Abstract
In bacteriophage (phage) therapy against Gram-positive bacteria, such as Staphylococcus aureus, Listeria monocytogenes, and Enterococcus faecalis, members of a genus of SPO1-like viruses are typically employed because of their extreme virulence and broad host spectrum. Phage φEF24C, which is a SPO1-like virus infecting E. faecalis, has previously been characterized as a therapeutic phage candidate. In addition to the phage itself, phage endolysin is also recognized as an effective antimicrobial agent. In this study, a putative endolysin gene (orf9) of E. faecalis phage φEF24C was analyzed in silico, and its activity was characterized using the recombinant form. First, bioinformatics analysis predicted that the open reading frame 9 (ORF9) protein is N-acetylmuramoyl-l-alanine amidase. Second, bacteriolytic and bactericidal activities of ORF9 against E. faecalis were confirmed by zymography, decrease of peptidoglycan turbidity, decrease of the viable count, and morphological analysis of ORF9-treated cells. Third, ORF9 did not appear to require Zn2+ ions for its activity, contrary to the bioinformatics prediction of a Zn2+ ion requirement. Fourth, the lytic spectrum was from 97.1% (34 out of 35 strains, including vancomycin-resistant strains) of E. faecalis strains to 60% (6 out of 10 strains) of Enterococcus faecium strains. Fifth, N-acetylmuramoyl-l-alanine amidase activity of ORF9 was confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and the subsequent MALDI-postsource decay (PSD) analyses. Finally, functional analysis using N- or C-terminally deleted ORF9 mutants suggested that a complete ORF9 molecule is essential for its activity. These results suggested that ORF9 is an endolysin of phage φEF24C and can be a therapeutic alternative to antibiotics.
Bacteriophage (phage) are an abundant and diverse life form, universally present in the environment. Tailed phage comprise 96% of isolated phage. Their life cycle involves adsorption, production of progeny phage, and the release of progeny phage by bacteriolysis. At the late infection phase, bacteriolysis is caused by two proteins called holin and endolysin that are encoded by phage genomes. It is thought that holins form pores to allow endolysin access to peptidoglycan and that endolysin degrades the peptidoglycan layer.
Along with the recent revival of phage therapy and the fast development of scientific research, in addition to phage itself, endolysin application has also been proven to be effective as an alternative antimicrobial agent against drug-resistant bacteria, and this is recognized as a part of phage therapy (4, 14). Endolysin is preferable in the treatment of intractable infections caused by drug-resistant Gram-positive bacteria, because it not only selectively destroys the target bacteria without disturbing normal microflora, but is also considered to have less chance of inducing resistant bacteria. One of the targeted bacteria in phage therapy using endolysin is Enterococcus faecalis. E. faecalis is recognized as an opportunistic pathogen and is the most commonly clinically isolated Enterococcus sp. In the worst case, it can develop drug resistance against vancomycin or linezolid, or both, and cause intractable infections (3).
Previously, we reported E. faecalis phage φEF24C, with high virulence and an extremely broad host spectrum, belonging to SPO1-like viruses, which are a group of virulent phage against Gram-positive bacteria (18, 19). It was characterized as a therapeutic phage candidate by in silico, in vitro, and in vivo analyses (19). Through a toxicogenomic study performed as part of an evaluation of therapeutic phage, the endolysin gene was hypothesized to be orf9 (19). In this study, endolysin ORF9 of phage φEF24C was characterized, using a recombinant protein produced by Escherichia coli.
MATERIALS AND METHODS
Bacterial strains, culture conditions, plasmids, and reagents.
All bacteria employed in this study are described in Table 1. Escherichia coli was propagated in Luria-Bertani medium. Staphylococcus aureus and E. faecalis strains were propagated in tryptic soy broth medium. All bacteria employed in this study were incubated in shaking culture at 37°C unless otherwise stated. The protein expression plasmid pCold III was purchased from Takara Bio (Kyoto, Japan). The medium was supplemented with ampicillin at a final concentration of 100 μg/ml for cloning of the gene and overexpression of the protein in E. coli. Culture media and chemicals were purchased from Becton Dickinson and Company (Sparks, MD) and Nacalai Tesque (Kyoto, Japan). Phosphate-buffered saline (PBS) at pH 7.2 was used in the study.
TABLE 1.
Bacterial strains employed in this study
| Bacterium | Strain | No. of strains | Description | Reference |
|---|---|---|---|---|
| E. coli | BL21 | 1 | Employed for protein expression | |
| DH5α | 1 | Employed for cloning/lytic-spectrum test | ||
| E. faecalis | EF1-EF30 | 30 | Clinical isolates | 18 |
| VRE1-VRE4 | 4 | Clinical vancomysin-resistant isolates | 18 | |
| 19433-U | 1 | Reference strain | 18 | |
| E. faecium | EFum1-10 | 10 | Clinical isolates | 18 |
| S. aureus | SA37, -46 | 2 | Clinical isolates | 15 |
| MR1, -3, -4, -5, -6, -7, -8 | 7 | Clinically isolated methicillin resistant strains | 15 | |
| 209P | 1 | Reference strain | 15 |
Bioinformatics analysis.
