Abstract
Surfaces made of copper or its alloys have strong antimicrobial properties against a wide variety of microorganisms. However, the molecular mode of action responsible for the antimicrobial efficacy of metallic copper is not known. Here, we show that dry copper surfaces inactivate Candida albicans and Saccharomyces cerevisiae within minutes in a process called contact-mediated killing. Cellular copper ion homeostasis systems influenced the kinetics of contact-mediated killing in both organisms. Deregulated copper ion uptake through a hyperactive S. cerevisiae Ctr1p (ScCtr1p) copper uptake transporter in Saccharomyces resulted in faster inactivation of mutant cells than of wild-type cells. Similarly, lack of the C. albicans Crp1p (CaCrp1p) copper-efflux P-type ATPase or the metallothionein CaCup1p caused more-rapid killing of Candida mutant cells than of wild-type cells. Candida and Saccharomyces took up large quantities of copper ions as soon as they were in contact with copper surfaces, as indicated by inductively coupled plasma mass spectroscopy (ICP-MS) analysis and by the intracellular copper ion-reporting dye coppersensor-1. Exposure to metallic copper did not cause lethality through genotoxicity, deleterious action on a cell's genetic material, as indicated by a mutation assay with Saccharomyces. Instead, toxicity mediated by metallic copper surfaces targeted membranes in both yeast species. With the use of Live/Dead staining, onset of rapid and extensive cytoplasmic membrane damage was observed in cells from copper surfaces. Fluorescence microscopy using the indicator dye DiSBaC2(3) indicated that cell membranes were depolarized. Also, during contact-mediated killing, vacuoles first became enlarged and then disappeared from the cells. Lastly, in metallic copper-stressed yeasts, oxidative stress in the cytoplasm and in mitochondria was elevated.
Copper is an essential nutrient in many organisms, including microbes, and enzyme-associated copper is a requirement for aerobic metabolism. On the other hand, excess accumulation of copper or intracellular release of free copper leads to severe toxicity. Aerobically, copper readily catalyzes reactions that result in the production of hydroxyl radicals through the Fenton and Haber-Weiss reactions (15, 16). The highly reactive oxygen intermediates are responsible for lipid peroxidation and oxidation of proteins and may be involved in damage to nucleic acids (16, 18, 35). Free copper ions are able to oxidize sulfhydryl groups, such as cysteine in proteins or the cellular redox buffer glutathione (20). Finally, copper might cause protein inactivation by damaging Fe-S clusters in proteins such as cytoplasmic hydratases (21). Damage to DNA caused by copper is controversial. Previously, it was believed that oxidative DNA damage contributed to copper ion toxicity (36). However, experiments with Escherichia coli demonstrated that under anaerobic conditions, the growth rate was even more strongly suppressed by copper ions than under aerobic conditions (31). More importantly, addition of copper ions to E. coli cells decreased oxidative DNA damage when cells were challenged with hydrogen peroxide (22).
Recently, the antimicrobial efficacy of metallic copper surfaces has been established for a variety of bacteria and fungi (25, 30). In these studies, cells in buffer were applied to copper surfaces, incubated under ambient conditions, and killed within hours. We recently established a method that mimics contact of microbes to dry copper touch surfaces. Under these conditions, most microbes are killed within minutes (11, 12). Studies by us and others have demonstrated that copper ions are released from metallic copper upon contact with cells, probably contributing to contact-mediated killing (12, 28). Further, extracellular supplementation with protective substances against oxidative stress, such as catalase, superoxide dismutase, or mannitol, a hydroxyl radical quencher, increased the time needed to kill copper surface-exposed E. coli cells (12). Currently, it is not known how microbes exposed to dry copper surfaces are rapidly inactivated, and we are only beginning to comprehend the molecular mode of action exerted by copper ions on microbes. The specific kinds of stress caused by metallic copper surfaces and the identity of sensitive cellular targets during contact-mediated killing have not been elucidated, yet. However, a recent study has suggested that DNA in bacteria exposed to copper surfaces might become degraded (37). Detailed understanding of the mode of action is required for a better understanding of why metallic copper alloys in hospital trials have proven to be excellent antimicrobial surfaces (6, 24, 26).
In this study, we investigated the mechanisms of contact-mediated killing on metallic copper (alloys) using two yeast model organisms, Candida albicans and Saccharomyces cerevisiae, including their mutants in copper ion homeostasis. Our results demonstrated that copper ions were accumulated by cells exposed to copper surfaces. The mode of action of contact-mediated killing did not involve genotoxicity through mutation, but instead, toxicity targeted the cells' membrane systems.
MATERIALS AND METHODS
Yeast strains and growth media.
The strains used in this work are listed in Table 1. C. albicans cells were grown to stationary phase in YPD medium (10 g of yeast extract, 5 g of peptone, and 20 g of glucose per liter) at 30°C for 24 h. Exponential-growth-phase cultures were grown as described above, but cells were diluted 1:100 from a 24-h culture into fresh YPD medium and harvested after 6 h. S. cerevisiae cells were grown in synthetic complete (SC) medium (2 g of yeast synthetic dropout medium supplement without uracil [Sigma-Aldrich], 20 g of dextrose, 6.7 g of yeast nitrogen base per liter), lacking uracil for plasmid selection (SC-ura), at 30°C for 48 h, unless otherwise stated. Solid medium was prepared by adding 1.5% Bacto agar (Difco BD). The S. cerevisiae plasmids were vector-only control p416-TEF (in the BY4741, ace1Δ, and S. cerevisiae ctr1Δ [Scctr1Δ] strains) and p416-TEF-Ctr1 and p416-TEF-Ctr1(300) (in the Scctr1Δ strain) (9, 41). An S. cerevisiae URA3 uracil prototroph, strain S288C, was used for the mutagenesis assay.
TABLE 1.
Strains used in this study
| Organism or strain | Relevant genotypic information | Reference |
|---|---|---|
| Candida albicans | ||
| SC5314 | Wild-type clinical isolate (parent of all Candida strains used) | 14 |
| CAF3-1 | Δura3::imm434/Δura3::imm434 | 13 |
| Cacrp1Δ strain | Δura3::imm434/Δura3::imm434 Δcrp1/Δcrp1 | 40 |
| Cacup1Δ strain | Δura3::imm434/Δura3::imm434 Δcup1/Δcup1 | 40 |
| Saccharomyces cerevisiae | ||
| S288C | MATα SUC2 gal2 mal mel flo1 flo8-1 hap1 ho bio1 bio6 | 29 |
| BY4741 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ | 4 |
| Scace1Δ strain | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ Δace1::KanMX6 | 2 |
| Scctr1Δ strain | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ Δcrt1::KanMX4 | 9 |
C. albicans strain SC5314 was employed to analyze killing kinetics at 23°C. To compare the role of impaired copper homeostasis systems, the wild-type strain used was CAF3-1, which is an ura3 auxotroph isogenic to strain SC5314.
