Abstract
Vibrio cholerae consists of pathogenic strains that cause sporadic gastrointestinal illness or epidemic cholera disease and nonpathogenic strains that grow and persist in coastal aquatic ecosystems. Previous studies of disease-causing strains have shown V. cholerae to be a primarily clonal bacterial species, but isolates analyzed have been strongly biased toward pathogenic genotypes, while representing only a small sample of the vast diversity in environmental strains. In this study, we characterized homologous recombination and structure among 152 environmental V. cholerae isolates and 13 other putative Vibrio isolates from coastal waters and sediments in central California, as well as four clinical V. cholerae isolates, using multilocus sequence analysis of seven housekeeping genes. Recombinant regions were identified by at least three detection methods in 72% of our V. cholerae isolates. Despite frequent recombination, significant linkage disequilibrium was still detected among the V. cholerae sequence types. Incongruent but nonrandom associations were observed for maximum likelihood topologies from the individual loci. Overall, our estimated recombination rate in V. cholerae of 6.5 times the mutation rate is similar to those of other sexual bacteria and appears frequently enough to restrict selection from purging much of the neutral intraspecies diversity. These data suggest that frequent recombination among V. cholerae may hinder the identification of ecotypes in this bacterioplankton population.
Homologous recombination can be an extremely important force of evolution in some microorganisms and has been implicated in the evolution of virulent Vibrio cholerae serotype O139 (12, 39). Transfer of DNA between genetically distant individuals can diversify otherwise clonal populations, while recombination tends to homogenize sequence divergence within groups that undergo frequent intragroup recombination (20). The frequency of recombination events varies broadly among and between bacteria of different species or genera (56) and leads to vastly different population structures. For instance, rare recombination among Pseudomonas syringae isolates promotes a highly clonal population structure (47), while frequent recombination within the species Neisseria gonorrhoeae or Helicobacter pylori leads to effectively panmictic structures that obscure any evolutionary relationships between isolates (40, 53). However, the effect of sampling bias is evident in the epidemic population structure of Neisseria meningitidis, where inclusion of many identical isolates due to expansion of a successful clone suggests a clonal structure but removal of redundant genotypes reveals a panmictic structure for the species as a whole (24, 50).
Despite the lack of consensus on a unifying framework for describing speciation in all bacteria, a number of useful models have been proposed for explaining evolutionary relationships between genetically similar bacterial isolates. The stable ecotype model (7) assumes that recombination is rare, sequences evolve by the accumulation of mutations, and diversity in all genes is periodically purged by selective sweeps. This model thus predicts that alleles will have significant linkage disequilibrium (nonrandom distribution among isolates), gene trees will have congruent phylogenies for different genes, and sequences will form distinct, cohesive clusters that correspond to ecotypes (groups of strains adapted to the same ecological conditions). The stable ecotype model predicts a primarily clonal population structure and is consistent with genetic data observed for populations of Staphylococcus aureus (15), Bacillus spp. (30), and Synechococcus spp. (58). Alternative models used to describe evolutionary relationships in bacteria are analogous to Mayr's biological species concept for eukaryotes (36). In most of these models, homologous recombination between closely related bacteria blur taxonomic boundaries and entangle evolutionary relationships within subpopulations (21, 22). Cohesive groups of bacteria are then maintained through leaky genetic isolation facilitated by a log-linear decline in recombination frequency with genetic distance (57). Several reviews provide a more thorough discussion of these models (7, 8, 44).
Although Vibrio cholerae is primarily known as the etiological agent of Asiatic cholera, only the O1 and O139 serotypes have been implicated in epidemic disease (11). Strains from the remaining 200-plus serotypes are autochthonous members of coastal aquatic communities around the globe (34). Numerous studies of DNA sequences from single or multiple loci have demonstrated strong clonality of the epidemic O1 and O139 lineages (9, 33), while uncovering broader genetic diversity among non-O1/O139 isolates (27, 31, 39). Recent genome sequences from several disease-causing V. cholerae isolates revealed an identical genome “backbone” in pandemic strains, with most of the intergenomic diversity generated by illegitimate recombination events (6). The observed disparity between clinical and nonpathogenic environmental isolates could be due to either differences in population structure between lineages or sampling bias. More phylogenetic information from nontoxigenic environmental V. cholerae isolates is needed to characterize the population structure of this species.
The detection of recombinant regions within gene sequences is important, because their presence can have a deleterious effect on the ability to accurately construct evolutionary relationships between bacterial genomes (48). Recombination can be detected in multilocus sequence analysis (MLSA) data using multiple techniques (43). These analyses primarily rely on contrasting evolutionary histories in different parts of the DNA sequences, leading to incongruence between gene trees at individual loci (14, 49), mosaic structure within gene sequences (35), or measurements of linkage disequilibrium between loci (50). In this study, we use a suite of methods to detect recombination among housekeeping genes in a population of Vibrio cholerae and other Vibrio isolates collected from central Californian and Hawaiian coastal environments. Results of these analyses are then compared to predicted outcomes for the stable ecotype model to assess the probability of identifying ecotypes among isolates from the V. cholerae population.
MATERIALS AND METHODS
Isolate selection.
