Abstract
Clostridium perfringens is a Gram-positive anaerobic pathogen which causes many diseases in humans and animals. While some genetic tools exist for working with C. perfringens, a tightly regulated, inducible promoter system is currently lacking. Therefore, we constructed a plasmid-based promoter system that provided regulated expression when lactose was added. This plasmid (pKRAH1) is an Escherichia coli-C. perfringens shuttle vector containing the gene encoding a transcriptional regulator, BgaR, and a divergent promoter upstream of gene bgaL (bgaR-PbgaL). To measure transcription at the bgaL promoter in pKRAH1, the E. coli reporter gene gusA, encoding β-glucuronidase, was placed downstream of the PbgaL promoter to make plasmid pAH2. When transformed into three strains of C. perfringens, pAH2 exhibited lactose-inducible expression. C. perfringens strain 13, a commonly studied strain, has endogenous β-glucuronidase activity. We mutated gene bglR, encoding a putative β-glucuronidase, and observed an 89% decrease in endogenous activity with no lactose. This combination of a system for regulated gene expression and a mutant of strain 13 with low β-glucuronidase activity are useful tools for studying gene regulation and protein expression in an important pathogenic bacterium. We used this system to express the yfp-pilB gene, comprised of a yellow fluorescent protein (YFP)-encoding gene fused to an assembly ATPase gene involved in type IV pilus-dependent gliding motility in C. perfringens. Expression in the wild-type strain showed that YFP-PilB localized mostly to the poles of cells, but in a pilC mutant it localized throughout the cell, demonstrating that the membrane protein PilC is required for polar localization of PilB.
Clostridium perfringens is an important bacterial pathogen, causing gas gangrene, food poisoning, necrotic enteritis, and other diseases in humans and many others in agriculturally important livestock (3, 11, 21, 22). Strains that are transformable by electroporation have been isolated and their genomes sequenced (18, 22, 25, 32), and many groups have developed effective methods to create site-directed and random mutants (7, 10, 14, 31). However, other genetic tools available for studying genes are limited or absent, including an inducible promoter system, although they have been developed for use in Clostridium acetobutylicum (6, 28). A well-regulated promoter system would be a valuable tool in C. perfringens for complementing mutants, controlling the expression of fusion-tagged proteins, and modulating the intracellular levels of proteins to near physiological conditions.
There are several characteristics which are desirable in an inducible promoter system. Ideally, a promoter system would work in a variety of C. perfringens strains, since different diseases are caused by strains that are genetically highly diverse (18). It would be easy to induce, not requiring a temperature shift, expensive chemicals, or specific media. It would have a large range of activity based on the concentration of inducer added and low activity in the absence of inducer. These characteristics would allow a protein to be highly expressed for potential purification or analysis of the effects of overexpression on the phenotype of the strain. Tight regulation allows the introduction of potentially lethal genes as well as observation of phenotypic changes that occur in the absence of expression of a gene that has been inactivated by mutation. Finally, a rapid response to the presence of the inducer would be desirable in studying short-term effects of the presence of the gene product on the cell. An example of a plasmid-based system that fulfills all of these criteria is the pBAD family of plasmids developed by Guzman and coworkers for use in E. coli (8). The plasmids employ the AraC protein to provide arabinose-inducible expression from the divergently transcribed promoter for the araBAD operon (8).