Metal surface contact-mediated-killing assay.
The metal surfaces used in this study were 2.5- by 2.5-cm copper coupons (pure copper [C11000; 99.9% copper], cupronickel [C75200; 62% maximum Cu, 18% maximum Ni, and 21% maximum Zn], and Nordic Gold [89% Cu, 5% Al, 5% Zn, and 1% Sn]) or stainless steel control coupons (AISI 304; approximately 67 to 72% Fe, 17 to 19.5% Cr, and 8 to 10.5% Ni). All copper alloy coupons were treated prior to each experiment to standardize the surface properties. Coupons were washed for 30 s in 3% (wt/vol) NaOH solution at 70°C and rinsed in distilled water. After transfer to 10% (vol/vol) sulfuric acid solution for 5 s at room temperature, coupons were immediately washed with distilled water. All coupons, copper and stainless steel, were disinfected and cleaned by immersion in ethanol and kept in a sterile container. To prevent surface reoxidation, cleaned coupons were not flamed after immersion in ethanol.
To study the survival of cells on dry metal surfaces, cultures were concentrated 10-fold in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, and 2 mM potassium phosphate monobasic, pH 7.4) and tested as described in reference 12, with minor changes. Samples of approximately 2.5 × 106 (Candida) or 1 × 106 (Saccharomyces) cells were streaked out on coupons using sterile cotton swabs. All samples dried completely within 5 s after contact with the surfaces. Unless indicated otherwise, this time point is considered “0” throughout this study. To avoid contamination from the laboratory environment, coupons were incubated in sterile petri dishes at 23°C for different times. Cells on surfaces were removed into a 50-ml conical falcon tube with 10 ml ice-cold TBSBX buffer (137 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl, pH 7.4, 100 mM BaCl2, and 0.1% Triton X-100) by vortexing the coupon for 1 min. Samples were diluted in PBS buffer and plated on YPD agar. Surviving yeasts were counted as numbers of CFU after 24 h (Candida) or 48 h (Saccharomyces) of incubation using an automatic counter (aCOLyte; Synbiosis) and the associated software (version 2.0.8).
We experienced severe difficulties removing yeast cells efficiently from copper surfaces, a factor not previously reported (25). Thus, we tested several different buffers for their usefulness. To easily assess the ability of buffers to remove yeast cells from the metal coupons, cells were grown as described above and stained with 50 μg × ml−1 of Calcofluor White (Fluorescent Brightener 28; Sigma-Aldrich) (excitation wavelength [λEx] = 365 nm; emission wavelength [λEm] = 435 nm). Calcofluor White is a fluorescent dye that nonspecifically binds to P-linked polysaccharide polymers (23). Stained cells were applied to metal coupons as described above and removed by vortexing them in buffer. To verify complete removal of yeast cells from the surface, coupons were immediately examined by fluorescence microscopy using a confocal laser scanning microscope with a LD405 laser (405 nm) and a 60× dry lens. Images were captured using the Fluoview 500 (Olympus) software program (verson 4.3 with TIEMPO). Only TBSBX buffer was able to completely remove cells from surfaces and was thus used throughout this study.
Mutagenicity assay.
The pyrimidine analogue 5-fluoroorotic acid (5-FOA) has been used to select for ura3− cells in S. cerevisiae (3). 5-FOA prevents the growth of the wild type but not of mutant yeast cells that have acquired a mutation in the URA3 gene. URA3 encodes orotidine 5-phosphate decarboxylase, which also converts 5-FOA to the toxic 5-fluorouracil, which causes cell death. Thus, it is possible to use 5-FOA to evaluate toxins for their mutagenicity leading to uracil-auxotrophic mutants. Here, we employed 5-FOA positive selection to investigate a possible role for genotoxicity as a mode of action of metallic copper surfaces. An advantage of this system is that any inactivation mutation (point, frameshift, deletion, or insertion) in uracil biosynthesis will generate 5-FOA-resistant cells that can be scored. A method (3), which provided a protocol for generating ura3 mutants in growing cells, was adapted for use with nongrowing surface-exposed cells on metallic surfaces. S. cerevisiae S288C cells, containing an intact URA3 gene, were used for this experiment. Cells were prepared as described above, applied to the surfaces for 5 s (a time period of exposure shorter than required for massive onset of lethal damage), removed as described above, concentrated, diluted, and spread onto solidified 5-FOA medium (7 g × liter−1 yeast nitrogen base, 1 g × liter−1 5-FOA [Oakland Products, Inc.], 50 mg × liter−1 uracil, 20 g × liter−1 glucose, 2% [wt/vol] agar) (3). For determination of total CFU, cells were plated in parallel onto the same medium containing no 5-FOA.
After 72 h of incubation at 30°C colonies originating from mutants able to grow on 5-FOA were counted. The frequency of appearance of mutants was calculated by dividing the number of 5-FOA CFU by the number of total CFU. As controls, cells were exposed for the same period of time on stainless steel or stainless steel with 0.9% (wt/vol) formaldehyde, a known mutagen (8). To assess if groups of data were statistically different from each other, a t test was performed for data of copper-exposed, stainless steel-exposed cells and formaldehyde exposed cells on stainless steel (positive control). The two-tailed probability values (P values) were ≤0.05 for stainless steel and copper surfaces compared to the level for the mutagen control.
ICP-MS analysis.
To determine the metal content of cells exposed to metal surfaces, cells were prepared and applied to the metal coupons as described above. Cells were removed at different time points by vortexing the coupon in 10 ml ice-cold TBSBX for 1 min. Cells were centrifuged for 5 min at 4,500 × g and resuspended in 5 ml of ice-cold PBS containing 20 μM EDTA to remove excess extracellularly bound ionic copper. Cells were pelleted again (5 min at 4,500 × g) and mineralized with concentrated 70% (wt/vol) HNO3 (trace metal grade; Mallinckrodt) at 70°C for 4 h. Samples were diluted to a final concentration of 5% (wt/vol) nitric acid. Gallium [as Ga(NO3)3] was added as internal standard at a final concentration of 50 ppb. Element analysis was performed via inductively coupled plasma mass spectrometry (ICP-MS) using an Agilent ICP-MS 7500cx instrument operating with a collision cell and flow rates of 3.5 ml × min−1 of H2 and 1.5 ml × min−1 of He. Data acquisition for each sample was accumulated in triplicate for 100 milliseconds. An external calibration curve was recorded with gallium in 5% (wt/vol) nitric acid. Samples were loaded onto 96-well plates prior to analysis, and an autosampler (Elemental Scientific) was used to inject samples.