The sequence of the putative endolysin gene (orf9) and that of its putative gene product (ORF9) were obtained from genomic sequence data (GenBank accession no. AP009390). ORF9 was bioinformatically analyzed using the protein-protein basic local alignment search tool (BLASTp) at the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Protein structure and function predictions were performed with I-TASSER (http://zhang.bioinformatics.ku.edu/I-TASSER/) (16).
Cloning and overexpression of the recombinant protein.
Phage φEF24C genomic DNA was obtained using a method described elsewhere (18, 19). All of the primers used in this study were designed based on the genome sequence (see Table S1 in the supplemental material). The coding sequence of orf9 was amplified by PCR (LaboPass SP-Taq kit; Hokkaido System Science, Hokkaido, Japan) with the appropriate primer sets (see Table S2 in the supplemental material), using φEF24C genomic DNA as a template, following the manufacturer's protocol. Subsequently, the terminal ends of the PCR product were digested with the restriction enzymes EcoRI and BamHI (Takara Bio) and were cloned into pUC18. The accurately cloned fragment in pUC18 was then excised with EcoRI and XbaI and recloned into the expression vector pCold III. The plasmids were transformed into E. coli strains DH5α and BL21 for cloning and protein expression, respectively.
To overexpress the recombinant protein, E. coli BL21 containing the appropriate plasmid was exponentially grown to an optical density at 600 nm (OD600) of 0.6 to 0.8 and then allowed to stand for 30 min at 15°C. The growth medium was supplemented with isopropyl-β-d-thiogalactopyranoside (IPTG) at 1 mM, and the bacteria were cultured aerobically for 24 h at 15°C. After centrifugation (6,000 × g; 10 min; 4°C), the supernatant was decanted, and cell pellets were obtained and then stored at −80°C until they were used. To check overexpression of the recombinant protein in E. coli, the cell pellets were dissolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and then boiled for 5 min. After centrifugation (20,000 × g; 4°C; 5 min), the supernatants of the IPTG-induced and non-IPTG-induced samples were separately electrophoresed in a 12.5% SDS-PAGE gel and stained using Coomassie brilliant blue R250 to confirm overexpression of the protein.
Purification of the recombinant protein.
Bacterial cells prepared from 250 ml culture were resuspended in 20 ml lysis buffer (100 mM sodium phosphate, 300 mM NaCl, pH 7.8) and sonicated (36 five-second pulses at five-second intervals) on ice. After centrifugation (10,000 × g; 20 min; 4°C), the supernatant was added to 1 ml Talon Metal Affinity Resin (Clontech Laboratories, Mountain View, CA) and gently mixed (90 min; 4°C). The resin was transferred into a column and eluted with 50 mM sodium phosphate, 300 mM NaCl, pH 6.0, followed by the same buffer supplemented with 5 mM imidazole. When 150 mM imidazole-supplemented buffer was applied, the eluate was collected. The collected fraction was dialyzed against PBS. The protein solution was analyzed on 12.5% SDS-PAGE gels using Coomassie brilliant blue R250 staining to determine the degree of recombinant protein purification. Protein was quantified using the Bradford reagent (Sigma-Aldrich, St. Louis, MO).
Preparation of bacteria and E. faecalis peptidoglycan.
E. faecalis, Enterococcus faecium, E. coli, and S. aureus strains, which were grown to mid-log phase, were washed three times with PBS. The bacteria were suspended in PBS and were used for a lysis assay and a host spectrum test.
SDS-treated E. faecalis was prepared for zymography. An exponentially growing culture (300 ml at an OD600 of 0.6) of E. faecalis strain EF24 was washed with PBS, and the cells were boiled in 4% SDS for 30 min. The cells were washed six times with deionized water and then freeze-dried. The bacterial powder was used for a zymographic analysis.
Turbidity assays and matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis used lyophilized E. faecalis STF-3 (ATCC 12984) cell walls (M3440; Sigma-Aldrich).
Zymography.
