Abstract
Over half of the bacterioplankton cells in ocean surface waters are capable of carrying out a demethylation of the phytoplankton metabolite dimethylsulfoniopropionate (DMSP) that routes the sulfur moiety away from the climatically active gas dimethylsulfide (DMS). In this study, we tracked changes in dmdA, the gene responsible for DMSP demethylation, over the course of an induced phytoplankton bloom in Gulf of Mexico seawater microcosms. Analysis of >91,000 amplicon sequences indicated 578 different dmdA sequence clusters at a conservative clustering criterion of ≥90% nucleotide sequence identity over the 6-day study. The representation of the major clades of dmdA, several of which are linked to specific taxa through genomes of cultured marine bacterioplankton, remained fairly constant. However, the representation of clusters within these major clades shifted significantly in response to the bloom, including two Roseobacter-like clusters and a SAR11-like cluster, and the best correlate with shifts of the dominant dmdA clades was chlorophyll a concentration. Concurrent 16S rRNA amplification and sequencing indicated the presence of Roseobacter, SAR11, OM60, and marine Rhodospirillales populations, all of which are known to harbor dmdA genes, although the largest taxonomic change was an increase in Flavobacteriaceae, a group not yet demonstrated to have DMSP-demethylating capabilities. Sequence heterogeneity in dmdA and other functional gene populations is becoming increasingly evident with the advent of high-throughput sequencing technologies, and understanding the ecological implications of this heterogeneity is a major challenge for marine microbial ecology.
Dimethylsulfoniopropionate (DMSP) is a ubiquitous phytoplankton metabolite that is degraded by marine microorganisms by at least two major pathways. The cleavage pathway involves degradation of DMSP by phytoplankton or bacteria to produce dimethylsulfide (DMS). DMS is the largest natural source of sulfur from the ocean to the atmosphere (2) and, upon oxidation, forms cloud condensation nuclei, hypothesized to affect climate on a global scale (1, 4, 19, 20). The alternative DMSP degradation pathway is carried out by bacteria alone and involves an initial demethylation to methylmercaptopropionate (MMPA), with some portion of the sulfur subsequently shunted to sulfur-containing amino acids (12, 14, 26).
The genes involved in the initial step of both DMSP degradation pathways have recently been discovered (7, 9, 31). Three different genes that each encode the first step of the DMSP cleavage pathway (dddD, dddL, and dddP) have been identified (7, 31, 32), although they are in low abundance in bacteria from surface ocean waters; an estimated 0.1% (10), 3.0% (10), and 3.2% (31) of cells contain these genes, respectively. Conversely, the only gene found thus far that encodes the first step in the DMSP demethylation pathway (dmdA) is highly abundant in surface waters; an estimated 58% of cells contain this gene (9, 10).
Five clades of DmdA, designated A through E, have been found in cultured marine bacteria or marine metagenomic sequences thus far. Clade A sequences include genes from various marine Roseobacter and Rhodospirillales species. Clade B dmdA sequences are represented by the only sequenced marine SAR116 group member, “Candidatus Puniceispirillum marinum” (22). Clade C sequences include a dmdA gene from SAR11 isolate Pelagibacter ubique HTCC7211 (34). Clade D sequences include genes from SAR11 isolates P. ubique strains HTCC1002 and HTCC1062, as well as a second homolog in P. ubique HTCC7211. Clade E sequences include a gene from marine gammaproteobacterium HTCC2080 in the OM60 clade (9, 10, 34). Each of these recognized clades has been further grouped into up to four distinct subclades that contain clusters of closely related sequences, including A1 to A3, B1 to B4, C1 and C2, D1 to D3, and E1 and E2 (34).
Across many open and coastal ocean surface water habitats, the distribution of genes across dmdA clades is surprisingly consistent (10). Generally, clade D sequences are found in the highest abundance (∼40% of cells) and clade E sequences are found in the lowest abundance (∼0.6% of cells) (9, 10). These clade and subclade distributions, however, have not been studied over time in a single environment, particularly as ecological conditions shift. Because phytoplankton blooms typically create substantial changes in DMSP concentration and flux (8, 33, 35, 39), and these might favor certain DmdA proteins or the organisms that carry them, this study tracked the composition and diversity of the dmdA gene pool in an experimental phytoplankton bloom induced by nutrient additions to Gulf of Mexico seawater. Relative changes in dmdA clade abundance were tracked by deep sequencing of amplicons using a universal dmdA primer set and interpreted in the context of simultaneous changes in DMSP concentration and fate, bacterial production, chlorophyll a (Chl a) concentration, and bacterial and archaeal community composition.
MATERIALS AND METHODS
Sample collection.
Seawater was collected from oligotrophic surface waters (<1 m deep) in the Gulf of Mexico (30° [03.041], 87° [59.708]; 27°C, salinity = 34) in October 2006. Water was filtered through a 200-μm mesh into six 20-liter extensively preleached Cubitainers (Fold-A-Carrier; Reliance Products, Ltd.) leaving negligible headspace to minimize volatile organic sulfur partitioning. Three experimental Cubitainer microcosms were amended with sodium nitrate (10 μM) and potassium phosphate (0.6 μM) to induce bloom conditions; three control microcosms received no nutrient amendments. The microcosms were incubated at 27°C with a 12-h-on/12-h-off light cycle (200 μmol quanta m−2 s−1) for the duration of the experiment.