One hundred sixty-five environmental and four clinical isolates (see Table S1 in the supplemental material) were chosen to be included in this study from a larger collection of putative Vibrio cholerae isolates (based on PCR from the 16S-23S rRNA intergenic spacer as described by Keymer et al. [28]). Representative isolates from the majority (>94%) of the enterobacterial repetitive intergenic consensus (ERIC)-PCR genotypes defined in Keymer et al. (29) were selected to include 45 isolates previously characterized by comparative genome hybridization (CGH) (28, 38), sediment and water column isolates collected contemporaneously, and four tropical environmental isolates collected from Hanalei Bay on the Hawaiian island of Kauai. These tropical isolates were cultivated from water column samples and processed as described previously (29).
MLSA.
Genomic DNA from selected isolates was extracted using the DNeasy blood and tissue kit (Qiagen) with the recommended manufacturer's protocols for Gram-negative bacterial cells. DNA sequences from seven housekeeping genes were PCR amplified in 50-μl reaction volumes containing 2.5 U HotStar HiFidelity DNA polymerase (Qiagen), 1× HotStar HiFidelity PCR buffer, and 1 μM each primer. More degenerate primers were needed to amplify sequences from a small number of isolates for each locus (9 to 14 isolates per locus). The degenerate primers required a less stringent proofreading enzyme and were added at 1 μM concentrations to 50 μl PCR mixture containing 2.5 U HotStarTaq polymerase (Qiagen) and 1× HotStar PCR buffer. The primer sequences, along with additional Mg2+ concentrations and annealing temperatures for each primer set, are provided in Table 1. Thermal cycling conditions included 30 to 35 cycles of 45 s of denaturation at 94°C, 1 min of annealing, and a 1-min extension at 72°C, followed by a final 10-min extension at 72°C. New primers were designed using Primer Express 2.0 (PE Applied Biosystems). PCR products of the appropriate size were purified using the MinElute gel extraction kit (Qiagen) and sequenced in both directions on an ABI 3730XL capillary sequencer (PE Applied Biosystems).
TABLE 1.
Primer pairs used to amplify MLSA gene sequences, with resulting amplicon lengths, annealing temperatures, and Mg2+ concentrations used in PCRa
| MLSA locus | Primer ID | Sequence (5′ to 3′) | Amplicon length (bp) | Anneal temp (°C) | Extra Mg2+ concn (mM) |
|---|---|---|---|---|---|
| gyrB | gyrB116b | TGGTTTTTGAGGTGGTGGATA | 1,214 | 60 | 0.5 |
| gyrB1330b | CGCTTGATTCTTACGGTTACG | 1,214 | 60 | 0.5 | |
| gyrB58 | CGYAAGCGTCCRGGTATGTA | 1,450 | 55 | 0.5 | |
| gyrB1507 | CGACRTCVGCATCGGTCA | 1,450 | 55 | 0.5 | |
| mdh | mdh540c | GTTTGACGGTCGGATACACC | 1,039 | 60 | 0.5 |
| mdh541c | AGAGCGGTATTTTCCAATGC | 1,039 | 60 | 0.5 | |
| mdh83 | CATYGGTCAAGCCCTDGC | 678 | 55 | 0.5 | |
| mdh760 | GCWAGACCRAARCGACAKGC | 678 | 55 | 0.5 | |
| recA | recA884c | TGGACGAGAATAAACAGAAGGC | 1,089 | 60 | 0.5 |
| recA885c | AACCTCTTTGCATTCAGCCC | 1,089 | 60 | 0.5 | |
| recA147 | ACCTGAGAGTGACTATCCGG | 1,004 | 55 | 0.5 | |
| recA1150 | GCATTTCACGCAGTTTTTTA | 1,004 | 55 | 0.5 | |
| recA172 | GGYYTACCAATGGGHCGTAT | 748 | 55 | 0.5 | |
| recA919 | TYGCTTTACCTTGRCCRA | 748 | 55 | 0.5 | |
| idh | idh4965b | TTCATTACTGCCGATTATTC | 1,063 | 55 | 0.5 |
| idh4966b | TTTGGTGTCTTTCTGCTTAC | 1,063 | 55 | 0.5 | |
| idh37 | ACTGATGAAGCMCCRGCGYT | 1,065 | 55 | 0.5 | |
| idh1101 | DCCCCACATYTGGCCWGA | 1,065 | 55 | 0.5 | |
| asd | asd414d | CCTTTGGCTAAACTCGG | 985 | 50 | 0.75 |
| asd416d | GTTATCCGCCACTACCC | 985 | 50 | 0.75 | |
| asd1051 | TGGGATMGHCGYGARTACAGY | 811 | 55 | 1 | |
| asd1862 | RCGKACRCACGTTGGGTT | 811 | 55 | 1 | |
| dnaE | dnaE713c | GATTTCTCTATGGTGGATGG | 1,111 | 60 | 0.5 |
| dnaE714c | ATTCCAGCGGATCAAGGTCG | 1,111 | 60 | 0.5 | |
| dnaE34 | CACAGTGATTTYTCDATGGTGG | 1,204 | 55 | 0.5 | |
| dnaE1237 | GRTCACGYTTATCCATACA | 1,204 | 55 | 0.5 | |
| nagB | nagB4 | AGACTTATCCCACTGAAAGCG | 787 | 55 | 0.5 |
| nagB790 | CGATGTTTTTGGCTTCTAACTC | 787 | 55 | 0.5 | |
| nagB32a | AAGTWGGYAARTGGGCRGC | 657 | 55 | 0.5 | |
| nagB688 | GMAGWGCAGARACKGTCCA | 657 | 55 | 0.5 |
DNA sequence analysis.