IPTG-induced and non-IPTG-induced E. coli BL21(pColdIII ORF9-His) (1.0 ml) was pelleted by centrifugation, and then the bacterial pellets were suspended in 100 μl SDS-PAGE sample buffer. After a 5-min boil, the samples were electrophoresed in a 12.5% SDS-PAGE gel containing 0.3 to 0.4 mg/ml of SDS-treated E. faecalis strain EF24. After the electrophoresis, the gel was washed three times in deionized water for 10 min each time. The gel was incubated in renaturation buffer (25 mM Tris-HCl, pH 7.8) overnight. To obtain better resolution of the clear band, the gel was stained using Coomassie brilliant blue R-250 and destained overnight with 7.5% acetic acid containing 10% methanol at room temperature. The gel was visualized using a GT-X800 scanner (Seiko Epson, Nagano, Japan).
Measurements of ORF9 peptidoglycan-degrading and bacteriolytic activities.
The turbidity of peptidoglycan or bacterial cells suspended in PBS was measured at 595 nm, using a Multiskan JX spectrophotometer (Thermo Labsystems, Stockholm, Sweden). The suspension (100 μl) was loaded in wells of a sterile, uncoated polystyrene 96-well plate. All plates were incubated with shaking at 37°C. Six replicates were prepared for each treatment group. In all of the lytic activity assays, PBS that did not contain the purified protein was also tested as a negative control. The dose-response and time-dependent activities were analyzed using GraphPad Prism 4 software (GraphPad Software, La Jolla, CA), and statistical analysis of the data was conducted with the GraphPad InStat 3 software (GraphPad Software). Lytic activity was considered to be present when the P value was <0.05. The initial turbidity of suspensions was set from about 0.2 to 0.3, except for E. faecalis STF-3 peptidoglycan, which was set as 0.1.
First, serial dilutions of the purified ORF9-His were added to E. faecalis STF-3 peptidoglycan in PBS. Turbidity measurements were conducted at 5, 15, 30, and 60 min. Next, serial dilutions of the purified ORF9-His were added to PBS-washed E. faecalis strain EF24 in PBS. Turbidity measurements were conducted at 15-min intervals up to 3 h. At the same time, bacterial suspensions incubated with various concentrations of ORF9-His for 3 h were plated on tryptic soy broth and then incubated at 37°C overnight. The colonies were counted. Along with the two experiments mentioned above, negative-control experiments were conducted to check the influence of protein on peptidoglycan and bacterial viability. Peptidoglycan and bacteria were treated with bovine serum albumin (BSA) at 0, 10−3, 10−2, 10−1, 1, and 10 mg/ml. Third, E. faecalis strain EF24 suspended in PBS with or without 50 mM EDTA was treated with 0.1 mg/ml of ORF9-His. The turbidity was monitored at 30-min intervals up to 90 min. Fourth, E. faecalis, E. faecium, S. aureus, or E. coli strains suspended in PBS were treated with 0.1 mg/ml of ORF9-His. The turbidity of the suspensions was monitored at 60-min intervals up to 180 min.
Finally, to test the lytic activity of one-side-deleted ORF9 mutant proteins, the deleted ORF9 mutant proteins in a soluble form (0.1 mg/ml) were added to E. faecalis strain EF24 suspended in PBS. ORF9-His was used as a positive control. The turbidity of the suspensions was monitored at 15-min intervals up to 60 min.
Microscopic observation.
PBS-washed E. faecalis strain EF24 was treated with ORF9-His (0.1 mg/ml) or PBS (total volume of 100 μl) and shaken at 37°C for 1 h. After incubation, 2% glutaraldehyde (TAAB Laboratories Equipment) was added to stop the reaction. The cell pellet was coated with 1% agarose, rinsed with PBS that contained 5% sucrose, and postfixed with 1.5% osmium tetroxide in 0.1 mol/liter phosphate buffer containing 5% sucrose at 4°C for 1 h. After dehydration through a graded series of ethanol concentrations, the fixative was replaced with propylene oxide, and the samples were embedded in epoxy resin (TAAB 812). Ultrathin sections were prepared, stained with uranyl acetate and lead citrate, and observed using an H7100 transmission electron microscope (Hitachi, Tokyo, Japan).
Analysis of the ORF9 peptidoglycan cleavage site.
The preparation of high-performance liquid chromatography (HPLC) samples generally followed the procedure of Yoshimura et al. (24). Briefly, E. faecalis STF-3 peptidoglycan (0.8 mg) was treated with either ORF9-His (0.2 mg/ml) or mutanolysin (0.14 mg/ml; Sigma-Aldrich) overnight at 37°C. The digested peptidoglycan was adjusted to pH 4.0 by adding 20% H3PO4 and then boiled for 5 min to stop the reaction. Insoluble material was removed by centrifugation (14,000 × g; 15 min; room temperature). The pH was adjusted using 1.5 M sodium borate buffer (pH 9.0), sodium borohydride was added, and the mixture was incubated for 15 min at room temperature before 20% H3PO4 was added. For deacetylation of muropeptides, the pH of the solution was brought to 12.0 with 4 N NaOH, and the reaction mixture was incubated for 90 min at 37°C. Subsequently, 4 N HCl was added to adjust the pH to 2.5. After centrifugation (14,000 × g; 15 min; room temperature), the supernatant was filtered to 0.22 μm. The muropeptides were separated on a Hypersil ODS (5-μm; 250- by 4.6-mm) column (Supelco, Bellefonte, PA) by HPLC (class LC-10 system; Shimadzu, Japan) using a 0.5-ml/min flow rate at 45°C. A linear gradient of 5% methanol in 50 mM NaH2PO4 (pH 2.5) to 30% methanol in 50 mM NaH2PO4 (pH 2.8) was applied over 180 min. The eluted compounds were detected by monitoring absorbance at 206 nm, and the eluates at peak were collected.