Chemical analysis and bacterial activity measurements.
At the initial time point (T = 0), the full contents of one control and one experimental microcosm (20 liters each) were sacrificed for sampling. Thereafter, samples were collected daily from each of the other four microcosms (two control and two experimental) over the next 6 days. The Cubitainers collapsed as water volume was removed, resulting in relatively constant headspace volume throughout the experiment. For chemical and activity measurements, ∼500 ml of water was collected and immediately subdivided as follows. Chlorophyll a (Chl a) samples were collected by filtration (50 ml) on Whatman GF/F filters. The filters were extracted in 90% acetone for 24 h at −20°C, and Chl a in the extracts was quantified by fluorometry (23). Samples for dissolved DMSP (DMSPd) were collected by small-volume drip filtration through GF/F filters (15). Total DMSP (dissolved plus particulate) was collected by acidifying whole seawater with 5 μl ml−1 of 50% H2SO4, similar to the method described by Curran et al. (6). Dissolved and total DMSP were quantified as DMS after alkaline hydrolysis. Bacterial production was measured by [3H]leucine incorporation using the method developed by Kirchman et al. (17) and modified by Smith and Azam (28); live and killed (5% trichloroacetic acid [TCA]) controls were incubated with [3H]leucine (20 nM final concentration) for 1 h in the dark. [35S]DMSP was synthesized from l-[35S]methionine by enzymatic decarboxylation/deamination with l-amino acid oxidase and subsequent methylation of 3-methiolpropionate with acidic methanol. Purification of the [35S]DMSP was by high-performance liquid chromatography (HPLC) and ion exchange using the procedures outlined in the work of Kiene et al. (13). Rate constants for DMSPd consumption were obtained by measuring the loss of tracer [35S]DMSP from the dissolved pool over time. At each time point, subsamples of seawater were filtered though 0.2-μm nylon membranes and the filtrate was immediately acidified for preservation. [35S]DMSP remaining in the filtrate was assayed as described in the work of Slezak et al. (27). The rate of DMSPd consumption (in nM day−1) was obtained by multiplying the DMSPd concentration by the rate constant. The fraction of [35S]DMSP assimilated into microbial macromolecules was determined after 24 h of incubation by filtering water samples through 0.2-μm nylon filters, rinsing the filters with 5% TCA, and counting the 35S activity remaining on the filter.
DNA extraction.
For DNA samples, water from each time point was sequentially filtered through 8-μm (293-mm-diameter) and 3-μm (47-mm-diameter) prefilters, and then particles were collected on 0.2-μm (47-mm)-pore-size polycarbonate filters (Poretics) until the filters clogged (∼250- to 500-ml volume, depending on the sample). The 0.2-μm filters were flash frozen in liquid nitrogen and then stored at −20°C until extraction. DNA was extracted using the PowerMax soil DNA isolation kit (MoBio Laboratories, Inc.) following manufacturer's instructions, and concentrations were estimated by absorbance on a NanoDrop spectrophotometer (Thermo Scientific).
Primer design.
The universal dmdA primer set, dmdAUF160 (3′-GTICARITITGGGAYGT-5′) and dmdAUR697 (3′-TCIATICKITCIATIAIRTTDGG-5′), was designed as described previously (34). Briefly, dmdA sequences from cultured organisms and the Global Ocean Sampling (GOS) metagenome (10) were aligned in Geneious Pro 3.5.6 (Geneious v4.7; A. J. Drummond et al., 2009) using the ClustalW algorithm. The primer set was modified with degenerate and inosine bases to target as many dmdA sequences as possible while excluding related gcvT and aminomethyltransferase gene sequences (34).
PCRs.
DNA extracted from duplicate control and experimental samples (2 treatments × 2 replicates × 7 time points; 28 total samples) was used as template for PCRs, followed by pyrosequencing of amplicons (21). PCRs were carried out with Platinum High-Fidelity Taq DNA polymerase (1/2 U; Invitrogen) in the native 10× PCR buffer with 8 ng (for dmdA amplifications), 5 ng (for bacterial 16S rRNA amplifications), or 10 ng (for archaeal 16S rRNA amplifications) of template DNA. Universal dmdA primers were modified from the work of Varaljay et al. (34) to include 454 Life Sciences A or B adaptor sequences and 14 tetranucleotide “key” sequences for sample differentiation during sequence analysis (11, 29). The dmdA cycling conditions were as follows: 94°C for 2 min, followed by 40 cycles of 94°C for 35 s, 41°C for 35 s, and 68°C for 60 s, with a final extension step of 68°C for 10 min (34). 16S rRNA bacterial and archaeal primers covering the V6 hypervariable region (developed by Sogin et al. [29] and modified by Huber et al. [11]; these are a combination of 5 forward and 4 reverse bacterial 16S rRNA primers and 1 forward and 2 reverse archaeal 16S rRNA primers) were made with 14 distinct tetranucleotide keys, as described above. Bacterial or archaeal primers were mixed equally from 10 μM stock solutions, and 2 μl of this pooled solution was used in each 25-μl PCR mixture (11). The 16S rRNA gene cycling conditions were as follows: 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 30 s, with a final extension step of 68°C for 10 min.