Raw sequence files were trimmed, aligned, and manually inspected using Sequencher 4.7 (Gene Codes, Ann Arbor, MI). Aligned sequences varied in length and position within the coding sequence: gyrB (1,119 bp, positions 160 to 1278), mdh (644 bp, positions 175 to 818), recA (690 bp, positions 349 to 1038), idh (926 bp, positions 121 to 1278), asd (690 bp, positions 16 to 705), dnaE (1,009 bp, positions 111 to 1119), and nagB (631 bp, positions 44 to 674). Unique alleles for each locus were identified with the NRDB program at http://www.mlst.net, and sequence types (STs) were defined for each unique combination of alleles. Standardized indices of association (IAS) and ratios of nonsynonymous to synonymous changes were computed in START2 (26) as measures of linkage disequilibrium and purifying selection, respectively. Tajima's D, a statistic testing alleles for departure from neutral evolution, was calculated using DnaSP 4.50.3 (45).
Recombination events were detected in concatenated DNA sequences using the RDP 3.0 software package (35) with the following settings: general (linear sequences, highest P value of 0.05, Bonferroni correction), RDP (no reference, window size of 8 polymorphic sites, 100% sequence identity range), Geneconv (scan triplets, G-scale of 1), Bootscan (window size of 500 bp, step size of 20 bp, 70% cutoff, F84 model, 200 bootstrap replicates, binomial P value), MaxChi (scan triplets, fraction 0.2 variable sites per window), Chimaera (scan triplets, window size equal to 20% of variable sites), SiScan (window of 500 bp, step size of 20 bp, use 1/2/3 variable positions, nearest outlier for 4th sequence, 1,000 P value permutations, 100 scan permutations).
Maximum likelihood (ML) trees were inferred with PhyML version 3.0 (19) using the general time reversible (GTR) substitution rate model with empirically estimated equilibrium frequencies, proportion of invariable sites, and gamma shape parameter with eight rate categories. Use of the GTR substitution model with invariable sites and gamma distribution was verified with Modeltest 3.7 (42). The likelihood congruence (LC) test was executed as described in Feil et al. (14), with 200 random trees generated in PAUP* version 4.0b10 (Sinauer Associates, Inc., Sunderland, MA). The approximately unbiased and Shimodaira-Hasegawa tests were performed with CONSEL (49). Population scale rates of recombination were estimated with LDhat 2.1 (37) using an average θW of 0.02455 per site per generation.
Concatenated nucleotide sequences for all Vibrio cholerae STs were used to construct a NeighborNet split network with the EqualAngle algorithm in the SplitsTree4 program (25). Distances were computed using the GTR substitution rate model with empirical frequencies.
Pairwise Hamming distances between individual and concatenated nucleotide sequences were determined with PAUP* version 4.0b10. Nonmetric multidimensional scaling plots of the Hamming distances were generated using Matlab 7.4 (MathWorks, Natick, MA).
Demarcation of ecotypes was performed automatically for each of the individual MLSA loci with the ecotype simulation program (30). To evaluate demarcations for different loci, all pairwise combinations of STs were scored as having the same or different ecotype identities. Summary statistics were generated to assess the frequencies that loci agreed (same or different ecotype) for any pair of STs. Ecotype assignments are provided in Table S2 in the supplemental material.
Biochemical analysis.
Biochemical differentiation of a subset of isolates was performed. Twenty-four of the 169 isolates were subjected to 24 different tests. As will be discussed in Results, a few of the putative V. cholerae isolates were deemed to be more similar to other Vibrio species. All 13 isolates of these non-V. cholerae species were included in the set of 24 for biochemical differentiation. In addition, 11 isolates that spanned the range in sequence divergence among the V. cholerae STs were included in the subset. Tests were performed as previously described (4, 10), using ATCC strains of Escherichia coli, Enterobacter aerogenes, Hafnia alvei, Salmonella typhimurium, and Staphylococcus aureus and Vibrio cholerae O1 El Tor strain N16961 as appropriate positive and negative controls. Bioluminescence was determined visually using LM medium described by Baumann and Baumann (2). All media were incubated at 25 to 30°C.
Nucleotide sequence accession numbers.
GenBank accession numbers for all dnaE sequences except the four Hawaiian isolates were provided by Keymer et al. (29), and all other sequences were deposited in the GenBank database under the accession numbers HM009380 to HM010397.
RESULTS
Taxonomy of analyzed isolates.