The peak fractions were analyzed by MALDI-TOF MS and MALDI-postsource decay (PSD). The individual peaks were first concentrated in a centrifugal vacuum evaporator and then desalted using ZipTipC18 pipette tips (Millipore, Billerica, MA). The peaks dissolved in 50% acetonitrile-0.1% trifluoroacetic acid were cospotted with an equal volume of the matrix (α-cyano-4-hydroxycinnamic acid dissolved in a mixture of 50% acetonitrile-0.1% trifluoroacetic acid in a saturated solution) onto a sample plate. The peaks were analyzed by MALDI-TOF or MALDI-PSD mass spectrometry, or both, using a Biflex IV mass spectrometer (Bruker Daltonik, Germany).
RESULTS
In silico analysis of ORF9.
According to the NCBI BLASTp analysis, the conserved protein domain, peptidoglycan recognition protein (PGRP) (amino acids 27 to 157, with an E value of 3e−15) or N-acetylmuramoyl-l-alanine amidase (amino acids 17 to 157, with an E value of 5e−12), and amino acid residues for substrate binding, Zn2+ binding, and amidase catalysis were predicted for ORF9 (Fig. 1). In contrast, no functionally important protein domains were detected in the C-terminal region (from amino acid 200 to 289), although this region is moderately homologous to those of prophage lysins of E. faecalis and Lactococcus phage asccφ28 lysin.
FIG. 1.
Bioinformatics analysis of the ORF9 amino acid sequence. The N-acetylmuramoyl-l-alanine amidase domain predicted by BLASTp is surrounded by a gray line. The amino acid residues for substrate binding (His-35, Asn-36, Ala-54, Ala-58, Val-71, Phe-78, His-79, Asn-95, His-145, Thr-149, Thr-151, Ala-152, and Cys-153), Zn2+ binding (His-34, His-145, and Cys-153), and amidase catalysis (His-34, Ala-58, His-145, Thr-151, and Cys-153) were also predicted in the N-acetylmuramoyl-l-alanine amidase domain predicted by BLASTp. According to the structure-based bioinformatics analysis by I-TASSER, ligand binding amino acid residues were predicted (shown in boldface and underlined).
To revalidate the bioinformatics prediction indicated above, a structure-based bioinformatics analysis was performed using I-TASSER. The N-acetylmuramoyl-l-alanine amidase functions were likewise structurally predicted. However, substrate binding amino acid residues scattered from the N- to the C-terminal region of the ORF9 sequence were estimated (Fig. 1). Thus, from this prediction, the entire ORF9 sequence seemed to be essential.
Peptidoglycan degradation and cell lysis activities of ORF9.
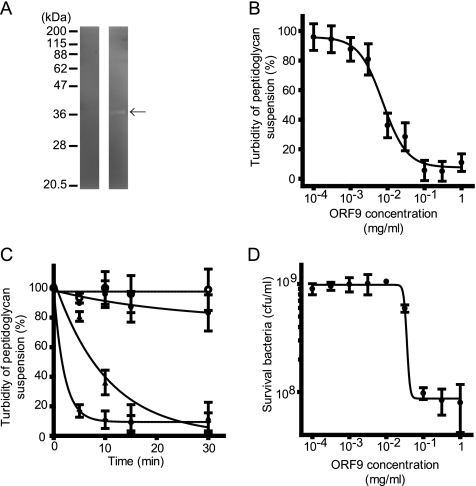
To confirm lytic activity, cell extracts of E. coli BL21 overexpressing the recombinant ORF9, ORF9-His, were analyzed by zymography (Fig. 2 A). A clear band of recombinant ORF9 expressed in E. coli was detected on the zymogram. Moreover, an E. faecalis (STF-3) peptidoglycan was incubated with various concentrations of the purified ORF9-His (0, 0.0001, 0.0003, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, and 1 mg/ml). After 10 min, the E. faecalis peptidoglycan treated with various concentrations of ORF9-His showed dose-dependent turbidity change (Fig. 2B), and a concentration of more than 0.1 mg/ml of ORF9-His showed the strongest lytic activity. In the presence of ORF9-His, the turbidity of the peptidoglycan suspension decreased with time elapsed (Fig. 2C). In the negative-control experiment, peptidoglycan turbidity was not different in the presence of BSA at any time point. Thus, the enzyme was considered to be able to degrade E. faecalis peptidoglycan.