We used the genome sequence of C. perfringens strain 13 to identify regulators and promoters that, like the pBAD plasmids, would likely fulfill the desired requirements. We identified a lactose-inducible regulatory system, consisting of a gene encoding a transcriptional regulator, bgaR, and the promoter of a divergently transcribed regulated gene, bgaL, and constructed a plasmid-based system for controlled gene expression that functions in three different pathogenic strains of C. perfringens. We used the system in one of these strains to express a yellow fluorescent protein (YFP)-tagged type IV pilus-associated ATPase (PilB) and showed that cellular localization of this protein was dependent on a membrane-bound partner protein, PilC.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains, plasmids, and primers used in this study are listed in Table 1. E. coli was grown in Luria-Bertani (LB) medium supplemented with antibiotics as needed (400 μg/ml erythromycin, 100 μg/ml ampicillin, or 20 μg/ml chloramphenicol) and agar for plates. C. perfringens was grown in a Coy anaerobic chamber in brain heart infusion (BHI) medium (Difco) or on BHI plates with the appropriate antibiotics as indicated (30 μg/ml erythromycin or 20 μg/ml chloramphenicol). PY medium (30 g/liter proteose peptone, 10 g/liter yeast extract, 1 g/liter sodium thioglycolate) was used to grow C. perfringens for β-glucuronidase assays.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description, relevant phenotype or genotype, or sequence (5′ to 3′) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | F−mcrA Δ(mrr-hsdRMS mcrBC) φ80dlacZΔM15 lacX74 deoR recA1 araD139 Δ(ara, leu)7697 galU galK ΔrpsL endA1 nupG | Gibco/BRL |
| C. perfringens | ||
| 13 | Gangrene strain | C. Duncan |
| SM101 | Acute food poisoning strain | 32 |
| JGS4143 | Chicken necrotic enteritis strain | J. G. Songer |
| AH1 | bglR::pAH4, from strain 13 | This study |
| AH2 | bgaR::pAH6, from strain 13 | This study |
| SM126 | pilC mutant derived from strain 13 | 30 |
| Plasmids | ||
| pGEM-T Easy | Ampicillin resistance | Promega |
| pSM240 | Contains cpe-gusA; chloramphenicol resistance | 9 |
| pAH1 | Mutated bgaR with PbgaL in pSM240 | This study |
| pAH2 | pSM240 with bgaR and PbgaL upstream of cpe-gusA | This study |
| pKRAH1 | Contains bgaR-PbgaL and polylinker; chloramphenicol resistance | This study |
| pSM300 | Suicide plasmid in C. perfringens; erythromycin resistance | 29 |
| pAH3 | Contains a fragment of strain 13 bglR in pGEM-T Easy | This study |
| pAH4 | Contains a fragment of strain 13 bglR in pSM300 | This study |
| pAH5 | Contains a fragment of strain 13 bgaR in pGEM-T Easy | This study |
| pAH6 | Contains a fragment of strain 13 bgaR in pSM300 | This study |
| pAH7 | Contains strain 13 bgaR and PbgaL in pGEM-T Easy | This study |
| pSW4-YFP | Vector containing a low GC codon-optimized yfp gene | 23 |
| pAH11 | Contains yfp-pilB in pGEM-T Easy | This study |
| pKRAH-Y | pKRAH1 with the yfp gene | |
| pKRAH-YB | pKRAH1 with the yfp-pilB fusion gene | |
| Primers | ||
| OAH1 | GTTAACCTCGAGATGAAAAGTATTAGGGC | |
| OAH2 | GCAGTTCTGCAGTATCTTCATGGTATTC | |
| OAH5 | GGAGAGGTACCAGTTGAACATAAGGGAGG | |
| OAH6 | CATGGAGCTCTCTTATCTCTAGAAACTAATTC | |
| OAH13 | CCGAGCTCTTACCCCCCGGGATC | |
| OAH14 | CCGCGGTAAATAACAAAAAGGAGAACGCATAATGTCAAAAGGAG | |
| OAH36 | CACACGGAATGGATGAATTATATAAGATGAAATATACCATTAAAGAT ATAGATATGAAGC | |
| OAH37 | CTTCATATCTATATCTTTAATGGTATATTTCATCTTATATAATTCATCCATTCCGTGTGTAATTCC | |
| OAH38 | GAAGCGAGCTCTTTAAACCCAAACATCTAAC | |
| OKR055 | GAATCTAGAATCCGCGGTAGTCGACATCCATGGTTGAGCTCATGGATCCTAA | |
| OKR056 | AGCTTTAGGATCCATGAGCTCAACCATGGATGTCGACTACCGCGGAT TCTAGATTCTGCA | |
| OAH56 | GGTACCCAAGTTGAGTATGTGGCTTCTATTGATG | |
| OAH57 | GAGCTCTTCTGCTTAAGTTTACATAATCAGC |
Electroporation of E. coli and C. perfringens.
Standard protocols were used for electroporation of plasmids into E. coli strain DH10B (5). Electroporation of C. perfringens was done, based on a previously published method (1), as follows: cells (3 ml) were grown anaerobically overnight in BHI, resuspended, and washed twice in sucrose electroporation buffer (333 mM sucrose, 6.7 mM NaHPO4 [pH 7], 1 mM MgCl2). Plasmid DNA and 400 μl of cell suspension were placed in a 4-mm cuvette on ice, and electroporation was done in an anaerobic chamber using an ECM 630 Electro Cell Manipulator (BTX, Harvard Apparatus). After electroporation, cell suspensions were incubated in BHI anaerobically at 37°C for 3 h, spread onto BHI plates with the appropriate antibiotics, and incubated anaerobically at 37°C until colony formation was observed.
Plasmid construction.