Nucleotide sequences were obtained for housekeeping loci from 169 presumptive Vibrio cholerae isolates chosen from a larger collection of primarily nontoxigenic (CTX negative by PCR [data not shown]) environmental isolates. One hundred sixty-five of these were previously analyzed by ERIC-PCR genomic fingerprinting (29); the four others are new isolates collected from Hanalei Bay in March 2008 on the island of Kauai. Seven loci were selected based on spatial separation from each other as well as from dispensable genes identified by comparative genome hybridization (28, 38). Six of the loci have been used successfully in previous studies of V. cholerae phylogeny (5, 9, 17, 27, 31, 46, 51). These six loci (asd, dnaE, gyrB, idh, mdh, and recA) are located on the large chromosome. The seventh locus (nagB), used for the first time in the present study, is located on the small chromosome. This gene was chosen due to its conserved role in chitin metabolism. A total length of 5,709 bp was analyzed using numerous methods to assess the presence and rate of recombination and its effect on the evolutionary structure of an environmental population of V. cholerae.
First, we examined pairwise nucleotide similarities and phenotypic characteristics to determine whether the isolates group as cohesive clusters that might represent species or ecotypes. In spite of the intended characterization of solely V. cholerae isolates, other Vibrio species were detected among our collection of isolates based on nucleotide divergence and phenotypic differences. When concatenating the sequences from all 7 housekeeping genes, we identified five distinct clusters of isolates with >93.6% DNA similarities among isolates of the same cluster (Table 2). In contrast, isolates from different clusters were typically ≤80% identical, although two pairs of clusters had intercluster similarities between 86.7% and 91.2%. The concatenated sequence identity cutoff used in our study (93.6%) is similar to the ones established empirically in other MLSA studies using known Vibrio species, including V. cholerae and Vibrio mimicus (54, 55). Applying this cutoff, we divided the isolates studied here into 156 V. cholerae isolates and 13 isolates of other Vibrio species designations (see Table S1 in the supplemental material). Eight of the non-V. cholerae Vibrio species corresponded to Vibrio aestuarianus (one isolate), Vibrio alginolyticus-Vibrio sp. Ex25 (four isolates), and Vibrio parahaemolyticus (three isolates) based on best-match Blastn similarities (data not shown) and phenotypic characteristics (see Table S3 in the supplemental material). The remaining cluster of five isolates was taxonomically unidentified; member isolates had concatenated nucleotide similarities of ≤91.2% with V. cholerae, indicating separate species, with even lower similarity to other Vibrio species, including V. mimicus. However, these isolates were phenotypically very similar to both V. cholerae and V. mimicus (see Table S3 in the supplemental material) and are here referred to as V. cholerae-like isolates. These species designations were employed in differentiating recombination events detected within and across Vibrio species by the RDP 3.0 software. All other sequence analyses are reported only for the V. cholerae isolates.
TABLE 2.
Ranges of pairwise concatenated sequence similarities within and between clusters of similar isolatesa
| Species | % similarity of isolate |
No. of isolates | ||||
|---|---|---|---|---|---|---|
| Vibrio cholerae | V. cholerae-like sp. | Vibrio parahaemolyticus | Vibrio alginolyticus | Vibrio aestuarianus | ||
| V. cholerae | 94.1-100 | 156 | ||||
| V. cholerae-like sp. | 89.4-91.2 | 93.6-100 | 5 | |||
| V. parahaemolyticus | 79.5-80.0 | 79.6-79.9 | 98.8-100 | 3 | ||
| V. alginolyticus | 79.1-80.0 | 79.1-79.7 | 86.7-87.0 | 94.7-98.6 | 4 | |
| V. aestuarianus | 79.2-79.6 | 78.8-79.0 | 78.9-79.0 | 78.5-79.0 | NA | 1 |
Five cohesive clusters of isolates, corresponding to the named Vibrio species, were detected among the isolates analyzed by MLSA. Intraspecies similarities were ≥93.6%, while interspecies similarities ranged from 78.5% to 80.0%, with the exception of 86.7 to 87.0% similarities between V. parahaemolyticus and V. alginolyticus and 89.4 to 91.2% similarities between V. cholerae and the V. cholerae-like species. NA, not applicable.
Recombination among Vibrio cholerae isolates.
To detect evidence of recombination in our collection of isolates, we analyzed DNA sequences for mosaic patterns suggestive of recombination. Recombination events were detected by three or more methods in most (72%) of the V. cholerae sequence types (n = 113) analyzed with the RDP 3.0 software package (see Table S4 in the supplemental material). Seventy-six of the total 107 events were identified by more than one detection method (P < 0.05, with Bonferroni correction), and 38 events were detected with between 4 and 7 methods (see Table S5 in the supplemental material), increasing confidence that these patterns are due to bona fide recombination events instead of statistical overcalling. Recombination events were evenly detected in five loci; fewer detected events in the mdh and nagB loci are most likely accounted for by the lower number of nucleotide differences in these sequences (see Results and Table S6 in the supplemental material).
Of the 56 recombination events that were statistically significant for three or more detection methods, intraspecific recombination among V. cholerae strains accounted for 43 events (77%), while four events involved only non-V. cholerae Vibrio species. The remaining nine events (16%) involved interspecific recombination between V. cholerae and other Vibrio spp., with V. cholerae isolates identified as recombinants in four events and donors in the other five events (see Figure S1 in the supplemental material). Interspecific recombination events involving V. cholerae were detected in gyrB (five events), recA (one event), idh (two events), and asd (one event).