FIG. 2.
Lytic activity of ORF9 against E. faecalis peptidoglycan and cells. (A) Zymography of ORF9-His-overexpressing E. coli. In the left and right lanes, E. coli BL21(pColdIII ORF9-His) without and with IPTG induction, respectively, is shown. Only E. coli BL21(pColdIII ORF9-His) with IPTG induction (right lane) showed a clear band, which was detected at the expected molecular size (arrow). (B) Dose-response curve of ORF9-His against E. faecalis peptidoglycan, summarizing the data at 10 min after the turbidity assay. Concentration-dependent ORF9 effects were observed. (C) Time course assay of ORF9-His treatment against E. faecalis peptidoglycan. Various concentrations of ORF9-His were treated with E. faecalis peptidoglycan; limited concentrations of ORF9-His treatment are presented. The ORF9-His concentrations at 0 (○), 10−4 (•), 10−2 (▴), and 1 (⧫) mg/ml are shown. The turbidity decreased time dependently. (D) Dose-response bactericidal effect of ORF9 against E. faecalis cells. Three hours after ORF9 treatment of E. faecalis cells, the viable count was measured. The error bars indicate the SD.
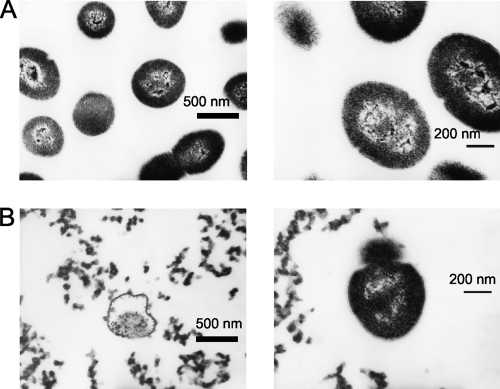
To prove bactericidal and bacteriolytic activity of ORF9 by peptidoglycan degradation, E. faecalis strain EF24 was treated with various concentrations of ORF9-His, and the bacterial viable count was measured at 3 h (Fig. 2D). In the negative-control experiment, the viable bacterial count was not different in the presence of BSA. The viable count also showed a dose-dependent bactericidal effect. The viability could be reduced to 9.6% ± 2.7% (mean ± standard deviation [SD]) after ORF9-His treatment at more than 0.1 mg/ml compared with the untreated control group. In addition, the morphology of the E. faecalis cells under either ORF9-His or PBS treatment was examined by electron microscopy (Fig. 3), which showed bacteriolysis and eruption of the cell contents. Therefore, ORF9 was considered to degrade the bacterial cell wall and kill the bacteria.
FIG. 3.
Electron micrograph of PBS-treated (A) and ORF9-His-treated (B) E. faecalis. In both panels, the right and left images show ×13,000 and ×22,000 magnification, respectively. Scale bars are also shown.
To examine the requirement for Zn2+ for ORF9-His activity, E. faecalis EF24 cells were treated with ORF9-His in the presence or absence of 50 mM EDTA in PBS. ORF9 was bioinformatically predicted to have N-acetylmuramoyl-l-alanine amidase activity, which requires a Zn2+ ion. However, the activity of ORF9-His was not statistically different in the presence or absence of 50 mM EDTA. Moreover, zymographic analysis did not indicate any cationic ion requirement for its lytic activity, because no cationic ions were included in the renaturation buffer. Thus, Zn2+ ions did not seem to be essential for ORF9 activity.
Bacteriolytic spectrum of ORF9.
To screen ORF9 bacteriolytic specificity against different bacterial species and strains, the turbidity changes of 35 E. faecalis strains (30 clinical isolates, 4 clinical vancomycin-resistant isolates, and 1 ATCC strain), 10 clinical E. faecium strains, 9 clinical (7 methicillin-resistant isolates and 2 methicillin-sensitive isolates) and 1 laboratory S. aureus strains, and E. coli strain DH5α between ORF9-His (0.1 mg/ml) and PBS treatments were assessed (Table 2). The lytic spectrum was from 97.1% (34/35) against E. faecalis to 60% (6/10) against E. faecium. On the other hand, ORF9-His did not show lytic activity against any S. aureus and E. coli strains.
TABLE 2.
Lytic spectrum of ORF9
| Tested bacterium | Lytic spectrum [% (no. of lysed/tested strains)] |
|---|---|
| E. faecalis | 97.1 (34/35) |
| E. faecium | 60 (6/10) |
| S. aureus | 0 (0/10) |
| E. coli | 0 (0/1) |
Site of peptidoglycan cleavage by ORF9.
