolization source. Although the dynamics of airborne viral transmission are poorly understood (29), appropriate ventilation practices should reduce airborne dissemination. Finally, a qPCR protocol was effectively adapted to detect lactococcal phages. However, it is not known whether these phages were active or inactive or whether they were inactivated by the sampling/elution procedure. The detection level and the limit of detection are the most important characteristics to consider when choosing a sampler for field studies.
Acknowledgments
We are grateful to M. Lamoureux and J. Monast for their collaborative work in cheese factories. We thank S. Simard for his assistance with statistical analysis and D. Tremblay for her technical assistance.
C. Duchaine and S. Moineau received funding from the Natural Sciences and Engineering Research Council of Canada (NSERC). C. Duchaine is also the recipient of an FRSQ Junior 2 scholarship and of a CCHSA time release program and is a member of the FRSQ Respiratory Health Network. This work was funded by a joint grant from FQRNT-NOVALAIT-MAPAQ and Agriculture and Agri-Food Canada.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1.Atamer, Z., et al. 2009. Screening for and characterization of Lactococcus lactis bacteriophages with high thermal resistance. Int. Dairy J. 19:228-235. [Google Scholar]
- 2.Bertin Technologies. Coriolis brochure. Bertin Technologies, Montigny-le-Bretonneux, France.
- 3.Binetti, A. G., M. L. Capra, M. A. Alvarez, and J. A. Reinheimer. 2008. PCR method for detection and identification of Lactobacillus casei/paracasei bacteriophages in dairy products. Int. J. Food Microbiol. 124:147-153. [DOI] [PubMed] [Google Scholar]
- 4.Bissonnette, F., S. Labrie, H. Deveau, M. Lamoureux, and S. Moineau. 2000. Characterization of mesophilic mixed starter cultures used for the manufacture of aged cheddar cheese. J. Dairy Sci. 83:620-627. [DOI] [PubMed] [Google Scholar]
- 5.Blachere, F. M., et al. 2009. Measurement of airborne influenza virus in a hospital emergency department. Clin. Infect. Dis. 48:438-440. [DOI] [PubMed] [Google Scholar]
- 6.Burton, N. C., S. A. Grinshpun, and T. Reponen. 2007. Physical collection efficiency of filter materials for bacteria and viruses. Ann. Occup. Hyg. 51:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandry, P. S., S. C. Moore, J. D. Boyce, B. E. Davidson, and A. J. Hillier. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26:49-64. [DOI] [PubMed] [Google Scholar]
- 8.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 9.del Rio, B., et al. 2007. Multiplex PCR for the detection and identification of dairy bacteriophages in milk. Food Microbiol. 24:75-81. [DOI] [PubMed] [Google Scholar]
- 10.del Rio, B., M. C. Martín, N. Martínez, A. H. Magadán, and M. A. Alvarez. 2008. Multiplex fast real-time PCR for quantitative detection and identification of cos- and pac-type Streptococcus thermophilus bacteriophages. Appl. Environ. Microbiol. 74:4779-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deveau, H., S. J. Labrie, M. C. Chopin, and S. Moineau. 2006. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72:4338-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupont, K., F. K. Vogensen, and J. Josephsen. 2005. Detection of lactococcal 936-species bacteriophages in whey by magnetic capture hybridization PCR targeting a variable region of receptor-binding protein genes. J. Appl. Microbiol. 98:1001-1009. [DOI] [PubMed] [Google Scholar]
- 13.Emond, E., and S. Moineau. 2007. Bacteriophages and food fermentations, p. 93-124. In S. McGrath and D. van Sinderen (ed.), Bacteriophage: genetics and molecular biology. Horizon Scientific Press/Caister Academic Press, Norwich, United Kingdom.
- 14.Hejnowicz, M. S., M. Golebiewski, and J. Bardowski. 2009. Analysis of the complete genome sequence of the lactococcal bacteriophage bIBB29. Int. J. Food Microbiol. 131:52-61. [DOI] [PubMed] [Google Scholar]
- 15.Hinrichs, J. 2001. Incorporation of whey proteins in cheese. Int. Dairy J. 11:495-503. [Google Scholar]
- 16.Josephsen, J., et al. 1994. An ecological study of lytic bacteriophages in Lactococcus lactis subsp. cremoris isolated in a cheese plant over a five year period. Int. Dairy J. 4:123-140. [Google Scholar]
- 17.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labrie, S. J., and S. Moineau. 2007. Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J. Bacteriol. 189:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K. W., and M. Ramamurthi. 1993. Filter collection, p. 179-205. In K. Willeke and P. A. Baron (ed.), Aerosol measurement: principles, techniques, and applications. John Wiley and Sons, Inc., New York, NY.