To construct pAH2, we amplified the bgaR-PbgaL region from strain 13 by PCR using the primers OAH1 and OAH2 and Phusion polymerase (New England Biolabs). Adenine residues were added to the ends of the PCR product using Taq polymerase, and the PCR product was ligated to pGEM-T Easy (Promega) to create plasmid pAH7. The insert was sequenced to determine if any mutations had occurred during amplification. We digested pAH7 with XhoI and PstI, and the bgaR-PbgaL fragment was ligated to XhoI- and PstI-digested pSM240 (9) to create pAH2. Plasmid pAH1 was constructed in the same manner but was discovered to have a frameshift mutation in bgaR (within codon 119 out of 279), causing translation to terminate after the addition of 14 aberrant amino acids. As such, it provides a useful negative control for experiments testing plasmid bgaR activity. To construct pKRAH1, we designed complementary primers (OKR055 and OKR056) that, when annealed, would contain several restriction recognition sequences rarely found in the C. perfringens chromosome and would have ends compatible with HindIII- and PstI-digested DNA. We ligated the annealed OKR055/OKR056 primers to pAH2 digested with PstI and partially digested with HindIII to avoid cutting at a second HindIII site internal to the bgaR gene. To construct pKRAH-Y, the yfp gene in plasmid pSW4-YFP (23) was amplified using PCR with primers OAH13 and OAH14, which contain a SacI and SacII site, respectively, and ligated to SacI- and SacII-digested pKRAH1. To construct pKRAH-YB, the yfp gene in plasmid pSW4-YFP was amplified with primers OAH14 and OAH36, and we then amplified the pilB gene from strain 13 chromosomal DNA by using primers OAH37 and OAH38. Using these partially overlapping PCR products as the template, we amplified yfp-pilB using OAH14 and OAH38 and ligated the product into pGEM-T Easy as described above, creating plasmid pAH11. pAH11 and pKRAH1 were then digested with SacII and SacI and ligated together to form pKRAH-YB.
Mutation of the bglR and bgaR genes in strain 13.
An 890-bp fragment internal to bglR was amplified from strain 13 by using the primers OAH5 and OAH6 and Phusion polymerase. Adenine residues were added to the ends of the PCR product by using Taq polymerase, and the PCR product was ligated into pGEM-T Easy to create plasmid pAH3. Plasmid pAH3 was digested with KpnI and SacI to release the bglR gene fragment, which was ligated to pSM300, forming pAH4. Plasmid pSM300 is a suicide plasmid carrying an ermBP resistance gene (9). Plasmid pAH4 was transformed into C. perfringens by electroporation and erythromycin-resistant (Ermr) recombinants selected using BHI plates with erythromycin. An Ermr transformant (strain AH1) was confirmed to have an insertion in the bglR gene by PCR amplification using primers that annealed within pAH4 and to an area of the chromosome outside the homologous region used for integration. Strain 13 chromosomal DNA was screened as a negative control. PCR using primers designed to amplify the bglR wild-type gene produced a very faint band in the bglR mutant strain AH1, which did not disappear after several days of growth in BHI, which suggested that there was a very small percentage of wild-type bacteria present, probably due to infrequent recircularization of the integrated plasmid via the regions of homology. A mutation was introduced into the bgaR gene in the same way, except a 389-bp fragment of bgaR was amplified from strain 13 with primers OAH56 and OAH57, cloned into PGEM-T Easy, creating pAH5, and then ligated into pSM300, creating pAH6. The mutant (strain AH2) was constructed and screened as described above for strain AH1.
β-Glucuronidase assays and β-galactosidase assays.
β-Glucuronidase assays were performed on mid-log-phase cells, and specific activity was calculated according to the protocol previously described by Melville et al. (16). β-Galactosidase assays were performed the same way, except that o-nitrophenyl-β-d-galactopyranoside (ONPG) was used as the substrate. For the timed induction assays, the cells were subcultured into 500 ml of prewarmed PY without inducer and grown to mid-log phase. The inducer was then added, and samples were removed at different time points and placed onto dry ice. The rest of the assay followed the same protocol as that described by Melville et al. (16).
Fluorescence microscopy.
For localization of YFP, 2% agarose pads with BHI, 0.1 mM lactose, and 20 μg/ml chloramphenicol were placed on slides as previously described (27). The slides were placed into a Coy anaerobic chamber, and bacteria were spread on them. After 2 h of growth, the slides were removed from the chamber and exposed to aerobic conditions for at least 20 min, and then a coverslip was placed on top and sealed with nail polish. Cells were viewed on an Olympus IX71 microscope using the Applied Precision deconvolution SoftWorx program. For localization of fluorescent proteins, images of >270 individual cells were examined.
RESULTS
Construction of a lactose-inducible promoter plasmid.