Evaluation of cytoplasmic membrane damage.
A Live/Dead staining technique (Live/Dead BacLight viability kit; Invitrogen) was employed to differentiate cells with undamaged and damaged permeable membranes on copper and stainless steel control surfaces. This kit employs two nucleic acid stains: green fluorescent SYTO 9 stain and red fluorescent propidium iodide stain. These stains differ in their ability to penetrate healthy yeast cells. When used alone, SYTO 9 stain labels DNA of both live and dead cells. In contrast, propidium iodide penetrates only cells with damaged membranes. Thus, live cells with intact membranes fluoresce green, while cells with damaged membranes fluoresce red. Cells were applied and removed from surfaces as described above. For staining, cells were suspended in 100 μl of 0.9% NaCl and 1 μl of the staining mixture (1 μl SYTO 9, 1 μl propidium iodide in 60 μl dimethyl sulfoxide [DMSO]) was added. Cell suspensions were incubated at 23°C in the dark for 15 min, then transferred onto glass slides, and immediately examined by fluorescence microscopy (λEx = 488/543 nm; λEm = 522/590 nm) under oil immersion using a confocal microscope. For SYTO 9, the laser used was an argon laser (488 nm), and for propidium iodide, a HeNe G laser (543 nm) was used. The image capture software was Fluoview 500 (Olympus).
The membrane depolarization indicator dye DiSBaC2(3) [bis-(1,3-diethylthiobarbituric acid) trimethine oxonol)] (Invitrogen) was also used to assess membrane damage. Surface-exposed cells were removed as described above and stained with 5 μM DiSBaC2(3) for 30 min at 23°C in the dark. Stained cells were analyzed with a BD FACSCanto II flow cytometer, equipped with a 15-mW, 488-nm argon ion laser (λEx = 535 nm; λEm = 560 nm). Signals were acquired for 2 × 104 cells for each sample. Fluorescence values were recorded using a 585-nm PE-A channel. Data were analyzed using the associated BD FACSDiva software program. Fluorescence values were normalized using the values of the unstained live control.
Assessment of vacuolar damage after exposure to metallic surfaces.
To analyze the effect of dry metallic surfaces on yeast vacuole integrity, cells from stationary-phase cultures were washed in 50 mM sodium citrate buffer (pH 5), containing 2% glucose and loaded with 10 μM the vacuolar marker 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (carboxy-DCFDA) for 30 min at 23°C. Upon uptake into the cell, carboxy-DCFDA is activated by hydrolysis of its acetate ester group via cellular esterase and the dye accumulates in the vacuoles due to their low pH. Vacuole-damaged cells exhibited either a very low, diffuse fluorescence or no fluorescence at all (34). Cells were centrifuged and concentrated 10-fold in PBS. Cells were streaked onto metal coupons and removed after different time periods with TBSBX as described above. Samples were visualized using a confocal microscope with an argon (488-nm) laser and performing Z-series sectioning. Images were captured using the Fluoview 500 (Olympus) software program and analyzed using the software program ImageJ (version 1.42q) (1). Cells were scored as vacuolated if they showed intact fluorescent vacuoles.
Visualization of labile intracellular Cu(I) pools.
Coppersensor-1 {CS1; 8-[N,N-bis(3′,6′-dithiaoctyl)-aminomethyl]-2,6-diethyl-4,4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene} (27, 42) was synthesized and employed to investigate changing intracellular Cu(I) concentrations. CS1 is a membrane-permeable fluorescent dye which, after binding to Cu(I), increases its red fluorescence by 10-fold. The dye binds Cu(I) stably and selectively over other metal cations in aqueous solution. The apparent Kd (dissociation constant) for binding of Cu(I) to CS1 is 3.6 × 10−12 M (42). CS1 is not a ratiometric dye, but higher copper ion concentrations result in increasing red fluorescent signals as long as the concentration of CS1 is higher than that of Cu(I). As such, CS1 can be used to monitor changes in cellular labile copper levels (27). To visualize intracellular labile Cu(I), surface-exposed cells were removed as described above and stained on ice with 2 μM CS1 in the dark for 20 min according to reference 42. After incubation, excess dye was removed by centrifugation and the pellet was resuspended in ice-cold PBS. Cells were kept on ice until they were analyzed under the microscope. Relative copper accumulation within cells was examined under oil immersion (λEx = 543 nm; λEm = 555 to 600 nm) with a confocal fluorescence microscope using the image capture software program Fluoview 500 (Olympus). The laser used was a HeNe G laser (543 nm).
Determination of cytoplasmic and mitochondrial ROS stress.
The fluorescent dyes dihydroethidium (DHE; Sigma-Aldrich) and MitoSOX Red (Invitrogen) were employed to assess reactive oxygen species (ROS), generated in the cytoplasm and mitochondria, respectively, during exposure to metal surfaces. DHE can freely permeate cell membranes and is used to monitor superoxide generation. DHE interacts with superoxide anions to form 2-hydroxyethidium, which fluoresces (λEx = 500 to 530 nm; λEm = 590 to 620 nm) when bound to DNA and is very well retained by the cells (5). MitoSOX Red is an indicator for mitochondrial superoxide. When localized within mitochondria, MitoSOX Red is oxidized by superoxide and the oxidation product becomes highly fluorescent upon binding to nucleic acids (λEx = 510 nm; λEm = 580 nm) (19). Surface challenged cells were removed as described above and incubated in the dark for 15 min with DHE (10 μM for S. cerevisiae and 20 μM for C. albicans) at 30°C or with 5 μM MitoSOX Red (both yeasts) at 23°C. Excess dye was removed by centrifugation, and cell samples were resuspended in PBS. Stained cells were analyzed with a BD FACSCanto II flow cytometer, equipped with a 15-mW, 488-nm argon ion laser. Signals from 2 × 104 cells were acquired for each sample and recorded using a 585-nm band-pass filter. Data were analyzed using the associated BD FACSDiva software program. Fluorescence values were standardized using the fluorescence values of the unstained control.
RESULTS
Candida and Saccharomyces cells exposed to dry copper (alloy) surfaces are rapidly killed.