Next, a robust estimate of the relative contributions of recombination and mutation to generating diversity in the Vibrio cholerae concatenated sequence alignments was obtained using LDhat 2.1 (37). This method is insensitive to different evolutionary models and deviation from the infinite sites assumption (43, 52). Using an average mutation rate (θ) computed as 21.0 mutations per locus per generation, the population recombination rate (ρ) was estimated at 136.5 recombinations per locus per generation. Therefore, the relative rate of recombination to mutation (ρ/θ) is approximately 6.5:1. This value is similar to our estimates based on the number of polymorphisms in single- and double-locus variants (≥4:1 or ≥8:1) (see Results and Table S7 in the supplemental material) and within the range expected for a sexual population (14, 16).
Effects of recombination on phylogeny and structure.
Frequent recombination can disrupt vertical evolutionary relationships between genes on the same genome, resulting in linkage equilibrium and incongruence of topologies constructed for individual loci. Despite the detection of recombination events in most of the isolates, significant linkage disequilibrium was detected for the Vibrio cholerae isolates, indicating some clonal population structure. A standardized index of association (IAS) (23) significantly different from zero was observed for all 156 V. cholerae isolates (IAS = 0.439) and remained significant when only unique sequence types (STs) were considered (IAS = 0.143). Thus, sampling bias alone does not explain the observed linkage disequilibrium. All indices were significantly nonzero for P values of <0.05.
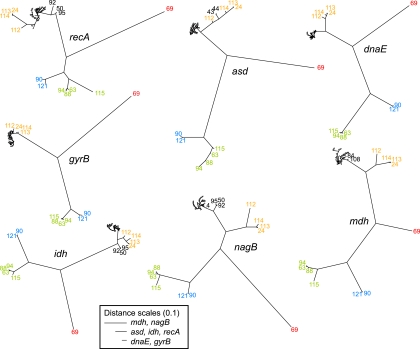
Topological congruence among gene trees can range along a spectrum from complete congruence to complete incongruence. At one end of the spectrum, the Shimodaira-Hasegawa and approximately unbiased tests identify deviations from perfect congruence. All possible pairs of seven loci trees for all V. cholerae STs were significantly incongruent (P < 0.001) according to the Shimodaira-Hasegawa and approximately unbiased tests (see Table S8 in the supplemental material). At the other end of the spectrum, the likelihood congruence test (14) assesses whether locus trees are more topologically similar than trees of random topology by comparing log likelihoods. According to this test, all possible pairs of locus trees were significantly (P < 0.01) more congruent than pairings with random trees (see Table S9 in the supplemental material). In contrast to the incongruence observed within trees for the V. cholerae STs, branching patterns in gene trees for other Vibrio isolates were highly congruent (Fig. 1).
FIG. 1.
Maximum likelihood trees for each locus. Trees include all Vibrio sequence types (STs). Most V. cholerae sequences fall into the dense sequence clusters, but branches are labeled for divergent sequence types (including other putative Vibrio species). Despite incongruence in topologies among V. cholerae STs across different loci, the topologies among other Vibrio species are largely congruent. V. cholerae STs are labeled in black, V. aestuarianus-like STs in red, V. alginolyticus-like STs in green, V. parahaemolyticus-like STs in blue, and the V. cholerae-like STs in orange. The scales of branch lengths vary for different loci and are provided in the legend box.
Incongruent phylogenies for different loci were also evident in the split network diagram constructed for all V. cholerae STs (see Fig. S2 in the supplemental material). The abundance of splits between the interior branches of the tree shows that recombination events have a significant impact on evolutionary relationships among isolates and restrict the ability to resolve a consensus phylogeny for this population.
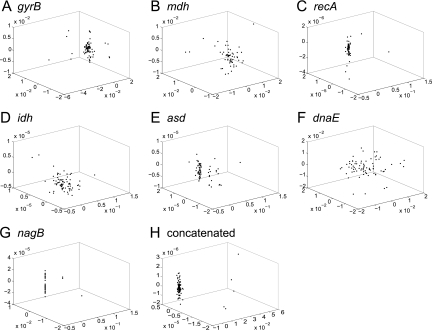
Nonmetric multidimensional scaling plots were constructed for pairwise Hamming distances between V. cholerae gene sequences. This ordination technique allows for visualization of cluster formation among sequence entries based on the computed nucleotide distances. If recombination is rare, as assumed in the stable ecotype model, sequences should form clusters corresponding to ecotypes. Moreover, ecotypes experiencing more recent periodic selection events should display less cluster dispersion. We observed little or no formation of clusters among the V. cholerae sequences. For the individual MLSA loci (Fig. 2 A to G), only plots for gyrB, asd, and perhaps recA appeared that they might contain multiple clusters. For the remaining loci and the concatenated sequences (Fig. 2H), the data formed a single diffuse cluster containing environmental and clinical isolates, with the exception of a few genetically distant outliers. In addition, ≥92% of the V. cholerae STs had identical amino acid sequences for the mdh, recA, and nagB loci, indicating that much or most of the nucleotide diversity in these sequences can be considered functionally neutral. Together, the data illustrate the maintenance of neutral nucleotide sequence diversity within the V. cholerae species cluster and no clear formation of subclusters that may correspond to ecotypes.
FIG. 2.