We analyzed the genome of strain 13 for putative regulators of carbohydrate metabolism that were oriented in the opposite direction from the genes they regulated, similar to araC and the araBAD operon in E. coli (8). An essential enzyme in most lactose metabolic pathways is β-galactosidase (19). C. perfringens strain 13 possesses four genes that have been annotated to encode β-galactosidases (assigned gene designation): CPE0167 (pbg), CPE0771 (bgaL), CPE0831 (also bgaL), and CPE1266 (none) (25). Of these β-galactosidase-encoding genes, only CPE0771 (bgaL) had a divergently oriented transcriptional regulator associated with it, CPE0770 (Fig. 1A). CPE0770 was annotated as encoding an AraC family transcriptional regulator (25). Because CPE0770 regulates bgaL in a lactose-dependent manner (see below), we renamed it bgaR (i.e., regulator of bgaL). Also, because there are two genes named bgaL in strain 13, we recommend that CPE0771 and CPE0831 be renamed bgaL and bgaM, respectively. Upstream and divergent to bgaR is a gene named gutA (Fig. 1A), which was annotated as a probable sugar transport protein (25) and may function as a transporter of lactose. The gutA, bgaR, and bgaL genes and their synteny are highly conserved in all nine C. perfringens strains with completely sequenced (18, 25) and partially sequenced (NCBI database) genomes.
FIG. 1.
(A) Gene order surrounding the bgaR (CPE0770) gene, an AraC family transcriptional regulator, in C. perfringens strain 13 (25). gutA encodes a probable sugar transport protein, and bgaL encodes a β-galactosidase. (B) Genes in the operon that includes bglR, a probable β-glucuronidase in strain 13. Gene CPE0145 is annotated as encoding a hypothetical protein, CPE0146 a 2-keto-3-deoxygluconate kinase, CPE0148 a putative transcriptional regulator, CPE0149 a putative oxidoreductase, CPE0150 a 2-dehydro-3-deoxyphosphogluconate/4-hydroxy-2-oxoglutarate aldolase, uxuA a d-mannonate dehydrolase, uxaC a glucuronate isomerase, uidB a glucuronide permease, and CPE0154 a putative beta-hexosamidase A.
The entire bgaR gene and the putative promoter and N-terminal region of bgaL (bgaR-PbgaL) were amplified by PCR and ligated into plasmid pSM240 to create plasmid pAH2 (Fig. 2). This placed the bgaL promoter immediately upstream of a cpe-gusA reporter gene fusion present in pSM240 (Fig. 2), which consists of the cpe ribosomal binding site and the first 13 codons fused in frame to the entire gusA gene of E. coli (16). The cpe gene from C. perfringens strain SM101 encodes the enterotoxin (CPE) responsible for acute food poisoning and has a ribosome binding site that likely provides a high level of translation of the protein since it is expressed at very high levels in strain SM101 (16). The E. coli gusA gene, encoding a β-glucuronidase (12), has been used successfully as a reporter in C. perfringens (16, 32), Clostridium acetobutylicum (6), Clostridium beijerinckii (6), and Clostridium difficile (15).
FIG. 2.
Relevant features of plasmids described in this report. Plasmid pSM240 was briefly described previously (9). T1(4) denotes four tandem terminators (26).
bgaR-PbgaL expression in three pathogenic strains of C. perfringens.
We transformed plasmid pAH2 into three different strains of C. perfringens and assayed β-glucuronidase activity at increasing concentrations of lactose (Fig. 3). Strain SM101, an acute food poisoning-associated strain, has a low endogenous β-glucuronidase activity of ∼4 to 8 U (16). We observed about an 80-fold increase in expression at maximum induction (10 mM lactose) and just 2-fold-increased expression above background levels in the absence of inducer (Fig. 3A). JGS4143, a strain isolated from an outbreak of chicken necrotic enteritis (G. Songer, personal communication), had 50 U of endogenous β-glucuronidase activity but showed lactose-inducible expression with pAH2 up to 10 times that of the background level (Fig. 3B). Strain 13, the source of bgaR-PbgaL1, also has significant (45 to 70 U) background β-glucuronidase activity (Fig. 3C) (17), but with pAH1 present, we could detect about 4-fold-increased β-glucuronidase activity as the lactose concentration increased (Fig. 3C).
FIG. 3.
β-Glucuronidase activity in C. perfringens at different concentrations of lactose. (A) Strain SM101; (B) strain JGS4143; (C) strain 13; (D) strain AH1. Strains containing no plasmid (squares), pSM240 (diamonds), pAH2 (circles), and pAH1 (triangles). Except for the sample at 0 mM lactose, lactose concentrations are shown on a log scale.
bglR in strain 13 encodes a β-glucuronidase.
Strain 13 is the most commonly used strain in C. perfringens pathogenesis and gene regulation studies (20, 21). Due to endogenous β-glucuronidase activity (Fig. 3C), it is difficult to gauge the precise levels of induction from the bgaR-PbgaL construct. Therefore, we located a gene that was annotated to encode a putative β-glucuronidase, bglR, in strain 13 (25). The bglR gene is the second gene in a nine-gene region that likely forms an operon (Fig. 1B). Based on comparison to the E. coli genes involved in glucuronide metabolism (19), the C. perfringens strain 13 operon appears to encode enzymes involved in metabolizing β-glucuronides. Using NCBI BLAST analyses with genes from strain 13, the genes associated with β-glucuronide metabolism are conserved in C. perfringens strains F4969, JGS1721, JGS1495, JGS1987, and NCTC 8239 but absent in strains ATCC 13124, SM101, and ATCC 3626.