Sequencing preparation.
PCR products were run on 1% (dmdA amplicons) or 2% (bacterial and archaeal 16S rRNA amplicons) agarose gels, followed by purification using the QIAquick gel extraction kit (Qiagen). As a final DNA purification, products were cleaned using the Ampure system (Agencourt Bioscience Corporation), with modifications to the volume of purified PCR products (30 μl) and AMPure beads (50.4 μl). After final purification, DNA concentrations were determined as described above and products (61 control and 60 experimental samples) were pooled in equal amounts, except that dmdA amplicons were added to the mixtures at double the amount of the 16S rRNA amplicons. Two separate pools were assembled, each of which used the distinct identification keys only once, and these were sequenced on separate halves of the PicoTiterPlate (454 Life Sciences). A total of ∼1,300 ng DNA was used in Roche GS FLX LR70 pyrosequencing at the University of South Carolina Environmental Genomics Facility (Columbia, SC).
Sequence analysis.
For reads of high quality (those that contained full and correct forward primer sequences and no uncalled bases and had average quality scores of >21), adaptor and key sequences were removed and sequences were clustered using the CD-HIT program (18) with ≥90% similarity for dmdA clusters (34) and ≥99% similarity for 16S rRNA clusters. The average read length was 190 bp for dmdA amplicons, 70 bp for bacterial rRNA amplicons, and 72 bp for archaeal rRNA amplicons after adaptor, key, and primer sequences were trimmed. For analysis of dmdA amplicons, reference sequences (defined as the longest read in the cluster) were analyzed via BLASTX against an in-house DmdA database assembled from cultured marine bacteria and the Global Ocean Sampling (25) metagenomic sequences (http://roseobase.org/DmdApaper) and augmented with a number of paralogous, non-DmdA sequences (10). Sequences were considered to be valid dmdA sequences if the top hit in the database had a bit score of ≥30. The sequences were assigned to the clade of the top hit (either A, B, C, D, or E or unclassified). These dmdA sequences were further assigned to protein subclades via a tree-building method using known DmdA subclade sequences as anchors (34). For taxonomic analysis of bacterial 16S rRNA amplicons, reference sequences were aligned against a marine bacterial 16S rRNA database with representatives of all major marine taxa (3). Amplicons with ≥90% similarity and an overlap of ≥70% with one of the representative marine sequences were assigned to that taxon. For taxonomic analysis of archaeal 16S rRNA amplicons, the SIMO RDP Agent (www.simo.marsci.uga.edu) was used to compare amplicon sequences against archaeal type species housed in the Ribosomal Database Project (RDP) database (5), using similarity cutoffs of 100% for species, >95% for genus, >92% for family, >91% for order, >85% for class, and >75% for phylum assignments. Because of the poor coverage of archaeal diversity among type species, most sequences were identified only to the phylum level or remained unclassified.
Statistical analysis.
Clusters were analyzed via multidimensional scaling (MDS) using Primer 5 for Windows software (Plymouth Marine Laboratory, Plymouth, United Kingdom). A Bray-Curtis similarity matrix (4th root transformed to deemphasize the contribution of any one particular dominant cluster) was constructed from the clusters from each replicate microcosm (two control and two nutrient amended) over time (6 days). For dmdA sequences, only the top 100 clusters (which contained 70% of the amplicons) were included in the statistical analysis. For bacterial 16S rRNA sequences, all clusters were analyzed (9,588 clusters representing 55 marine taxa). dmdA and 16S rRNA clusters were also evaluated with a SIMPER analysis (Primer 5), which determines the clusters contributing most to the differences between samples. Reference sequences from the dmdA clusters that cumulatively created 50% of the dissimilarity between control and bloom microcosms were analyzed in a maximum-likelihood tree created using RAxML 7.0 with 100 bootstrap samplings. dmdA and 16S rRNA clusters were evaluated with both BIO-ENV and BVSTEP routines (Primer 5) to determine the measured environmental variables that correlated with changes in these clusters (BIO-ENV does a full search while BVSTEP does a stepwise search to determine the variable[s] best explaining the cluster structure changes). A rarefaction curve of all dmdA sequences clustered at the 90% similarity level was created using EcoSim 7.0 (EcoSim: null models software for ecology, version 7; N. J. Gotelli and G. L. Entsminger, 2008, Acquired Intelligence Inc. and Kesey-Bear, Jericho, VT) with 1,000 resamplings.
Nucleotide sequence accession numbers.
rRNA sequences and accompanying metadata (compiled according to Minimum Information for Metagenomic Sequences [MIMS] standards) have been deposited in the Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis (CAMERA) database with project identification (ID) CAM-PROJ_DICE (sample CAM_S_A001 for experimental microcosms and CAM_S_A002 for control microcosms). dmdA amplicon sequences have been deposited in the NCBI Short Read Archive with project ID 49967.
RESULTS
Phytoplankton bloom dynamics.