Nonmetric multidimensional scaling plots were constructed for pairwise distances between gene sequences from Vibrio cholerae isolates. Unique sequences are represented as closed circles, and the three-dimensional distances between coordinates approximate Hamming distances between sequences. The first seven plots represent sequences from individual loci, gyrB (A), mdh (B), recA (C), idh (D), asd (E), dnaE (F), and nagB (G); the last plot (H) displays distances for concatenated sequences from all MLSA loci.
Ecotype simulation.
To test whether the sequence data are compatible with the stable ecotype model, an ecotype simulation program (30) was used to demarcate ecotypes for each of the individual MLSA loci (see Table S2 in the supplemental material). On average for any two loci, there was a 63% probability (range, 44 to 83%) that ecotype predictions from a pair of STs were in agreement. Only 14% of the time were two STs confidently assigned to the same ecotype or different ecotypes for all of the seven loci. If the criterion is relaxed to six of seven genes in agreement, there is a 43% chance of being able to confidently assign two STs. For comparison, the Bacillus simplex and Bacillus subtilis-Bacillus licheniformis populations analyzed by Koeppel et al. (30) had 95% and 98% probabilities of two STs having ecotype demarcations in agreement for two loci, respectively. All three loci in that study produced consistent ecotype assignments for >93% of the STs. In contrast, for any three loci from the V. cholerae MLSA data set, consistent ecotype assignments are predicted only for 44% of the STs on average (range, 28 to 72%).
DISCUSSION
Genetic cohesion of species or species-like clusters is usually attributed to the evolutionary forces of natural selection and recombination (7). Stable ecotype models have been used to explain intraspecies patterns in diversity for bacteria with low to moderate levels of recombination (30, 58). The model predicts that periodic selection in such systems produces discrete sequence clusters with different ecological specifications. It is difficult to reconcile the MLSA data from the Vibrio cholerae isolates presented herein with the stable ecotype model. First, sequences from the isolates appear to form discrete clusters only at the species level and produce diverse, incongruent topologies at finer genetic resolution. Second, despite significant linkage disequilibrium between alleles, the recombination rate in V. cholerae is within the range of those of other sexual bacteria. Third, high genomic and phenotypic diversity within portions of the V. cholerae population implies potential ecotypes (28, 29); however, these potential ecotypes, including disease-causing clinical isolates, do not form distinct sequence clusters. Finally, there is only a 1 in 7 chance that ecotype demarcations for all seven genes agree that any two different V. cholerae isolates belong to the same ecotype or not. Given the data, the ecotype model is not a convincing fit for the V. cholerae population described here. A model similar to Mayr's biological species concept may be a more appropriate fit for these isolates and should be explicitly applied to the data in future work.
The propagation and persistence of nucleotide diversity within the V. cholerae species could be explained by weak or inefficient natural selection that fails to purge diversity through selective sweeps and/or frequent recombination that uncouples genes from selection at the genome level (41). The former explanation would result in neutral sequence and gene content diversity accumulating in all loci. In contrast, the latter explanation would produce a number of adaptive alleles purged of neutral diversity, while that diversity is maintained in many other genes, including core housekeeping genes typically used for phylogenetic analyses (1, 32). In a previous study, we examined gene content differences using CGH among a subset of the isolates included in the present study. We found significant correlations between the presence of certain genes and presumably favorable environmental conditions (28). The apparent gene-level selection among these isolates suggests that frequent recombination and not necessarily weak selection may be responsible for the extensive intraspecies diversity.
The explanations for persistent diversity outlined above present a problem for identifying ecotypes in the population. This is because they either are not clearly separated from other ecotypes or can be identified only in adaptive genes that are traditionally excluded from phylogenetic analyses. While much research has focused on the genes and phenotypes of ecotypes involved in causing human disease, we know relatively little about the genes and phenotypes that are useful and important for V. cholerae in environmental systems. Phenotypic tests commonly used to classify Vibrio species could not adequately distinguish isolates with 90% or lower concatenated nucleotide identity (see Table S3 in the supplemental material). This lack of specificity highlights that these tests are relatively uninformative for environmental bacteria and more environmentally relevant phenotypes should be investigated. Without knowledge of potentially niche-adaptive genes or phenotypes in coastal ecosystems, we will be unable to put observations of genetic or phenotypic diversity into the context of V. cholerae ecology and evolution.
Previous work by Miller et al. (38) demonstrated that representative isolates from this V. cholerae population become naturally transformable when grown on chitin surfaces. While all known bacteria with panmictic population structures are capable of natural transformation, Smith et al. (50) showed that Haemophilus influenzae has maintained a primarily clonal structure despite being transformable. Therefore, Vibrio cholerae might satisfy the biological requirements to become a freely recombining species, but some factors in its ecology restrict the breakdown in linkage disequilibrium. Low population densities typically observed in coastal water samples may limit the spatial proximity required for transformation, but more work is needed to understand V. cholerae occurrence and behavior in surface-attached environmental communities. The mechanism of recombination in these environmental bacteria is still poorly understood and warrants further study.