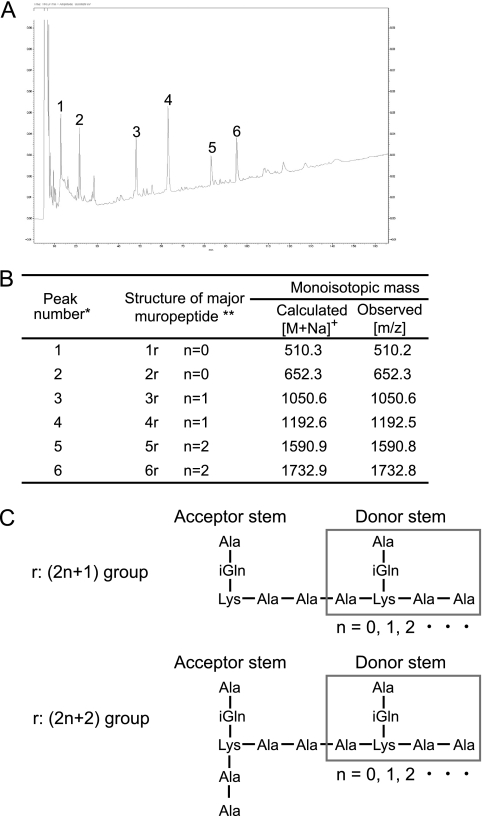
To verify the peptidoglycan cleavage site for ORF9 activity, a structural analysis of ORF9-His-digested E. faecalis peptidoglycan was performed. The HPLC chromatograms of ORF9-His- and mutanolysin-treated E. faecalis peptidoglycan showed different separation patterns, so ORF9 was considered to have enzymatic activity different from that of mutanolysin. Six major peaks of ORF9-His-treated peptidoglycan were collected and then analyzed by MALDI-TOF MS for molecular mass elucidation (Fig. 4 A and B). As a result, a peptide that was detached from peptidoglycan was considered to be a major component. Subsequently, these HPLC fractions were further analyzed by MALDI-PSD for structural elucidation (data not shown), and the structure of the ORF9-His-treated peptidoglycan was confirmed (Fig. 4C). Thus, ORF9 was confirmed to have N-acetylmuramoyl-l-alanine amidase activity, consistent with the prediction from bioinformatics analysis.
FIG. 4.
Structural analysis of ORF9-His-treated E. faecalis peptidoglycan. (A) Reversed-phase HPLC separation of ORF9-His-treated E. faecalis peptidoglycan. The numbers from 1 to 6 above the peaks indicate the isolated fractions. (B) Structures and molecular masses of the isolated fractions. *, peak numbers refer to the HPLC chromatograms; **, 1r, Ala-iGln-Lys(ɛ)-Ala-Ala; 2r, Ala-iGln-Lys[(ɛ)-Ala-Ala]-Ala-Ala; iGln, isoglutamine. (C) Determined structure of the ORF9-His-treated E. faecalis peptidoglycan.
Construction of ORF9 deletion mutants and their lytic activities.
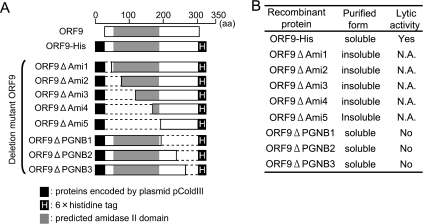
ORF9 function was identified as N-acetylmuramoyl-l-alanine amidase, as described above. We investigated the functionally important region of ORF9 using N- and C-terminally deleted ORF9 mutants. Eight defective mutant ORF9 proteins that had deletions in either their N-terminal or C-terminal ends were constructed (Fig. 5 A), and their lytic activities against E. faecalis cells were measured in a turbidity assay. Unfortunately, only three C-terminally deleted ORF9 mutants, ORF9ΔPGNB1, ORF9ΔPGNB2, and ORF9ΔPGNB3, were able to be purified in a soluble form. Their lytic activities against E. faecalis strain EF24 were measured. The turbidity after treatment with all three C-terminally deleted ORF9 mutants was no different from that after treatment with PBS (i.e., the negative control), although the turbidity after ORF9-His treatment was significantly decreased. These results (Fig. 5B) suggest that the N- and C-terminal portions are significantly important for ORF9 solubilization and stability of the protein and essential for lytic activity, respectively. Therefore, we believe that a complete ORF9 protein is important for enzymatic stability and activity.
FIG. 5.