We introduced a mutation into the bglR gene of strain 13 via homologous recombination of a nonreplicating plasmid (see Materials and Methods). The mutant strain, AH1, exhibited an 89% decrease in the β-glucuronidase activity at 0 mM lactose (Fig. 3D), suggesting that bglR encoded an enzyme responsible for the majority of β-glucuronidase activity. In strain AH1, pAH2 produced lower activity in the absence of inducer than did pAH2 in strain SM101 (compare Fig. 3D to A).
Also, we tried substituting a 10 mM concentration of the gratuitous inducer isopropyl β-d-1-thiogalactopyranoside (IPTG) in place of lactose as an inducer of PbgaL but did not observe an increase in activity (data not shown), suggesting that IPTG either is not an inducer or is not transported into the cell.
Expression from bgaR-PbgaL begins 5 to 10 min after the addition of an inducer.
To determine the kinetics of induction of the bgaR-PbgaL construct, we added 1 mM lactose to two strains, AH1(pAH2) and SM101(pAH2), in mid-log phase and measured the induction of activity over time (Fig. 4). The β-glucuronidase activity began to increase between 5 and 10 min after the addition of lactose and reached maximum levels between 1 to 2 h after lactose was added (Fig. 4).
FIG. 4.
Induction of β-glucuronidase activity (squares) at 1 mM lactose with strain AH1 (bglR mutant) (A) and SM101 (B) containing pAH2. OD (optical density at 600 nm) is shown as filled circles. Each chart shows a representative sample from two independent trials. The insets show an enlarged image of the first 20 min after induction.
Glucose has no significant effects on induction from bgaR-PbgaL1.
We performed all our assays in PY, a low-carbohydrate medium, as carbohydrate-inducible promoters in E. coli have exhibited repression in the presence of glucose (8). To determine if glucose has an effect on bgaR-PbgaL expression, we tested the effects of adding 10 or 110 mM glucose (the amount present in PGY media [16]) to strains AH1(pAH2) and SM101(pAH2). Neither 110 mM glucose nor 10 mM glucose had a significant effect on bgaR-PbgaL expression (data not shown).
bgaR is the activator responsible for the lactose-induced activity.
It was possible that the results described above could be due to the activity of a transcriptional regulator other than BgaR on the bgaL promoter. To ensure that BgaR was the protein responsible for regulating the bgaL promoter, we introduced a mutation into the chromosomal copy of the bgaR gene (see Materials and Methods) to create strain AH2. We then individually transformed pAH2 and pAH1 into this strain. Plasmid pAH1 has the same overall structure as pAH2, but the bgaR gene contains a frameshift mutation at residue 119 (out of 279) that leads to a prematurely terminated protein of 133 residues in length that is inactive. The β-glucuronidase expression in strain AH2(pAH2) upon addition of lactose was similar to that seen with strain 13(pAH1) (Fig. 5). However, strain AH2(pAH1), which lacks any wild-type copy of the bgaR gene, showed a complete lack of induction in the presence of lactose, indicating that BgaR is responsible for lactose-mediated induction. Since the absence of BgaR leads to loss of induction (Fig. 5), it is likely that BgaR functions as a transcriptional activator and not a repressor in response to lactose.
FIG. 5.
β-Glucuronidase activity in response to lactose concentration. Strain AH2 (open symbols) with pAH1 (triangles) and pAH2 (circles). For comparison, strain 13 with pAH1 (same data as that shown in Fig. 3C) is shown by filled triangles.
The bgaR mutant strain was examined to see if the mutation affected the total amount of the β-galactosidase activity, measured by hydrolysis of ONPG, in the cell. The level of β-galactosidase activity was relatively low in the wild-type strain (10 to 14 U) and showed limited induction in the presence of lactose (Fig. 6). However, there was a significantly reduced level of enzyme activity in the bgaR mutant strain at each concentration of lactose except 0.1 mM, and the limited amount of lactose-dependent induction was lost (Fig. 6). Because the activity of ONPG hydrolysis was relatively low, we performed a control where the E. coli lacZ gene, encoding β-galactosidase, under the control of the cpe gene promoter from strain SM101, was placed on a plasmid which was transformed into strain 13. This strain exhibited a significant increase in ONPG hydrolysis (data not shown), indicating that there were no limitations to measuring ONPG hydrolysis in our assays.
FIG. 6.
β-Galactosidase activity in response to lactose concentration. Strain 13 (squares) and strain AH2 (bgaR mutant) (diamonds).