Using a robust likelihood-based estimator of recombination rate (LDhat 2.1) produced a ratio of 6.5 for the relative rate of locus divergence by recombination compared to mutation. An additional rudimentary pairwise comparison of isolates differing at two or fewer loci estimated that recombination was at least 45 times as likely as mutation to be responsible for nucleotide divergence in clonal genotypes (recombination/mutation ≥ 45) (see Results in the supplemental material). These values are similar or higher than those estimated for other opportunistic pathogens with sexual characteristics, such as E. coli (20) and N. meningitidis (13), as well as frequently recombining aquatic bacteria, including V. parahaemolyticus and Vibrio vulnificus (3, 18, 56). However, highly similar isolates persist within the V. cholerae population, and the index of association for this population suggests that recombination is not frequent enough to completely disrupt linkage disequilibrium. Future work should focus on the promiscuity of recombination in V. cholerae and assess whether linkage disequilibrium and nonrandom topologies observed here reflect patterns in positive selection, targeted recombination, or bias associated with isolate selection.
Previous studies of Vibrio cholerae isolates by MLSA tend toward low to intermediate levels of recombination and clonal to weakly clonal evolutionary relationships (5, 17, 31, 46), but these studies have been strongly biased toward clinical and environmental isolates from regions with epidemic disease. In this study, we used a suite of methods to detect recombination among housekeeping genes in a population of Vibrio cholerae and other Vibrio isolates collected from central Californian and Hawaiian coastal environments. Significant linkage disequilibrium was detected among V. cholerae sequence types, but the population recombination rate was 6.5 times greater than the mutation rate and produced significant incongruence among trees from individual loci. These results implicate recombination as an important factor in the evolution of gene sequences in environmental V. cholerae. Our data, which provide the most thorough examination of recombination and population structure in environmental V. cholerae isolates to date, imply that frequent recombination may impede the identification of V. cholerae ecotypes by multilocus sequence analysis. Additional isolates from other areas of the world's coastal ocean should be analyzed to confirm this finding.
Supplementary Material
Acknowledgments
This work was funded by NOAA Oceans and Human Health Initiative grant NA04OAR4600195, NSF grant OCE-0742048, and the Gerhard Casper Stanford Graduate Fellowship (D.P.K.).
We thank several colleagues, particularly Rachel J. Whitaker, for their helpful comments on the manuscript. Nick de Sieyes, Tim Julian, Blythe Layton, Alyson Santoro, Kevan Yamahara, and Lilian H. Lam provided valuable support in the laboratory.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S., V. Klepac-Ceraj, D. Hunt, C. Pharino, I. Ceraj, D. Distel, and M. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., and L. Baumann. 1981. The marine Gram-negative eubacteria: genera Photobacterium, Alteromonas, Pseudomonas, and Alcaligenes, p. 1302-1331. In M. P. Starr, H. Stolp, H. G. Truper, A. Balows, and H. Schlegel (ed.), The prokaryotes: a handbook on habitats, isolation, and identification of bacteria, 1st ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 3.Bisharat, N., D. I. Cohen, M. C. Maiden, D. W. Crook, T. Peto, and R. M. Harding. 2007. The evolution of genetic structure in the marine pathogen, Vibrio vulnificus. Infect. Genet. Evol. 7:685-693. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, D. J., and J. J. Farmer III. 2005. Family I. Enterobacteriaceae, p. 587-850. In D. J. Brenner, N. R. Krieg, J. T. Staley, G. M. Garrity, D. R. Boone, P. de Vos, M. Goodfellow, F. A. Rainey, and K.-H. Schleifer (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 5.Byun, R., L. D. Elbourne, R. Lan, and P. R. Reeves. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun, J., C. J. Grim, N. A. Hasan, J. H. Lee, S. Y. Choi, B. J. Haley, E. Taviani, Y.-S. Jeon, D. W. Kim, J.-H. Lee, T. S. Brettin, D. C. Bruce, J. F. Challacombe, J. C. Detter, C. S. Han, A. C. Munk, O. Chertkov, L. Meincke, E. Saunders, R. A. Walters, A. Huq, G. B. Nair, and R. R. Colwell. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 106:15442-15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457-487. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle, W. F., and O. Zhaxybayeva. 2009. On the origin of prokaryotic species. 19:744-756. [DOI] [PubMed] [Google Scholar]
- 9.Farfan, M., D. Minana-Galbis, M. C. Fuste, and J. G. Loren. 2002. Allelic diversity and population structure in Vibrio cholerae O139 Bengal based on nucleotide sequence analysis. J. Bacteriol. 184:1304-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer, J. J., III, and J. M. Janda. 2005. Family. I. Vibrionaceae, p. 491-555. In D. J. Brenner, N. R. Krieg, J. T. Staley, G. M. Garrity, D. R. Boone, P. de Vos, M. Goodfellow, F. A. Rainey, and K.-H. Schleifer (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 11.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque, S. M., D. A. Sack, R. B. Sack, R. R. Colwell, Y. Takeda, and G. B. Nair. 2003. Emergence and evolution of Vibrio cholerae O139. Proc. Natl. Acad. Sci. U. S. A. 100:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 14.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. U. S. A. 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, C., W. P. Hanage, and B. G. Spratt. 2007. Recombination and the nature of bacterial speciation. Science 315:476-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg, P., A. Aydanian, D. Smith, J. G. Morris, Jr., G. B. Nair, and O. C. Stine. 2003. Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emerg. Infect. Dis. 9:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Escalona, N., J. Martinez-Urtaza, J. Romero, R. T. Espejo, L.-A. Jaykus, and A. DePaola. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 20.Guttman, D. S., and D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380-1383. [DOI] [PubMed] [Google Scholar]