(A) Schematic overview of ORF9-His and eight deletion mutants of ORF9-His. ORF9ΔAmi1, ORF9ΔAmi2, ORF9ΔAmi3, ORF9ΔAmi4, and ORF9ΔAmi5 are N-terminal deletions of ORF9-His. ORF9ΔPGNB1, ORF9ΔPGNB2, and ORF9ΔPGNB3 are C-terminal deletions of ORF9-His. (B) Purified forms and lytic activities of ORF9-His and the deletion mutants. Lytic activity was tested against E. faecalis strain EF24. Only the soluble deletion mutant ORF9s and ORF9-His were subjected to lysis assay. N.A., not applicable; yes, lysed; no, not lysed.
DISCUSSION
Some N-acetylmuramoyl-l-alanine amidases have been reported to have Zn2+-dependent peptidoglycan recognition domains (21, 24). ORF9 was also identified as the N-acetylmuramoyl-l-alanine amidase in this study. However, despite the bioinformatics prediction of Zn2+ binding sites in the N-terminal protein domain by BLASTp, Zn2+ did not seem to be essential in its activity, because ORF9 showed lytic activity in the turbidity assay in the presence of EDTA and in the zymographic resolution after renaturation without any ion supplementation.
Moreover, although the C-terminal region did not seem to be essential in the BLASTp analysis, the C-terminally deleted ORF9 mutant did not show any lytic activity. This observation was consistent with the structure-based bioinformatics analysis (Fig. 1). Even the longest ORF9 mutant with a C-terminal deletion, ORF9ΔPGNB3, which was deficient in the last three essential amino acid residues for ligand binding, Asp-257, Ile-258, and Pro-259, as predicted by structure-based bioinformatics analysis, did not show lytic activity. Thus, the C-terminal portion of ORF9 appears to be essential for N-acetylmuramoyl-l-alanine amidase activity.
A similar phenomenon, in which lytic activity of an endolysin was abolished by any deletion of the protein, was previously reported in the well-studied Listeria phage endolysins Ply118 and Ply500 (12). Both endolysins are similar in molecular size to ORF9. Interestingly, while the N-terminally deleted proteins (i.e., C-terminal regions) of both Ply118 and Ply500 showed cell wall binding activity without lytic activity, the C-terminally deleted protein (i.e., the N-terminal region) showed neither lytic activity nor binding activity. Based on this evidence, the C-terminal region of ORF9 may also have a binding function.
The level of the lytic activity of ORF9 against E. faecalis varied from strain to strain (data not shown). This strain-to-strain difference in lytic activity may be due to different levels of peptidoglycan modification. For example, O-acetylated E. faecalis peptidoglycan is known to be resistant to lysozyme (6). ORF9 also showed weak lytic activity against some E. faecium strains, suggesting that a difference in the peptidoglycan interpeptide bridge between E. faecalis and E. faecium (l-Ala-l-Ala and d-Asp, respectively) is not essential for the activity of ORF9 (20).
Phage endolysin generally has a broader host spectrum in bacterial species than its original phage, which is also consistent with phage φEF24C and its endolysin, ORF9. However, interestingly, ORF9 could not lyse the only E. faecalis strain, EF9, although phage φEF24C can multiply in the strain. This phenomenon is very unusual, because phage multiplication requires endolysin to release progeny phage at the late phase of infection. In phage φEF24C, more than half of the hypothetical genes have not been functionally predictable against the present database. Recently, different types of endolysin from the holin-endolysin system have been reported (8, 23). Therefore, φEF24C may possibly have another type of lytic mechanism for progeny phage release, in addition to the ORF9-associated holin-endolysin system.
In terms of the genomic location and protein domain structure, two types of endolysin seem to be present in SPO1-like viruses, including φEF24C. First, endolysins of Lactobacillus plantarum phage LP65 and S. aureus phage K, G1, Twort, and 812 are predicted to have more than two protein domains, and the genes are located at the ends of the tail protein genes, in the middle of DNA replication-associated genes, or in both locations (2, 7, 13). Some of the endolysin genes have introns. On the other hand, among phage φEF24C and Listeria monocytogenes phage A511 and P100, the endolysin gene, with only one N-acetylmuramoyl-l-alanine amidase domain, is unusually located in the head-neck protein gene region (1, 9). Fortunately, the L. monocytogenes phage A511 endolysin ply511 is well characterized as an endolysin (5, 10, 11, 17). Therefore, ORF9 is considered to be an endolysin, belonging to the same group as L. monocytogenes phage A511 in terms of the location of the gene in the genome and the primary protein structure of its gene product.
A recent study of human phage therapy against Pseudomonas chronic otitis showed that a reduction of bacteria to approximately 20% within 21 days results in successful treatment (22). Considering that approximately 90% of viable-count reduction by ORF9 treatment occurs within 3 h in vitro, ORF9 may be applicable to such intractable infections. However, despite good lytic activity of ORF9 against nongrowing E. faecalis cells, the conditions required to halt cell growth completely have not yet been found (data not shown). Therefore, screening of enhancing factors or optimal conditions for ORF9 lytic activity will be important for its practical use.