Replacement of the cpe-gusA reporter with a multiple cloning site allows controlled expression of desired proteins.
After determining that pAH2 works well for controlled induction, we replaced the cpe-gusA gene fusion with a multiple cloning site containing restriction enzyme recognition sites that are rarely found in C. perfringens. The resulting plasmid, pKRAH1 (Fig. 2), allows lactose-induced controlled expression of any gene desired.
YFP-PilB localizes to the pole in C. perfringens strain 13 but not in a pilC mutant strain.
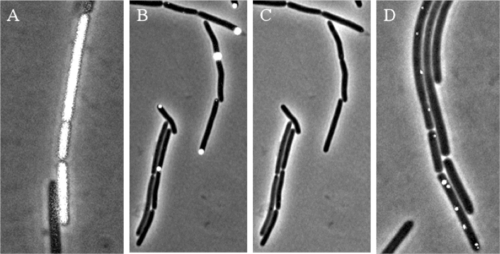
We have previously shown that type IV pili (TFP) have a role in C. perfringens gliding motility (30). One of the gene clusters involved in TFP motility was an operon containing the genes pilB, pilC, CPE1842, and CPE1841. The pilB gene product is likely an ATPase responsible for assembly and extension of TFP (30). The pilC gene product is a membrane protein that we showed was necessary for TFP gliding motility in C. perfringens by mutagenesis of the gene (30). PilC homologs have been shown to aid in localization of PilB to the poles of cells in Gram-negative bacteria with TFP, including Pseudomonas aeruginosa (2). To determine whether PilC performed the same in the Gram-positive C. perfringens, we inserted genes encoding YFP and a YFP-PilB fusion protein, in which YFP was attached to the N terminus of PilB, into pKRAH1. In medium without lactose, there was no visible fluorescence (data not shown). While expression of the yfp gene alone led to diffusion of the YFP protein throughout the cytoplasm of the bacteria (Fig. 7 A), the YFP-PilB protein localized to the pole in 73% of the cells (Fig. 7B and C). In strain SM126, the previously constructed nonmotile pilC mutant derived from strain 13 (30), the fluorescence was still punctate, but only 42% was on the cell poles, and the fluorescent signals were less intense than the polar fluorescence in strain 13 (Fig. 7D). These results suggest that the PilC protein is at least partly responsible for polar localization of PilB in C. perfringens.
FIG. 7.
Cellular localization of YFP and the YFP-PilB fusion protein. Strains were grown on agarose pads with BHI and 0.1 mM lactose before images of cells were collected with phase-contrast microscopy and fluorescence microscopy (where indicated). (A) Strain 13 with pKRAH-Y, showing YFP fluorescence (white coloration in cells) distributed evenly throughout the cell. (B) Strain 13 with pKRAH-YB, in which the fluorescence (white spots) is distributed mostly to the poles. (C) Same image as that in panel B but with phase contrast only to distinguish the location of cell poles. (D) Strain SM126, a pilC mutant derived from strain 13, with pKRAH-YB. Fluorescence intensity levels were adjusted to the same minimum and maximum levels for the images in panels B and D so they could be directly compared to each other.
DISCUSSION
Two gaps in the genetic tools available for C. perfringens were the absence of a vector in which promoters from genes of interest could be studied in the native C. perfringens host and a readily usable vector for highly controlled gene expression. Plasmid pSM240, which fulfills the first criterion, was briefly described previously, where it was used to analyze the expression patterns of promoters from the sigK and spoIIGA genes from strain SM101 (9), but a detailed description of its utility was not provided. Because it has a polylinker between a region containing four tandem terminators and the cpe-gusA reporter gene fusion (Fig. 2), it can be used to study the regulation of almost any promoter found in C. perfringens. The effectiveness of the terminators can be seen in Fig. 3A, B, and D, where the strains containing pSM240 show almost no additional β-glucuronidase activity over that of the wild-type strain lacking any plasmids. The cpe-gusA reporter in pSM240 is more useful in a C. perfringens strain 13 background if the bglR mutant strain, AH1, is used as the host. Since the endogenous β-glucuronidase activity was reduced from ∼70 U to ∼8 U in strain AH1, it will make possible the analysis of all but weak promoters.
We used a regulator of a putative β-galactosidase-encoding gene and its adjacent promoter as the basis for constructing a vector to fulfill the second need for highly controlled gene expression. We deliberately searched for sugar metabolism genes with a divergent regulator and promoter, because this orientation is more likely to avoid read-through transcription from an upstream gene in the same orientation. This has been clearly demonstrated with the pBAD family of expression vectors developed by Guzman and coworkers (8). In the case of the pBAD plasmids, the promoter could be repressed by the addition of glucose to the medium, giving a larger range of regulation (8), but the bgaR-PbgaL in pAH2 was not. Although this may limit the range of transcriptional regulation, it can also be viewed as a positive feature since C. perfringens can be grown in rich medium with high levels of glucose without repression, which would be valuable for obtaining high levels of synthesis of a target protein for purification or other uses.