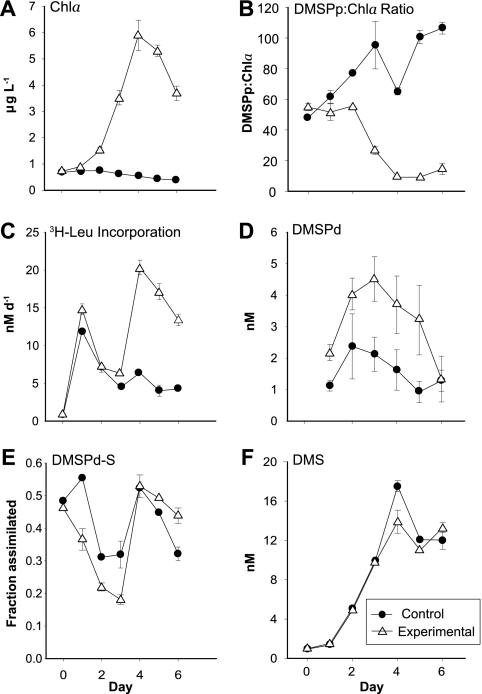
While control microcosms maintained low Chl a levels throughout the experiment (<1 μg liter−1), the nutrient-amended experimental microcosms had variable and higher Chl a concentrations that peaked at day 4 (6 μg liter−1 [Fig. 1 A]), indicating a phytoplankton bloom. Changes in rates of bacterial secondary production suggested increased substrate availability after day 3 in the experimental microcosms, consistent with bloom conditions (Fig. 1C).
FIG. 1.
Biological and chemical properties of control and experimental microcosms during the 6-day study. Panels show concentrations of chlorophyll a (A), the ratio of particulate DMSP concentration to chlorophyll a concentration (B), rates of heterotrophic bacterial protein production (C), the concentration of dissolved DMSP (D), the fraction of the dissolved DMSP consumed by bacteria that was assimilated into macromolecules (E), and the concentration of DMS (F). Each point represents the mean and standard deviation of three independent determinations from two replicate microcosms.
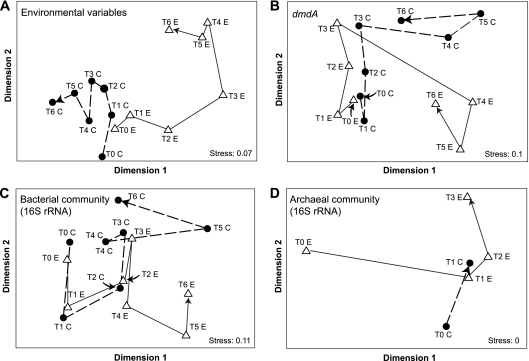
The fraction of consumed dissolved DMSP (DMSPd) degraded to DMS (maximally 14% [data not shown]) and the fraction of DMSPd-sulfur assimilated into macromolecules (maximally 55% [Fig. 1E]) followed the same trend in both control and experimental microcosms. However, the ratio of particulate DMSP (DMSPp) to Chl a, a proxy for the relative amount of DMSP produced per phytoplankton cell (35), decreased over the course of the induced bloom in the experimental microcosms (Fig. 1B). This pattern suggests that the phytoplankton dominating the bloom were poor DMSP producers relative to those in the initial and control communities, although their higher numbers nonetheless caused substantial increases in DMSP flux. For all environmental variables except NO3− and PO43− (which were excluded because they were experimentally manipulated), the conditions and processes in the experimental microcosms were most dissimilar to the initial samples and the control microcosms on days 4 to 6 (Fig. 2 A).
FIG. 2.
Multidimensional scaling of the Bray-Curtis similarity matrix generated from measures of environmental variables (principal-component analysis) (A), the relative abundance of the 100 largest dmdA clusters (B), the relative abundance of bacterial 16S rRNA gene clusters (C), and the relative abundance of archaeal 16S rRNA gene clusters (D) over time in control and experimental microcosms. Arrows show the time progression for control “C” (closed circles) and experimental “E” (open triangles) microcosms.
DMSP demethylase gene (dmdA) dynamics.
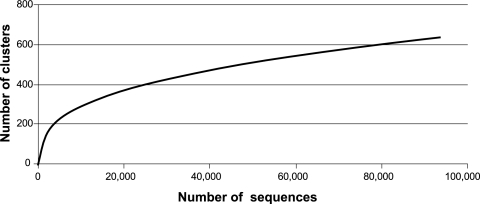
The 91,418 dmdA sequences obtained (from 28 samples representing two replicates per treatment for days 0 to 6) formed 578 clusters at 90% nucleotide sequence identity (average of 158 sequences per cluster). At this similarity level, the experiment-wide richness had not yet reached a plateau (Fig. 3). Given the error rate for FLX pyrosequencing (∼0.5% [21]) compared to the 10% sequence divergence within a cluster, sequencing artifacts played little or no role in cluster assignments. Non-dmdA sequences captured by the universal dmdA primer set were also found (20,761 sequences); these formed 647 clusters (average of 32 sequences per cluster) and were not considered further. Overall, 82% of the amplicons were identified as dmdA sequences, while 18% were identified as encoding GcvT proteins or other related sequences.
FIG. 3.