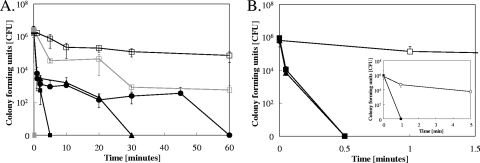
Survival of two yeast species, C. albicans and S. cerevisiae, was tested on different copper alloys and control stainless steel surfaces. The dry exposure method previously developed (12) was employed to mimic dry copper touch surfaces. When exposed to pure metallic copper, 106 stationary-growth-phase cells of Candida were completely killed after 5 min at 23°C (Fig. 1A). Saccharomyces was inactivated within 30 s, 10 times faster than Candida. In contrast, live cells of both species were recovered at the end of the experiment from stainless steel surfaces (Fig. 1A and B). Use of copper alloys such as cupronickel or Nordic Gold resulted in longer times needed, 30 and 60 min, respectively, before all exposed Candida cells were inactivated (Fig. 1A). At 4°C, all Candida cells were inactivated after 60 min on pure copper (Fig. 2A). Under these conditions, Saccharomyces was killed after 1 min on pure copper (Fig. 1B, inset) and after 20 min on cupronickel (Fig. 2B).
FIG. 1.
Yeasts are quickly killed on dry metallic copper surfaces. Cells of C. albicans (A) or S. cerevisiae (B) were exposed at 23°C to stainless steel (□), pure copper (▪), cupronickel (▴), or Nordic Gold (•) (panel A only) for the indicated times, removed, washed, and plated on solidified growth medium. The inset in panel B shows killing of S. cerevisiae on pure copper (▪) or stainless steel (□) at 4°C. Indicated in gray are the responses of exposed exponential-growth-phase cells of C. albicans. Survivors were counted as numbers of CFU after 24 h (Candida) or 48 h (Saccharomyces) of growth at 30°C. Shown are averages of results from triplicate experiments, with standard deviations (error bars).
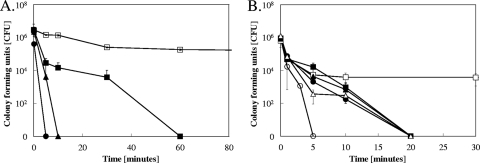
FIG. 2.
Mutations in yeast copper ion homeostasis influence survival time of cells on copper surfaces. Cells of C. albicans (A) or S. cerevisiae (B) were exposed (at 4°C to slow down killing kinetics) to stainless steel (□), pure copper (Candida), or cupronickel (Saccharomyces) (▪, •, and ▴) for the indicated times, removed, washed, and plated on solidified growth medium. Survivors were counted as numbers of CFU after 24 h (Candida) or 48 h (Saccharomyces) of growth at 30°C. Results are shown for wild-type C. albicans (□ and ▪), a Cacrp1Δ/Cacrp1Δ mutant lacking a copper efflux P-type ATPase (▴), or a Cacup1Δ/Cacup1Δ mutant lacking a copper metallothionein (•) (A) and wild-type S. cerevisiae (□ and ▪), an Scctr1Δ mutant lacking a copper uptake permease but harboring the empty control plasmid p416-TEF (▴), an Scctr1Δ mutant complemented with ScCTR1 (•), an Scctr1Δ mutant complemented with hyperactive Scctr1(300) (○), or an Scace1Δ mutant lacking the positive regulator AceI, necessary for metallothionein expression but harboring the empty control plasmid p416-TEF (▵) (B). Shown are averages of results from triplicate experiments, with standard deviations (error bars).
Exponential-growth-phase Candida cells were also tested for their ability to withstand metallic copper surfaces. These cells were much more susceptible to metallic copper than cells grown to stationary phase, with no survivors obtained at time point “0” (Fig. 1A, closed gray squares). In contrast, some of the exponential-phase Candida cells were able to survive up to 60 min on stainless steel control surfaces (Fig. 1A, open gray squares), demonstrating a specific effect of metallic copper in the killing process.
These results suggested rapid antifungal properties of dry copper surfaces, with pure copper being more efficient than alloys. Noteworthy, both Candida and Saccharomyces experienced slow decreases in cell numbers over time on stainless steel control surfaces, probably due to desiccation. However, viable cells were retrieved from stainless steel after 24 h of incubation (data not shown).
Mutant cells impaired in copper homeostasis are more susceptible to copper surface toxicity than wild-type cells.
To test whether genes involved in copper ion homeostasis influenced the survival of yeast cells on dry metallic copper (alloys), killing kinetics were monitored for Candida and Saccharomyces strains mutated in genes implied in copper homeostasis. Due to the fast killing kinetics at 23°C, the following experiments with Saccharomyces were performed at 4°C to slow down reactions and using cupronickel, which has a lower copper content than pure copper. Assays for Candida mutants were performed at 4°C using pure copper.
In contrast to S. cerevisiae, which lacks a copper efflux pump, C. albicans possesses both a cytoplasmic copper metallothionein and a copper-efflux P-type ATPase (40). We reasoned that the lack of these proteins would render Candida more susceptible to the toxicity exerted by copper surfaces, because copper ions were previously found to play a part in contact-mediated killing in bacteria (12, 28). Candida is a diploid organism, and the mutants used are lacking both copies of the respective gene. A null-mutant strain for the C. albicans Cup1 (CaCup1) Cu metallothionein was inactivated after 5 min at 4°C, and a strain deleted in the gene for the P-type ATPase CaCrp1 was killed after 10 min (Fig. 2A). In contrast, Candida wild-type cells were completely inactivated after 60 min.
Similarly, ScCtr1p is the primary copper uptake system for Saccharomyces, and cells lacking ScCtr1p experience copper deprivation (9). A mutant derivative of ScCtr1, the ScCtr1(300) mutant, lacks the last 106 amino acids in the protein's C-terminal domain and is deregulated for copper uptake (41). When tested on cupronickel, the Scctr1Δ mutant and the complemented (Scctr1Δ pScCTR1) mutant did not differ significantly from Saccharomyces wild-type cells in their killing kinetics, in that all strains were completely inactivated within 20 min of exposure (Fig. 2B). In contrast, the Scctr1Δ pScctr1(300) mutant, with a hyperactive copper transporter, was killed four times faster. After 5 min, no live cells were recovered from copper coupons (Fig. 2B). Saccharomyces harbors multiple copies of cup1 metallothionein genes (17). In order to prevent their expression, a strain deleted in their positive regulator ace1 was investigated. The ace1Δ strain was killed as fast as the wild-type or the ctr1Δ deletion strain. No survivors could be recovered after 20 min of exposure, indicating a very limited role for metallothioneins for resistance against copper surface toxicity in Saccharomyces in contrast to what was observed for Candida.