The bgaR-PbgaL construct in pAH2 was regulated efficiently by lactose in three different strains of C. perfringens that have three different pathogenesis profiles, gas gangrene (strain 13), acute food poisoning in humans (strain SM101), and necrotic enteritis in chickens (strain JGS4143) (Fig. 3). Since the gutA-bgaR-bgaL operon was found in all sequenced strains of C. perfringens, with GutA being the putative lactose permease, it suggests that this construct can be utilized in virtually any strain in this species. Although not tested yet, bgaR-PbgaL may also provide regulated expression in other Clostridium species as long as they can transport lactose into the cell.
Sequence alignment of the four potential β-galactosidase proteins in strain 13 reveals a remarkable degree of divergence in the sequences, since the pair with the highest level of similarity, BgaM and CPE1266, exhibit only 24.2% sequence identity (Table 2). A search for homologs of the four β-galactosidase enzymes indicates that they are found in all strains of C. perfringens with sequenced genomes (data not shown). Distinguishing characteristics of the β-galactosidases are listed in Table 2. For still unknown reasons, it appears that C. perfringens has evolved multiple mechanisms for facilitating the metabolism of lactose.
TABLE 2.
Characteristics of four putative β-galactosidases in C. perfringens strain 13
| β-Galactosidase | % sequence similarity to: |
Other bacteria with homology | Distinguishing characteristic(s) | |||
|---|---|---|---|---|---|---|
| BgaL (CPE0771) | BgaM (CPE0831) | Pbg (CPE0167) | CPE1266 | |||
| BgaL (CPE0771) | 17.3 | 10.4 | 15 | Firmicutes | Homologous proteins often annotated as glycoside hydrolases | |
| BgaM (CPE0831) | 17.3 | 10.2 | 24.2 | Agrobacterium-Sinorhizobium | None | |
| Pbg (CPE0167) | 10.4 | 10.2 | 10.2 | Firmicutes | Transcription shown to be induced by lactose (13) and cleaved X-Gal when cloned into an E. coli plasmid and activated by ONPG (24) | |
| CPE1266 | 15 | 24.2 | 10.2 | Bifidobacterium and pathogenic members of the Firmicutes | Potential secretion signal sequence at the N terminus; homologous proteins are surface anchored and contribute to avoidance of phagocytosis by inhibiting complement deposition on the surface of the bacterium (4) | |
Despite strain 13 having multiple putative β-galactosidase enzymes, we detected a relatively low level of enzyme activity (10 to 14 U) in strain 13 by using ONPG as the substrate, and this activity was only slightly affected by the presence of lactose (Fig. 6). Loss of bgaR function resulted in decreased activity, but 60% of the activity remained (Fig. 6). Since we have observed an 80-fold induction of the bgaL promoter by adding lactose (Fig. 3), it seems likely that the BgaL enzyme does not hydrolyze ONPG as a primary enzymatic activity.
Kobayashi et al. suggested that CPE0166 (ORF54) was a lactose transporter, likely a permease (13). The gutA gene product, the putative lactose permease divergent to the bgaR gene (Fig. 1A), has little sequence similarity with CPE0166 and was likely given the gutA designation because it has similarity to glucuronide transport proteins (25). No obvious transporter was found nearby either the bgaM gene or CPE1266, which encode the other two putative β-galactosidases in strain 13 (data not shown).
Another useful outcome of this study was the confirmation of the gene assignment that bglR encoded a β-glucuronidase in strain 13. This was done by mutagenesis of the bglR gene and by showing that the mutant had a significant decrease in β-glucuronidase activity (Fig. 3D). However, the method used for mutagenesis, insertion of a nonreplicating plasmid by homologous recombination, is likely to have polar effects on downstream genes in the locus, so we cannot exclude the possibility that one of these genes is also responsible for this phenotype. The remaining 11% of β-glucuronidase activity in the bglR mutant strain may be due to residual activity from another β-glucuronidase enzyme that cannot be detected by sequence homology to known β-glucuronidases or nonspecific activity of another hexuronidase. Strain SM101 lacks the bglR gene yet has low but detectable β-glucuronidase activity of 4 to 8 U (Fig. 3A), suggesting that other enzymes with this activity exist in C. perfringens.
To demonstrate the utility of the expression system, we used the pKRAH1-based vector to express a gene encoding a YFP-PilB protein fusion and showed that polar localization of PilB is due, at least in part, to the membrane-bound protein PilC (Fig. 7). This is similar to results seen in the Gram-negative bacterium P. aeruginosa and suggests that PilC functions as the membrane-bound partner for the PilB protein in C. perfringens. Expression of heterologous proteins, including the green fluorescent protein (GFP) family, in C. perfringens should provide the means to make significant discoveries related to gene and protein functions in this bacterium.