Rarefaction curve of dmdA sequences (all treatments and samples combined) clustered at the 90% similarity level.
dmdA cluster richness was highest in both treatments 24 h after the initiation of the experiment (day 1 [Table 1]), with 240 and 238 clusters, respectively, in experimental and control microcosms. Richness generally decreased after this point (normalized cluster richness in Table 1), but overall the temporal shifts were not large.
TABLE 1.
dmdA sequence number, cluster number (90% sequence identity), diversity, and coveragea
| Day | Control |
Experimental |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of reads | No. of clusters | Normalized no. of clusters | H′ | Coverage (%) | No. of reads | No. of clusters | Normalized no. of clusters | H′ | Coverage (%) | |
| 0 | 6,222 | 208 | 168 (152-185) | 3.85 | 66 | 4,842 | 206 | 178 (161-191) | 3.92 | 69 |
| 1 | 6,632 | 238 | 192 (175-206) | 4.01 | 68 | 5,254 | 240 | 203 (183-217) | 4.12 | 69 |
| 2 | 6,385 | 194 | 163 (149-176) | 3.86 | 76 | 6,348 | 201 | 170 (154-184) | 4.01 | 75 |
| 3 | 7,854 | 181 | 143 (125-162) | 3.58 | 70 | 6,765 | 197 | 158 (141-176) | 3.64 | 69 |
| 4 | 9,130 | 216 | 160 (141-175) | 3.69 | 67 | 7,202 | 205 | 161 (143-178) | 3.81 | 66 |
| 5 | 15,882 | 211 | 162 (148-175) | 3.81 | 86 | 6,851 | 245 | 193 (176-211) | 4.17 | 67 |
| 6 | 7,394 | 164 | 135 (118-148) | 3.49 | 76 | 6,167 | 185 | 158 (143-169) | 4.02 | 76 |
Normalized no. of clusters, cluster numbers estimated for a 4,800-sequence library to normalize for differences in number of reads across samples; the range of 1,000 bootstraps is given in parentheses. H′, Shannon-Wiener diversity index. Coverage, Good's coverage.
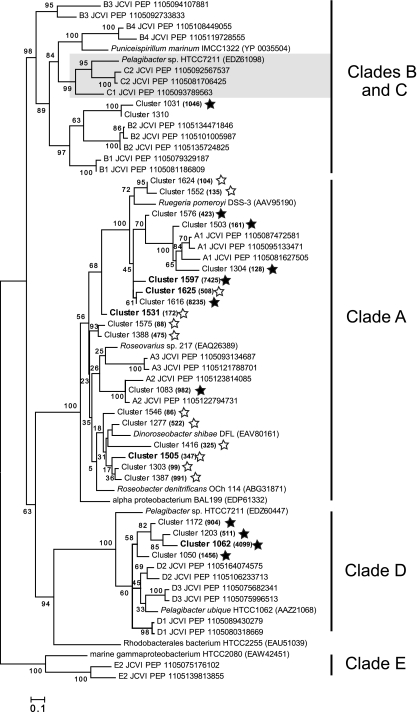
To track the temporal dynamics of the community dmdA pool at a finer scale, the 100 largest clusters (averaging 784 sequences per cluster and accounting for 70% of all dmdA sequences) were analyzed in more detail. This analysis assumes that PCR biases, to whatever extent they occurred, were consistent across samples. A multidimensional scaling (MDS) plot showed that cluster compositions on days 4 to 6 in the experimental microcosms were more similar to each other than to the control samples or the other experimental samples (Fig. 2B). Clusters contributing most to this pattern were from clades A, B, and D (24, 4, and 4 clusters, respectively) (Fig. 4). A group of 6 clusters related to Dinoroseobacter shibae dmdA (clade A) contained only sequences from the experimental microcosms, while 4 clusters related to SAR11 dmdA sequences (clade D) contained only sequences from the control microcosms (Fig. 4).
FIG. 4.
Maximum-likelihood tree of the translated dmdA clusters most responsible for the dissimilarity between control and experimental microcosms on days 4 to 6, as determined by SIMPER analysis (sequences named “Cluster xxxx”), along with representative dmdA sequences from the GOS metagenome (sequences named “xx JCVI PEP”) and cultured bacteria. Cluster sequences in bold are the five creating the greatest dissimilarity (9.5% of the total). Filled and open stars indicate clusters with more sequences in control or experimental microcosms, respectively, and numbers in parentheses indicate the number of sequences in that cluster. Gray shading indicates clade C, which is internal to clade B in this tree. The tree was created using RAxML 7.0, and values at the nodes show the number of times that the node appeared in 100 resamplings.
At higher-order groupings, the percentage of clade A sequences in the amplicon pools increased in experimental microcosms from 47% to 70% of the total dmdA sequences between days 3 and 4, while clade E sequences increased from 9 to 20% over the same time period. Sequences from clades C and D decreased in relative abundance (from 18 to 4% and from 26 to 6% of dmdA sequences, respectively; data not shown). There was evidence for shifts in relative abundance of sequences in the control microcosms over this same time interval, but to a smaller extent (clade A sequences decreased from 68 to 65% of total dmdA sequences; clades C, D, and E sequences increased from 4 to 6%, 10 to 20%, and 15 to 18% of total dmdA sequences, respectively). A BIO-ENV/BVSTEP analysis indicated that the observed changes in relative abundance of the 100 largest DmdA clusters were best correlated in both control and bloom microcosms with the change in Chl a concentrations (R = 0.813).