- 20.Lubbers, M. W., N. R. Waterfield, T. P. Beresford, R. W. Le Page, and A. W. Jarvis. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4349-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madera, C., C. Monjardin, and J. E. Suarez. 2004. Milk contamination and resistance to processing conditions determine the fate of Lactococcus lactis bacteriophages in dairies. Appl. Environ. Microbiol. 70:7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahony, J., et al. 2006. Sequence and comparative genomic analysis of lactococcal bacteriophages jj50, 712 and P008: evolutionary insights into the 936 phage species. FEMS Microbiol. Lett. 261:253-261. [DOI] [PubMed] [Google Scholar]
- 23.McIntyre, K., H. A. Heap, G. P. Davey, and G. K. Y. Limsowtin. 1991. The distribution of lactococcal bacteriophage in the environment of a cheese manufacturing plant. Int. Dairy J. 1:183-197. [Google Scholar]
- 24.Mistry, V. V., and F. V. Kosikowski. 1986. Influence of milk ultrafiltration on bacteriophages of lactic acid bacteria. J. Dairy Sci. 69:2577-2582. [DOI] [PubMed] [Google Scholar]
- 25.Moineau, S., et al. 1996. Isolation and characterization of lactococcal bacteriophages from cultured buttermilk plants in the United States. J. Dairy Sci. 79:2104-2111. [Google Scholar]
- 26.Moineau, S., J. Fortier, H. W. Ackermann, and S. Pandian. 1992. Characterization of lactococcal bacteriophages from Quebec cheese plants. Can. J. Microbiol. 38:875-882. [Google Scholar]
- 27.Moineau, S., and C. Lévesque. 2005. Control of bacteriophages in industrial fermentation, p. 285-296. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and applications. CRC Press, Boca Raton, FL.
- 28.Moineau, S., D. Tremblay, and S. Labrie. 2002. Phages of lactic acid bacteria: from genomics to industrial applications—dairy microbiologists seek new ways to protect milk-fermenting microorganisms against damaging phages. ASM News 68:388-393. [Google Scholar]
- 29.Morawska, L. 2006. Droplet fate in indoor environments, or can we prevent the spread of infection? Indoor Air 16:335-347. [DOI] [PubMed] [Google Scholar]
- 30.Neve, H., A. Berger, and K. J. Heller. 1995. A method for detecting and enumerating airborne virulent bacteriophage of dairy starter cultures. Kieler Milchw. Forsch. 47:193-207. [Google Scholar]
- 31.Neve, H., U. Kemper, A. Geis, and K. J. Heller. 1994. Monitoring and characterization of lactococcal bacteriophages in a dairy plant. Kieler Milchw. Forsch. 46:167-178. [Google Scholar]
- 32.Neve, H., A. Laborius, and K. J. Heller. 2003. Testing of the applicability of battery-powered portable microbial air samplers for detection and enumeration of airborne Lactococcus lactis dairy bacteriophages. Kieler Milchw. Forsch. 55:301-315. [Google Scholar]
- 33.Quiberoni, A., D. Tremblay, H.-W. Ackermann, S. Moineau, and J. A. Reinheimer. 2006. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J. Dairy Sci. 89:3791-3799. [DOI] [PubMed] [Google Scholar]
- 34.Raiski, A., and N. Belyasova. 2009. Biodiversity of Lactococcus lactis bacteriophages in the Republic of Belarus. Int. J. Food Microbiol. 130:1-5. [DOI] [PubMed] [Google Scholar]
- 35.Rousseau, G. M., and S. Moineau. 2009. Evolution of Lactococcus lactis phages within a cheese factory. Appl. Environ. Microbiol. 75:5336-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schouler, C., S. D. Ehrlich, and M.-C. Chopin. 1994. Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology 140:3061-3069. [DOI] [PubMed] [Google Scholar]
- 37.SKC Inc. BioSampler operating instructions, form #37084, rev. 1008. SKC Inc., Eighty Four, PA.
- 38.Suárez, V., S. Moineau, J. Reinheimer, and A. Quiberoni. 2008. Argentinean Lactococcus lactis bacteriophages: genetic characterization and adsorption studies. J. Appl. Microbiol. 104:371-379. [DOI] [PubMed] [Google Scholar]
- 39.Szczepańska, A. K., M. S. Hejnowicz, P. Kołakowski, and J. Bardowski. 2007. Biodiversity of Lactococcus lactis bacteriophages in Polish dairy environment. Acta Biochim. Pol. 54:151-158. [PubMed] [Google Scholar]
- 40.Verreault, D., S. Moineau, and C. Duchaine. 2008. Methods for sampling of airborne viruses. Microbiol. Mol. Biol. Rev. 72:413-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verreault, D., et al. 2010. Comparison of polycarbonate and polytetrafluoroethylene filters for sampling of airborne bacteriophages. Aerosol Sci. Technol. 44:197-201. [Google Scholar]
- 42.Zago, M., L. Rossetti, J. Reinheimer, D. Carminati, and G. Giraffa. 2008. Detection and identification of Lactobacillus helveticus bacteriophages by PCR. J. Dairy Res. 75:196-201. [DOI] [PubMed] [Google Scholar]