Wild-type cells on stainless steel experienced an initial decline in numbers but remained stable after 5 min at about 5 × 103 cells. Survival of Candida or Saccharomyces mutant cells exposed to stainless steel control surfaces did not significantly differ from that of wild-type cells (data not shown), and thus, only the wild type on stainless steel is indicated (Fig. 2A and B). Taken together, these data suggest that Saccharomyces and Candida cells unable to maintain proper copper ion homeostasis were more prone to copper surface toxicity but did not show altered survival on stainless steel, suggesting a role for copper ions in the contact-mediated killing process.
Copper ions accumulate inside cells exposed to copper surfaces.
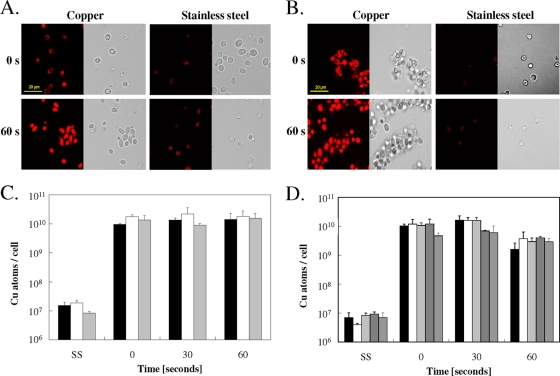
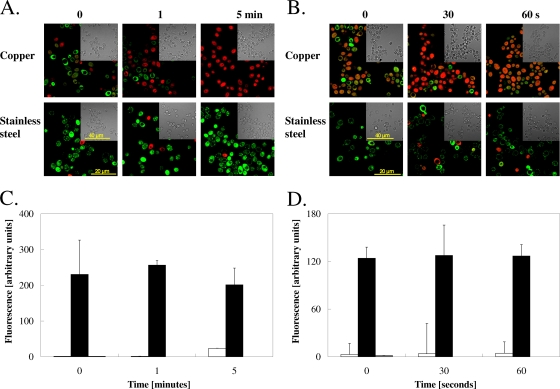
To further investigate the role of copper ions released from copper surfaces for inactivation of yeasts and potential copper ion uptake, we used a copper-specific fluorescent dye, coppersensor-1 (CS1) (42), to monitor Cu(I) ions inside exposed cells. Staining of cells of Candida or Saccharomyces with CS1 immediately (at “0” seconds, directly after the sample had dried up) after contact with copper at 23°C showed increased fluorescence, whereas only weak signals were observed from cells exposed to stainless steel (Fig. 3A and B). After 60 s of exposure, the signal from copper surfaces was very strong, indicating that yeast cells rapidly took up copper ions from metallic copper surfaces.
FIG. 3.
Copper uptake into cells exposed to copper surfaces. Cells of C. albicans (A, C) or S. cerevisiae (B, D) were exposed at 23°C to pure copper or to stainless steel (SS) for the indicated times, removed, washed, stained with the Cu(I)-specific fluorescent dye coppersensor-1, and subjected to fluorescence microscopy (A, B) or were mineralized and subjected to ICP-MS analysis for determination of cellular copper content (C, D). Bars represent wild-type C. albicans (black), a Cacrp1Δ/Cacrp1Δ mutant lacking a copper efflux P-type ATPase (gray), or a Cacup1Δ/Cacup1Δ mutant lacking a copper metallothionein (white) (C) and wild-type S. cerevisiae (black), an Scctr1Δ mutant lacking a copper uptake permease but harboring a control plasmid (white), an Scctr1Δ mutant complemented with ScCTR1 (light gray), an Scctr1Δ mutant complemented with hyperactive Scctr1(300) (dark gray), or an ace1Δ mutant not expressing metallothioneins (medium gray) (D). Shown are fluorescence (left) and representative phase contrast (right) microscopy images (A, B) or averages of results from triplicate experiments, with standard deviations (error bars) (C, D).
The numbers of copper atoms that enter cells exposed to the metal coupons were measured and quantified using ICP-MS analysis from EDTA-washed cells. Candida cells prior to exposure to copper surfaces contained (1.85 ± 0.60) × 106 atoms of copper per cell, but after a brief exposure (“0” seconds, equaling 5 s of exposure to the surface during the drying process), this number increased to (9.51 ± 0.78) × 109 (Fig. 3C). Longer exposure times (results for 60 s are shown in Fig. 3C; results for 5 and 10 min are not shown) did not further increase the cellular copper content. The copper contents of the Cacrp1Δ or Cacup1Δ mutant cells were quite similar to those of the wild-type cells, providing no clues on the benefits of the presence of copper homeostasis systems under these conditions.
Similar to Candida cells, cells of Saccharomyces experienced a large increase in intracellular copper after the shortest possible contact time (“0” seconds) on copper surfaces (Fig. 3D). In contrast to Candida cells, however, cells of Saccharomyces accumulated less copper at 60 s, (1.59 ± 1.04) × 109 atoms/cell, than at “0” seconds, (1.02 ± 0.17) × 1010 atoms/cell (Fig. 3D). This is likely caused by the extremely fast killing kinetics of Saccharomyces on copper surfaces. Cells after 30 s of exposure were lethally damaged (compare Fig. 1B) and failed to grow into colonies when transferred onto solid growth medium. At 30 s, the maximum amount of copper was accumulated in Saccharomyces cells, (1.60 ± 0.66) × 1010 atoms/cell. The amounts of copper accumulated were similar for wild-type cells or cells deleted in ScCTR1 or Sccup1. In contrast, cells producing the hyperactive ScCtr1(300) mutant protein contained slightly less cellular copper ([6.90 ± 0.37] × 109 atoms/cell) than wild-type cells. One explanation for this counterintuitive result is that by 60 s, the previously accumulated copper has leaked from the already dead Scctr1(300) mutant cells since the copper-induced killing was probably caused by structural membrane damage.
Cells of Candida and Saccharomyces exposed for 5 s to stainless steel control surfaces contained only (1.50 ± 0.46) × 107 and (6.90 ± 3.2) × 106 atoms of copper, respectively. These concentrations are higher than those seen in unchallenged cells, (1.85 ± 0.6) × 106 and (3.60 ± 0.85) × 105 atoms/cell, respectively. As a consequence, the copper concentration in copper-exposed cells (Fig. 3C and D) is likely overestimated by a similar factor.