Bacterial and archaeal communities.
The 180,801 bacterial 16S rRNA sequences obtained from control and experimental microcosms formed 9,588 taxonomic clusters at 99% nucleotide sequence identity (all time points considered together). Given the ∼0.5% error rate typical for 454 sequencing (21), some clusters may be influenced by sequencing artifacts. However, the 1% sequence divergence allowance means that amplicons with a single miscalled base will be placed in the same cluster as otherwise identical amplicons. The major bacterial groups represented by these clusters were Roseobacter, SAR11, and Flavobacteria (Fig. 5 A). The relative abundance of Flavobacteria amplicons increased over time in the experimental microcosms (from 12% to 34% of total sequences), while SAR11 abundance decreased (from 31% to 12% [Fig. 5A]). These changes were larger than those in the control microcosms, suggesting that they were linked to conditions associated with the development and decline of the phytoplankton bloom. Roseobacters accounted for a significant percentage of the 16S rRNA amplicons from the microcosm bacterial communities (17% ± 7% [Fig. 5A]) but showed no dynamics related to time point or treatment.
FIG. 5.
Relative abundances of bacterial (A and B) and archaeal (C and D) 16S rRNA gene sequences over time in control and experimental microcosms.
The distribution of 16S rRNA sequences among bacterial clusters diverged over days 4 to 6 in the experimental microcosms compared to the control microcosms and the initial sample (Fig. 2C). A SIMPER analysis indicated that the increases in Flavobacteria amplicons were most responsible for this difference. The environmental variable best correlated with the observed changes in bacterial community in control microcosms was DMS concentration (R = 0.796), while a combination of environmental variables were correlated in the experimental microcosms (DMSPp-to-Chl a ratio, DMSPp, and DMS; R = 0.886).
Archaeal 16S rRNA genes could be amplified from microcosm DNA only for days 0 to 3 for both control and experimental treatments, suggesting that Archaea abundance declined significantly after 3 days of incubation. The 6,578 archaeal 16S rRNA sequences formed 519 clusters at the 99% sequence identity level. The majority of archaeal 16S rRNA amplicons (∼83%) could not be classified to the phylum level since they were <75% identical to any type species (likely reflecting an underrepresentation of mesophilic Archaea among characterized isolates). Of the sequences that could be categorized to the phylum level, the majority were Euryarchaeota (averaging 16.6% of all archaeal sequences) regardless of treatment or time point (Fig. 5B), and a small number were Crenarchaeota (averaging 0.2% of archaeal sequences). Analysis of archaeal clusters, including those composed of unclassified sequences, showed a change in composition over the limited time course in which their 16S rRNA genes could be amplified (Fig. 2D). These observed changes in the archaeal community are best correlated in experimental microcosms with Chl a and DMSPd concentrations (R = 0.829). The control microcosms had too few sample points for statistical analysis.
DISCUSSION
In the context of an experimentally induced phytoplankton bloom, we asked whether shifting ecological conditions, including DMSP supply, influenced the relative composition of the community dmdA sequence pool. At least five clades and 14 subclades of DmdA are found in marine bacterioplankton communities (10, 34), yet little information is currently available on whether or not these groups represent proteins with differing kinetic properties or ecological roles. Because some of the sequence groups are associated with specific taxa, changes in relative DmdA abundance might alternatively be linked to changes in taxonomic composition.
Overall, 578 dmdA clusters were identified at a conservative definition of 90% nucleotide sequence similarity (34). Clusters unique to either experimental or control microcosms (29% of the total) averaged only 1.5 sequences each, indicating that a significant amount of rare diversity that would not have been captured in a shallower sequencing effort was present. Nonetheless, we still likely underestimated richness, based both on evidence from rarefaction analysis (Fig. 3) and on recognition that some dmdA sequences likely have mismatches to the universal primers (34).
Several dmdA subclades shifted in relative abundance in response to the phytoplankton bloom, with two groups in clade A (Roseobacter-like) and one in clade D (SAR11-like) showing the most significant changes (Fig. 4). All members of one of the Roseobacter-like groups showed a positive response to bloom conditions (Fig. 4), suggesting a “bloom” subclade. Conversely, the SAR11-like group members all showed a negative response to bloom conditions, suggesting a “nonbloom” subclade.
The DmdA composition at the clade level had smaller overall changes. Reisch et al. (24) found that purified proteins representing clade A (from Roseobacter isolate Ruegeria pomeroyi DSS-3) and clade D (from SAR11 isolate Pelagibacter ubique HTCC1062) had similar catalytic efficiencies, pH optima, and Km values. Some bacterioplankton may accumulate DMSP to high intracellular concentrations (e.g., 70 mM in R. pomeroyi [24]), which is typical for compounds used as an osmoprotectant and in agreement with field studies of natural bacterioplankton communities (30, 37). Accumulation of DMSP to high intracellular concentrations would reduce selective pressures for enzymes optimized to the low external DMSP concentrations (low nM), an idea consistent with the relatively invariant clade representation found previously across the GOS metagenome sites (10). While PCR amplifications are not quantitative in the same way as metagenomic data sets, the current study supports the idea that dmdA relative abundance is largely consistent at the clade level but dynamic at the subclade and cluster level (Fig. 2).