- 21.Hanage, W. P., C. Fraser, and B. G. Spratt. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanage, W. P., B. G. Spratt, K. M. E. Turner, and C. Fraser. 2006. Modelling bacterial speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2039-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haubold, B., M. Travisano, P. B. Rainey, and R. R. Hudson. 1998. Detecting linkage disequilibrium in bacterial populations. Genetics 150:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmes, E., R. Urwin, and M. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 25.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 26.Jolley, K. A., E. J. Feil, M. S. Chan, and M. C. Maiden. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230-1231. [DOI] [PubMed] [Google Scholar]
- 27.Karaolis, D. K., R. Lan, and P. R. Reeves. 1995. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177:3191-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keymer, D. P., M. C. Miller, G. K. Schoolnik, and A. B. Boehm. 2007. Genomic and phenotypic diversity of coastal Vibrio cholerae strains is linked to environmental factors. Appl. Environ. Microbiol. 73:3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keymer, D. P., L. H. Lam, and A. B. Boehm. 2009. Biogeographic patterns in genomic diversity among a large collection of Vibrio cholerae isolates. Appl. Environ. Microbiol. 75:1658-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koeppel, A., E. B. Perry, J. Sikorski, D. Krizanc, A. Warner, D. M. Ward, A. P. Rooney, E. Brambilla, N. Connor, R. M. Ratcliff, E. Nevo, and F. M. Cohan. 2008. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc. Natl. Acad. Sci. U. S. A. 105:2504-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotetishvili, M., O. C. Stine, Y. Chen, A. Kreger, A. Sulakvelidze, S. Sozhamannan, and J. J. G. Morris. 2003. Multilocus sequence typing has better discriminatory ability for typing Vibrio cholerae than does pulsed-field gel electrophoresis and provides a measure of phylogenetic relatedness. J. Clin. Microbiol. 41:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan, R., and P. R. Reeves. 2000. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 8:396-401. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. H., K. H. Han, S. Y. Choi, M. E. Lucas, C. Mondlane, M. Ansaruzzaman, G. B. Nair, D. A. Sack, L. von Seidlein, J. D. Clemens, M. Song, J. Chun, and D. W. Kim. 2006. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J. Med. Microbiol. 55:165-170. [DOI] [PubMed] [Google Scholar]
- 34.Lipp, E. K., A. Huq, and R. R. Colwell. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin, D. P., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 36.Mayr, E. 1942. Systematics and the origin of species from the viewpoint of a zoologist. Columbia University Press, New York, NY.
- 37.McVean, G., P. Awadalla, and P. Fearnhead. 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160:1231-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, M. C., D. P. Keymer, A. Avelar, A. B. Boehm, and G. K. Schoolnik. 2007. Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains. Appl. Environ. Microbiol. 73:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Shea, Y. A., F. J. Reen, A. M. Quirke, and E. F. Boyd. 2004. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 42:4657-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Losada, M., R. P. Viscidi, J. C. Demma, J. Zenilman, and K. A. Crandall. 2005. Population genetics of Neisseria gonorrhoeae in a high-prevalence community using a hypervariable outer membrane porB and 13 slowly evolving housekeeping genes. Mol. Biol. Evol. 22:1887-1902. [DOI] [PubMed] [Google Scholar]
- 41.Polz, M. F., D. E. Hunt, S. P. Preheim, and D. M. Weinreich. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2009-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 43.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 44.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 45.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 46.Salim, A., R. Lan, and P. R. Reeves. 2005. Vibrio cholerae pathogenic clones. Emerg. Infect. Dis. 11:1758-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarkar, S. F., and D. S. Guttman. 2004. Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70:1999-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schierup, M. H., and J. Hein. 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics 156:879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 50.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. U. S. A. 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stine, O. C., S. Sozhamannan, Q. Gou, S. Zheng, J. J. G. Morris, and J. A. Johnson. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infect. Immun. 68:7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stumpf, M. P. H., and G. A. T. McVean. 2003. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 4:959-968. [DOI] [PubMed] [Google Scholar]
- 53.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, F. L., D. Gevers, C. C. Thompson, P. Dawyndt, S. Naser, B. Hoste, C. B. Munn, and J. Swings. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson, C. C., F. L. Thompson, and A. C. P. Vicente. 2008. Identification of Vibrio cholerae and Vibrio mimicus by multilocus sequence analysis (MLSA). Int. J. Syst. Evol. Microbiol. 58:617-621. [DOI] [PubMed] [Google Scholar]
- 56.Vos, M., and X. Didelot. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199-208. [DOI] [PubMed] [Google Scholar]
- 57.Vulić, M., F. Dionisio, F. Taddei, and M. Radman. 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. U. S. A. 94:9763-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward, D. M., M. M. Bateson, M. J. Ferris, M. Kuhl, A. Wieland, A. Koeppel, and F. M. Cohan. 2006. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1997-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.