Acknowledgments
We thank Katherine Rodgers for assistance in constructing plasmid pKRAH1 and Stephen Leppla for plasmid pSW4-YFP.
This work was supported by USDA/NRICGP grant 2007-01543 and NIH grant 1R21AI088298-01 to S.B.M.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Allen, S. P., and H. P. Blaschek. 1990. Factors involved in the electroporation-induced transformation of Clostridium perfringens. FEMS Microbiol. Lett. 58:217-220. [DOI] [PubMed] [Google Scholar]
- 2.Chiang, P., M. Habash, and L. L. Burrows. 2005. Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collie, R. E., J. F. Kokai-Kun, and B. A. McClane. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe 4:69-79. [DOI] [PubMed] [Google Scholar]
- 4.Dalia, A. B., A. J. Standish, and J. N. Weiser. 2010. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect. Immun. 78:2108-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girbal, L., et al. 2003. Development of a sensitive gene expression reporter system and an inducible promoter-repressor system for Clostridium acetobutylicum. Appl. Environ. Microbiol. 69:4985-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillouard, I., T. Garnier, and S. T. Cole. 1996. Use of site-directed mutagenesis to probe structure-function relationships of alpha-toxin from Clostridium perfringens. Infect. Immun. 64:2440-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harry, K. H., R. Zhou, L. Kroos, and S. B. Melville. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific sigma factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 191:2728-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heap, J. T., O. J. Pennington, S. T. Cartman, G. P. Carter, and N. P. Minton. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452-464. [DOI] [PubMed] [Google Scholar]
- 11.Hogenauer, C., et al. 1998. Mechanisms and management of antibiotic-associated diarrhea. Clin. Infect. Dis. 27:702-710. [DOI] [PubMed] [Google Scholar]
- 12.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. Beta-glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. U. S. A. 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi, T., T. Shimizu, and H. Hayashi. 1995. Transcriptional analysis of the beta-galactosidase gene (pbg) in Clostridium perfringens. FEMS Microbiol. Lett. 133:65-69. [DOI] [PubMed] [Google Scholar]
- 14.Lanckriet, A., et al. 2009. Generation of single-copy transposon insertions in Clostridium perfringens by electroporation of phage Mu DNA transposition complexes. Appl. Environ. Microbiol. 75:2638-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mani, N., B. Dupuy, and A. L. Sonenshein. 2006. Isolation of RNA polymerase from Clostridium difficile and characterization of glutamate dehydrogenase and rRNA gene promoters in vitro and in vivo. J. Bacteriol. 188:96-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melville, S. B., R. Labbe, and A. L. Sonenshein. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 62:5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mendez, M., et al. 2008. Carbon catabolite repression of type IV pilus-dependent gliding motility in the anaerobic pathogen Clostridium perfringens. J. Bacteriol. 190:48-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myers, G. S., et al. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neidhardt, F. C., et al. (ed.). 1996. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 20.Rood, J. I. 1997. Genetic analysis in Clostridium perfringens, p. 65-72. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, Inc., San Diego, CA.
- 21.Rood, J. I. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333-360. [DOI] [PubMed] [Google Scholar]
- 22.Rood, J. I., and S. T. Cole. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55:621-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sastalla, I., K. Chim, G. Y. C. Cheung, A. P. Pomerantsev, and S. H. Leppla. 2009. Codon-optimized fluorescent proteins designed for expression in low-GC Gram-positive bacteria. Appl. Environ. Microbiol. 75:2099-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu, T., T. Kobayashi, W. Ba-Thein, K. Ohtani, and H. Hayashi. 1995. Sequence analysis of flanking regions of the pfoA gene of Clostridium perfringens: beta-galactosidase gene (pbg) is located in the 3′-flanking region. Microbiol. Immunol. 39:677-686. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu, T., et al. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sussman, J. K., C. Masada-Pepe, E. L. Simons, and R. W. Simons. 1990. Vectors for constructing kan gene fusions: direct selection of mutations affecting IS10 gene expression. Gene 90:135-140. [DOI] [PubMed] [Google Scholar]
- 27.Tran, P. T., A. Paoletti, and F. Chang. 2004. Imaging green fluorescent protein fusions in living fission yeast cells. Methods 33:220-225. [DOI] [PubMed] [Google Scholar]
- 28.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varga, J., V. L. Stirewalt, and S. B. Melville. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 186:5221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga, J. J., et al. 2006. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other clostridia. Mol. Microbiol. 62:680-694. [DOI] [PubMed] [Google Scholar]
- 31.Vidal, J. E., J. Chen, J. Li, and B. A. McClane. 2009. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One 4:e6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]