Biogeochemical data collected during the microcosm experiment provided a framework for interpreting the observed cluster-level shifts in relative abundance of dmdA. Measurements of DMSP and DMS concentrations and flux confirmed that the induced phytoplankton bloom resulted in higher concentrations of dissolved DMSP and DMS compared to the controls (Fig. 1D and F). However, the ratio of DMSPp to Chl a indicated that the phytoplankton responsible for the bloom were poor producers of DMSP on a per-cell basis (Fig. 1B). Microscopic analysis and removal of dissolved H3SiO4 (data not shown) indicated that diatoms, which are not strong DMSP producers, dominated the bloom. Thus, although higher in an absolute sense, DMSP was relatively less important as a bacterial substrate in the experimental microcosms compared to the controls. This more minor role for DMSP as a source of reduced carbon and sulfur is consistent with the increase in relative abundance of Flavobacteriaceae over the course of the bloom (Fig. 5A), a group which may not metabolize DMSP based on the absence of demethylation and cleavage genes in the existing genome sequences (10; however, see reference 36). Flavobacteriaceae are known to make use of plankton-derived extracellular polymers (polysaccharides and other high-molecular-weight compounds excreted from cells [16]), which may be more abundant toward the end of the bloom. Thus, the data are suggestive of a reduced bacterioplankton dependence on DMSP within the bloom community compared to the control.
In an ecosystem context, it is interesting that despite substantial differences between bloom and nonbloom conditions in the concentration and consumption of dissolved DMSP (Fig. 1B and D), there was little effect on the ratio of products from the two competing pathways for DMSP degradation (DMS production averaged 20% of DMSP assimilation in the experimental microcosms and 16% in the controls). Future experimental studies can address the important interplay between functional gene sequence heterogeneity and the pathways and rates of microbially mediated sulfur transformations in the ocean.
The dynamics of five clades and multiple subclades of dmdA would not have been detected without the deep coverage libraries made possible by high-throughput amplicon sequencing. Such dynamics were likely driven by a collection of factors that caused differential growth rates among bacterioplankton taxa (Fig. 1C) and shifts in taxonomic composition (Fig. 5). Factors driving these changes may have been directly related to DMSP dynamics: for example, if an ecological advantage was conferred by certain variants in translated demethylase sequences as environmental conditions changed. Alternatively, they may have been driven by factors not explicitly related to DMSP supply, such as bloom-related changes in organic carbon or nutrient availability or food web interactions. Among the suite of environmental factors measured in this study, the best correlate with shifts of the dominant dmdA clades was Chl a concentration, not concentration or flux of DMSP or its metabolites (Fig. 2). The dmdA population changes occurring in this case, therefore, may be an outcome of phytoplankton-driven taxonomic shifts rather than specifically linked to DMSP cycling. There have been few exhaustive analyses of the diversity of functional genes (34, 38). Even in this study, we are still scratching the surface of characterizing the diversity and understanding the ecological relevance of functional gene sequence variation.
Acknowledgments
We thank B. Tolar, E. Fichot, M. Coll-Lladó, A. Rellinger, C. Smith, S. Gifford, X. Mou, and A. Spaulding for assistance in sample collecting and processing; H. Luo and V. Varaljay for expertise in phylogenetic tree construction; and the captain of the R/V E. O. Wilson (Dauphin Island, AL) for help in sample collection.
This research was supported by NSF grant OCE-0724017 (to M.A.M. and R.P.K.) and by a grant from the Gordon and Betty Moore Foundation (to M.A.M.).