For comparison, we also quantified copper uptake into cells that were acutely challenged with elevated external copper ions in liquid medium. Increasing CuCl2 concentrations from 0.1 to 100 mM for 30 s resulted in about 108 to 1010 copper atoms per Saccharomyces cell as determined by ICP-MS. The same external copper concentrations (0.1 to 100 mM) for Candida increased cellular copper from 107 to 109, indicating that the cellular homeostasis systems of yeasts were overwhelmed under these high-extracellular-copper concentrations. Collectively, these results suggest that copper release and uptake from copper surfaces into yeast cells are fast, and cells were copper saturated within seconds of exposure. This confirms the limited capability of copper homeostasis factors for protecting cells under these conditions.
Exposure to copper surfaces does not lead to genotoxicity in yeast.
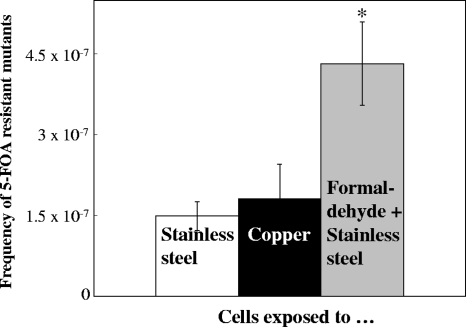
The mode of action and the molecular cellular targets of metallic copper toxicity are currently unknown for any microorganism, but a recent study reported DNA damage in Gram-positive enterococci as a result of contact with copper surfaces (37). Here, we used selection for 5-FOA resistance to investigate the potential mutagenicity of metallic copper for yeasts. Exposure to metallic copper did not increase the mutation rates in Saccharomyces (Fig. 4) compared to the level for a stainless steel control. Approximately the same frequencies of 5-FOA-resistant mutants arose after contact with both metals. There was, however, a significant, about-2-fold increase in the number of mutants when cells on stainless steel surfaces were additionally treated with 0.9% (wt/vol) formaldehyde, a known mutagen. We also observed a 2-fold increase in mutants after 2 h of exposure in liquid cultures to 65 or 130 μM formaldehyde, similar to what was previously reported for this mutagen (7). These observations clearly suggest that metallic copper is not genotoxic to yeasts, making it unlikely that copper surface-exposed cells are killed by the generation and accumulation of lethal mutations in the cells' DNA.
FIG. 4.
Exposure to metallic copper surfaces does not lead to increased mutations in S. cerevisiae. A total of approximately 7 × 107 S. cerevisiae cells were exposed to stainless steel (white column), copper surfaces (black column), or 0.9% (wt/vol) of the mutagen formaldehyde on stainless steel (gray column) surfaces for 5 s, removed, washed, concentrated, and spread onto solid medium containing 1 g × liter−1 of the pyrimidine analogue 5-FOA. After 72 h of incubation at 30°C, colonies were counted as originating from mutation events leading to resistance to 5-FOA. Shown are averages of results from triplicate experiments, with standard deviations (error bars). The asterisk denotes significantly (P ≤ 0.05) different values in formaldehyde-challenged cells.
Cells exposed to dry copper surfaces have damaged membranes.
We reasoned that damage to yeast cells occurs because of the proximity between the copper surface and cellular membranes. Cell membrane damage can be assessed using Live/Dead staining techniques that make use of dyes with different permeability properties. Propidium iodide enters cells and stains the cellular DNA only if the cytoplasmic membranes are damaged. Cells of Candida and Saccharomyces were exposed to copper coupons for “0” seconds to 5 min (Candida) or from “0” to 60 s (Saccharomyces), removed, Live/Dead stained, and examined by fluorescence microscopy. Figure 5A and B show that membranes of both yeast species accumulated substantial damage immediately after contact with metallic copper. Extended exposure times further increased the number of damaged cells. In contrast, much less damage was observed in cells from stainless steel surfaces (Fig. 5A and B), suggesting that the observed damage is copper specific and not related to desiccation.
FIG. 5.
Cells exposed to copper surfaces suffer membrane damage. Cells of C. albicans (A, C) or S. cerevisiae (B, D) exposed to copper or control surfaces or unchallenged cells were removed and stained with propidium iodide and SYTO9 (A, B) as indicators for physical membrane damage and visualized by fluorescence microscopy or stained with DiSBaC2(3), an indicator for membrane depolarization (C, D), and analyzed by flow cytometry. Live yeasts with intact membranes fluoresce green, while those with damaged membranes fluoresce red (A, B). Cells were stained with 5 μM DiSBaC2(3) for 30 min, after which excess dye was removed. Samples were subjected to flow cytometry and fluorescent signals recorded. Fluorescence values were standardized by dividing them by the fluorescence values for unstained live cells. White bars represent cells exposed to stainless steel, black bars cells exposed to copper surfaces, and gray bars unexposed control cells. Shown are representative micrographs (A, B) or averages with standard deviations (error bars) for the median fluorescence values from three independent flow cytometry experiments from individual cultures performed on multiple days (C, D).
We also monitored the response of the cells' membrane potentials upon contact with copper surfaces. The fluorescent dye DiSBaC2(3) is an indicator for membrane depolarization, and depolarized cells exhibit increased fluorescence signals. Candida and Saccharomyces cells from copper surfaces responded immediately and showed a strong signal at the shortest exposure times that could be measured (Fig. 5C and D). During the following 5 min (Candida) or 60 s (Saccharomyces), subsequent fluorescence increases were not observed, suggesting that depolarization is very fast, and longer exposure did not change the degree of depolarization reached at the beginning of the experiment. In contrast, cells from stainless steel were not depolarized and largely exhibited fluorescence signals at the same level as unchallenged control cells. The small increase in depolarization for Candida at 5 min (Fig. 5C) might have occurred because of the onset of desiccation stress in Candida.
Thus, copper surfaces cause membrane damage and depolarization. Potentially, this is a major mode of action leading to cell death in exposed yeast cells because of the extent of the damage observed.