Supplementary Material
Acknowledgments
We thank Russ Chess-Williams, Bond University, Australia, for his help with statistical analysis.
This study was supported by the Kochi System Glycobiology Center and the Center of Biomembrane Functions Controlling Biological Systems, Kochi University, as part of a project.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Carlton, R. M., W. H. Noordman, B. Biswas, E. D. de Meester, and M. J. Loessner. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43:301-312. [DOI] [PubMed] [Google Scholar]
- 2.Chibani-Chennoufi, S., M. L. Dillmann, L. Marvin-Guy, S. Rami-Shojaei, and H. Brüssow. 2004. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 186:7069-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eliopoulos, G. M. 2009. Microbiology of drugs for treating multiply drug-resistant Gram-positive bacteria. J. Infect. 59(Suppl. 1):S17-S24. [DOI] [PubMed] [Google Scholar]
- 4.Fischetti, V. A. 2008. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11:393-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaeng, S., S. Scherer, H. Neve, and M. J. Loessner. 2000. Gene cloning and expression and secretion of Listeria monocytogenes bacteriophage-lytic enzymes in Lactococcus lactis. Appl. Environ. Microbiol. 66:2951-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hébert, L., et al. 2007. Enterococcus faecalis constitutes an unusual bacterial model in lysozyme resistance. Infect. Immun. 75:5390-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahánková, J., V. Ruzicková, and J. Doskar. 2007. Genome rearrangements in host-range mutants of the polyvalent staphylococcal bacteriophage 812. Folia Microbiol. 52:331-338. [DOI] [PubMed] [Google Scholar]
- 8.Kakikawa, M., et al. 2002. Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage φg1e. Gene 299:227-234. [DOI] [PubMed] [Google Scholar]
- 9.Klumpp, J., et al. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190:5753-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loessner, M. J., A. Schneider, and S. Scherer. 1996. Modified Listeria bacteriophage lysin genes (ply) allow efficient overexpression and one-step purification of biochemically active fusion proteins. Appl. Environ. Microbiol. 62:3057-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loessner, M. J., G. Wendlinger, and S. Scherer. 1995. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol. Microbiol. 16:1231-1241. [DOI] [PubMed] [Google Scholar]
- 12.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 13.O'Flaherty, S., et al. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186:2862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Flaherty, S., R. P. Ross, and A. Coffey. 2009. Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33:801-819. [DOI] [PubMed] [Google Scholar]
- 15.Rashel, M., et al. 2007. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage φMR11. J. Infect. Dis. 196:1237-1247. [DOI] [PubMed] [Google Scholar]
- 16.Roy, A., A. Kucukural, and Y. Zhang. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner, M. S., F. Waldherr, M. J. Loessner, and P. M. Giffard. 2007. Antimicrobial activity of lysostaphin and a Listeria monocytogenes bacteriophage endolysin produced and secreted by lactic acid bacteria. Syst. Appl. Microbiol. 30:58-67. [DOI] [PubMed] [Google Scholar]
- 18.Uchiyama, J., et al. 2008. Isolation and characterization of a novel Enterococcus faecalis bacteriophage φEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 278:200-206. [DOI] [PubMed] [Google Scholar]
- 19.Uchiyama, J., M. Rashel, I. Takemura, H. Wakiguchi, and S. Matsuzaki. 2008. In silico and in vivo evaluation of bacteriophage φEF24C, a candidate for treatment of Enterococcus faecalis infections. Appl. Environ. Microbiol. 74:4149-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vollmer, W., D. Blanot, and M. A. de Pedro. 2008. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32:149-167. [DOI] [PubMed] [Google Scholar]
- 21.Ward, J. B., C. A. Curtis, C. Taylor, and R. S. Buxton. 1982. Purification and characterization of two phage PBSX-induced lytic enzymes of Bacillus subtilis 168: an N-acetylmuramoyl-L-alanine amidase and an N-acetylmuramidase. J. Gen. Microbiol. 128:1171-1178. [DOI] [PubMed] [Google Scholar]
- 22.Wright, A., C. H. Hawkins, E. E. Anggård, and D. R. Harper. 2009. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 34:349-357. [DOI] [PubMed] [Google Scholar]
- 23.Xu, M., D. K. Struck, J. Deaton, I. N. Wang, and R. Young. 2004. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc. Natl. Acad. Sci. U. S. A. 101:6415-6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura, G., et al. 2006. Identification and molecular characterization of an N-acetylmuraminidase, Aml, involved in Streptococcus mutans cell separation. Microbiol. Immunol. 50:729-742. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.