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Andreae, M. 1990. Ocean-atmosphere interactions in the global biogeochemical sulfur cycle. Mar. Chem. 30:1-29. [Google Scholar]
- 2.Andreae, M. O., and H. Raemdonck. 1983. Dimethyl sulfide in the surface ocean and the marine atmosphere: a global view. Science 221:744-747. [DOI] [PubMed] [Google Scholar]
- 3.Biers, E. J., S. Sun, and E. C. Howard. 2009. Prokaryotic genomes and diversity in the surface ocean: interrogating the Global Ocean Sampling metagenome. Appl. Environ. Microbiol. 75:2221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlson, R. J., J. E. Lovelock, M. O. Andreae, and S. G. Warren. 1987. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate. Nature 326:655-661. [Google Scholar]
- 5.Cole, J. R., et al. 2007. The Ribosomal Database Project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curran, M. A. J., G. B. Jones, and H. Burton. 1998. Spatial distribution of dimethylsulfide and dimethylsulfoniopropiate in the Australasian sector of the Southern Ocean. J. Geophys. Res. 103:16667-16689. [Google Scholar]
- 7.Curson, A. R. J., R. Rogers, J. D. Todd, C. A. Brearley, and A. W. B. Johnston. 2008. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine alpha-proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10:757-767. [DOI] [PubMed] [Google Scholar]
- 8.González, J. M., et al. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard, E. C., et al. 2006. Bacterial taxa that limit sulfur flux from the ocean. Science 314:649-652. [DOI] [PubMed] [Google Scholar]
- 10.Howard, E. C., S. Sun, E. J. Biers, and M. A. Moran. 2008. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 10:2397-2410. [DOI] [PubMed] [Google Scholar]
- 11.Huber, J. A., et al. 2007. Microbial population structure in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 12.Kiene, R. P. 1996. Production of methanethiol from dimethylsulfoniopropionate in marine surface waters. Mar. Chem. 54:69-83. [Google Scholar]
- 13.Kiene, R. P., L. P. Hoffman Williams, and J. E. Walker. 1998. Seawater microorganisms have a high affinity glycine betaine uptake system which also recognizes dimethylsulfoniopropionate. Aquat. Microb. Ecol. 15:39-51. [Google Scholar]
- 14.Kiene, R. P., L. J. Linn, J. M. Gonzalez, M. A. Moran, and J. A. Bruton. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65:4549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiene, R. P., and D. Slezak. 2006. Low dissolved DMSP concentrations in seawater revealed by small volume gravity filtration and dialysis sampling. Limnol. Oceanogr. 4:80-95. [Google Scholar]
- 16.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic systems. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 17.Kirchman, D. L., E. K'nees, and R. E. Hodson. 1985. Leucine incorporation and its potential as a measure of protein synthesis by baceria in natural aquatic systems. Appl. Environ. Microbiol. 49:599-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, W., and A. Godzik. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658-1659. [DOI] [PubMed] [Google Scholar]
- 19.Lovelock, J. E., R. J. Maggs, and R. A. Rasmussen. 1972. Atmospheric dimethyl sulphide and that natural sulphur cycle. Nature 237:452-453. [Google Scholar]
- 20.Malin, G., and G. O. Kirst. 1997. Algal production of dimethyl sulfide and its atmospheric role. J. Phycol. 33:889-896. [Google Scholar]
- 21.Margulies, M., et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh, H.-M., et al. 2010. Complete genome sequence of “Candidatus Puniceispirillum marinum” IMCC1322, a representative of the SAR116 clade in the Alphaproteobacteria. J. Bacteriol. 192:3240-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons, T. R., Y. Maita, and C. M. Lalli. 1984. A manual for chemical and biological methods for seawater analysis. Pergamon, New York, NY.
- 24.Reisch, C. R., M. A. Moran, and W. B. Whitman. 2008. Dimethylsulfoniopropionate-dependent demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 190:8018-8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rusch, D. B., et al. 2007. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:0398-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simó, R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological, evolutionary links. Trends Ecol. Evol. 16:287-294. [DOI] [PubMed] [Google Scholar]
- 27.Slezak, D., R. P. Kiene, D. A. Toole, R. Simó, and D. J. Kieber. 2007. Effects of solar radiation on the fate of dissolved DMSP and conversion to DMS in seawater. Aquat. Sci. 69:377-393. [Google Scholar]
- 28.Smith, D. C., and F. Azam. 1992. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107-114. [Google Scholar]
- 29.Sogin, M. L., et al. 2006. Microbial diversity in the deep sea and the unexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sunda, W., D. J. Kieber, R. P. Kiene, and S. Huntsman. 2002. An antioxidant function for DMSP and DMS in marine alga. Nature 418:317-320. [DOI] [PubMed] [Google Scholar]
- 31.Todd, J. D., A. R. J. Curson, C. L. Dupont, P. Nicholson, and A. W. B. Johnston. 2009. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some ascomycete fungi. Environ. Microbiol. 11:1376-1385. [DOI] [PubMed] [Google Scholar]
- 32.Todd, J. D., et al. 2007. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315:666-669. [DOI] [PubMed] [Google Scholar]
- 33.Turner, S. M., G. Malin, P. S. Liss, D. S. Harbour, and P. M. Holligan. 1988. The seasonal variation of dimethyl sulfide and dimethylsulfoniopropionate concentrations in nearshore waters. Limnol. Oceanogr. 33:364-375. [Google Scholar]
- 34.Varaljay, V. A., E. C. Howard, S. Sun, and M. A. Moran. 2010. Diversity of dimethylsulfoniopropionate demethylase genes in a marine ecosystem assessed by deep amplicon sequencing of dmdA primer sets. Appl. Environ. Microbiol. 76:609-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vila, M., et al. 2004. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl. Environ. Microbiol. 70:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vila-Costa, M., et al. 2010. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. doi: 10.1038/ismej.2010.62. [DOI] [PubMed]
- 37.Wolfe, G. V. 1996. Accumulation of dissolved DMSP by marine bacteria and its degradation via bactivory, p. 277-291. In R. P. Kiene, P. T. Visscher, M. D. Keller, and G. O. Kirst (ed.), Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, NY.
- 38.Yutin, N., M. T. Suzuki, and O. Beja. 2005. Novel primers reveal wider diversity among marine aerobic anoxygenic phototrophs. Appl. Environ. Microbiol. 71:8958-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zubkov, M. V., et al. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulfoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]