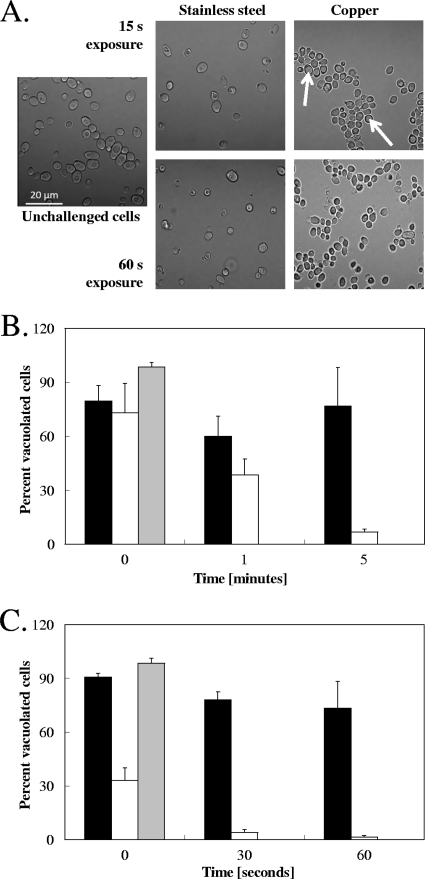
Vacuoles of yeasts exposed to dry copper surfaces are damaged.
Cells on copper surfaces suffer cytoplasmic membrane damage, as indicated by Live/Dead staining, but they also exhibit enlarged cell vacuoles after a brief exposure. The cells became opaque and appeared to lack vacuoles later (compare Fig. 6A and Fig. 3A and B). To quantify this observation, we used the vacuole fluorescent stain carboxy-DCFDA and Z-series sectioning to monitor the presence of vacuoles in cells from copper and stainless steel surfaces (Fig. 6B and C). In both Candida and Saccharomyces, exposure to copper surfaces resulted in a striking decline in the number of vacuolated cells observed, indicating vacuolar damage. After 5 min of exposure to metallic copper, 93% of all cells were nonvacuolated in C. albicans (Fig. 6B), and in S. cerevisiae, almost all cells had lost their vacuoles after 60 s, with only a small fraction (1.5%) retaining their vacuoles (Fig. 6C). Cells on stainless steel also suffered some vacuolar damage over time, but the degree was smaller than on copper in both Candida and Saccharomyces, with losses of about 25% after 5 min (Candida) and 60 s (Saccharomyces) (Fig. 6B and C). Overall, Saccharomyces was more sensitive to copper toxicity than Candida. This difference in onset of copper surface-mediated vacuole damage follows the kinetics of contact-mediated killing on copper, as shown in Fig. 1, which also shows Saccharomyces being more sensitive. Thus, contact-mediated killing immediately affects both cytoplasmic and internal vacuolar membranes.
FIG. 6.
Cells stressed by contact with metallic copper exhibit vacuolar damage. Cells of C. albicans (A, B) or S. cerevisiae (C) exposed to pure copper or stainless steel surfaces were removed, visualized by phase-contrast microscopy (A), or stained with the dye carboxy-DCFDA and visualized by fluorescence microscopy and Z-series sectioning (B, C). Cells were counted and scored as vacuolated if they presented a visible fluorescent vacuole. Black bars represent cells exposed to stainless steel, white bars cells exposed to copper surfaces, and gray bars unexposed cells serving as controls to verify proper loading of the dye. Shown are representative micrographs (white arrows indicate enlarged vacuoles) (A) or averages with standard deviations (error bars) from at least 3 different images from 2 individual experiments using independent cultures (B).
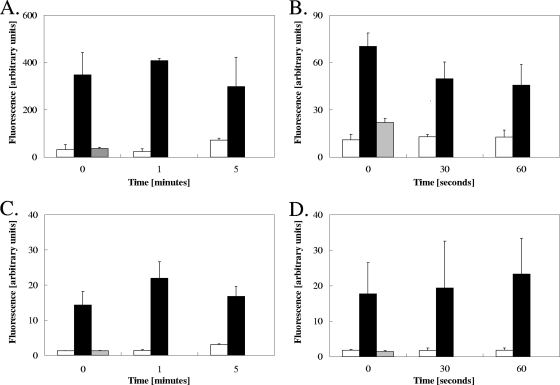
Exposure to metallic copper results in cytoplasmic and mitochondrial ROS stress.
Next, we investigated the degree of ROS stress in exposed cells to further define the copper surface mode of action against yeasts. Dihydroethidium (DHE) is an indicator for cytoplasmic ROS and upon stress increases its fluorescence signal. The response of DHE was very fast and led to strong signals in both Candida and Saccharomyces cells from copper surfaces at the shortest exposure time (Fig. 7A and B). In the more sensitive S. cerevisiae cells, the signals declined by about 35% after 1 min, which is likely related to the onset of cell death (compare Fig. 1) and concomitant leakage of dye from the cell. Fluorescence signals from unchallenged cells (Fig. 7A and B, gray bars) or cells from stainless steel (Fig. 7A and B, white bars) were much lower than those from copper-exposed cells (black bars). Yet, after 5 min, Candida from stainless steel experienced a slight increase in cytoplasmic ROS stress, probably indicating the onset of desiccation stress (Fig. 7A).
FIG. 7.
Exposure to metallic copper surfaces induces oxidative stress in yeast cells. Cells of C. albicans (A, C) or S. cerevisiae (B, D) exposed to copper or stainless steel surfaces were removed, stained with the dye DHE (A, B) or MitoSOX (C, D), and visualized by fluorescence microscopy. White bars represent cells exposed to stainless steel black bars cells exposed to copper surfaces, and gray bars unexposed cells serving as controls to verify proper loading of the dye. Fluorescence values were standardized by dividing them by the fluorescence values for unstained live cells. Shown are averages with standard deviations (error bars) of the median fluorescence values from three independent experiments.
Because yeasts experienced vacuolar damage (Fig. 6), we also tested if other intracellular compartments such as mitochondria were affected by metallic copper. The dye MitoSOX Red monitors the oxidative status in the mitochondrion, with superoxide stress causing a fluorescence signal increase. Similar to stress observed with the use of DHE in the cytoplasm (Fig. 6), mitochondria in cells of Candida and Saccharomyces from copper surfaces were also highly redox stressed, as indicated by rapid and strong fluorescence signals at the earliest time points that could be sampled (Fig. 7C and D). Longer times of exposure on copper did not result in further increases in fluorescence, suggesting that stress became maximum very rapidly after contact. In contrast, fluorescence in unexposed and stainless steel-exposed cells was low, about 10% (Candida) or 7% (Saccharomyces) of the fluorescence value from copper-exposed cells, indicating that stainless steel does not cause mitochondrial oxidative stress except after the onset of desiccation stress at 5 min (Fig. 7C).
DISCUSSION
Copper has been used as an antimicrobial agent for thousands of years, but the mode of action exerted by copper against microbes remains unknown. Today, nosocomial infections are a major health care threat and are responsible every year for tens of thousands of deaths and for billions of dollars spent in additional hospital costs in the United States alone (numbers for Pennsylvania are reported in reference 32). C. albicans has emerged as one of the most important invasive fungal agents for hospital-acquired infections (HAI) and has become more and more resistant against commonly used drugs (33). New tools are needed to support existing hygiene regimens to fight this and other microbial challenges. Recent hospital trials around the globe evaluating the antimicrobial efficacy of metallic copper surfaces have suggested that these materials efficiently kill exposed microbes (6, 24, 26). Reductions of the microbial loads of up to 100% compared to the levels for noncopper control surfaces were reported (6).
Most of the previous studies that validated the antimicrobial proprieties of copper (alloy) surfaces mainly focused on describing the killing kinetics of a variety of microbial species exposed to different alloys (25, 30, 39), without deeper analysis of the mechanisms that are responsible for cell inactivation. While it is becoming clear that metallic copper has excellent antimicrobial properties, the molecular mode of action exerted by copper surfaces and the sensitive cellular targets are still largely unknown.
Two different methodologies have been employed in recent studies to analyze the antimicrobial proprieties of copper surfaces. In the “moist” method, a droplet containing a suspension of microbes was applied to a metal surface: cells remained in a liquid suspension for a prolonged time and were usually inactivated within hours. A “dry” method was developed in our laboratory to better mimic the transfer of bacteria from hands to dry touch surfaces usually found in health care environments. With the use of this “dry” method, cells are directly in contact with the surface within a few seconds of application and the time of killing was greatly reduced compared to that obtained with the “moist” method (12).
Here, we investigated contact-mediated-killing mechanisms in the eukaryotic model organism S. cerevisiae and the nosocomial opportunistic pathogen C. albicans. Initial experiments measuring yeast survival on copper surfaces indicated extremely fast killing kinetics, and no Saccharomyces cells survived the initial brief exposure to metallic copper at room temperature. Closer examination of the surfaces of the copper coupons using cells stained with Calcofluor White and microscopy revealed that more than 20% of the applied cells remained on the copper coupon after the wash step. Extended wash steps including vigorous vortexing did not improve the situation. Cells were finally successfully removed (>99.99%) from the copper coupons, without affecting their viability, using an improved wash buffer containing barium chloride and Triton X-100. This problem of sticky cells did not occur with yeast cells on stainless steel surfaces and is thus most probably a unique feature of metallic copper.
To the best of our knowledge, this is the first study addressing the mechanisms leading to inactivation of yeasts on metallic copper surfaces. Two earlier studies demonstrated that fungi such C. albicans (25) and spores of Candida and the ascomycetes Aspergillus spp. or Fusarium spp. were readily killed on copper surfaces (with the exception of spores of Aspergillus niger) (38). In both of these studies, the “moist” exposure method was employed and the authors concentrated their efforts solely on the killing kinetics. No cellular targets of copper surface toxicity were investigated, and the mode of action was not investigated. The study of spore survival on copper (38) used a Live/Dead staining technique for comparing plating of survivors with direct estimation of stained metabolic active (i.e., live) cells on copper surfaces. However, plate count numbers and fluorescent-cell counts did not match up well. This problem might have been partly caused by internal structural damage in the vacuoles of the cells. This might be a problem that we too have observed in this study, since the dye Fun-1, used by others (38), accumulates in vacuoles.
Previous work has suggested a limited role for copper ion homeostasis factors in bacteria for survival on copper surfaces. Cells of Pseudomonas aeruginosa (10), Enterococcus hirae (28), or E. coli (12) deleted in copper ion defense systems died more rapidly on metallic copper than wild-type cells, but the survival times of mutants were not dramatically different from those of wild-type cells. Here, we demonstrated that similarly, in yeasts, proper copper ion homeostasis can slow down killing kinetics but does not prevent killing. Two open questions, however, remain. First, why were the intracellular copper concentrations in wild-type and mutant cells on copper surfaces virtually the same, when the killing kinetics were clearly different? Why do cells exposed to stainless steel apparently have increased copper contents over time? The latter is unlikely related to desiccation since drying would not change the absolute number within a cell. It might be because there is bioavailable copper on that surface even though the surfaces were extensively cleaned or because some other metal is interfering with the quantification.
Most of the insight into the process of contact-mediated killing on copper stems from work with bacteria, and very recently, two alternative models were suggested for explaining why bacterial cells are inactivated on metallic copper. One model proposes DNA damage as the underlying mechanism (37). Cells plated with the “moist” method experienced large-scale DNA fragmentation when genomic DNA was recovered from long-term-stressed cells. The other model sees the cellular membranes as the prime target of copper surface-mediated stress, with extended membrane damage occurring upon contact with copper surfaces, leading to physical destabilization of the cells, with subsequent lysis (11a). The DNA damage model can be challenged on the basis of several findings. We and others have shown that copper ions released from metallic copper surfaces play a role in contact-mediated killing (10, 12, 28). Copper ions themselves were previously shown not to cause genotoxicity in E. coli and even to protect the DNA when cells were additionally challenged with an oxidant such as hydrogen peroxide (22). A study performed in our laboratory recently indicated that the mutation rate of E. coli cells exposed to copper surfaces was not higher than that of cells on stainless steel, and DNA fragmentation was not observed (11a). More importantly, an organism known to be able to recover from massive DNA fragmentation events, Deinococcus radiodurans (8), was killed as fast as E. coli on metallic copper surfaces (11a), clearly indicating that the DNA damage hypothesis is nonvalid. Probably, the DNA fragmentation observed in reference 37 occurred postmortem, as a secondary phenomenon. In contrast, in our study of yeasts, and elsewhere, using bacteria as models (11a), we showed that membranes suffer damage from metallic copper exposure.
The integrity of the cell wall structure of Candida cells in contact with metallic copper surfaces was also monitored using high-resolution microscopy. Cells always appeared intact, and no major structural damage, such as breakage or leakage, was observed, even after 24 h of exposure (data not shown). However, cell debris was commonly visible in confocal microscopy, when cells removed from copper were investigated (data not shown). Similarly, no structural damage was reported for fungal spores when visualized under epifluorescence microscopy (38). Thus, the overall stability of the yeast cell walls becomes somewhat compromised, but damage becomes evident only if cells are exposed to physical stress during removal from surfaces after exposure.
An advantage of yeast as a model system is that yeast cells not only have cytoplasmic membranes but also have internal membranous compartments such as the vacuole and mitochondria. Unexpectedly, not only the cytoplasmic membrane but also these internal membrane systems exhibited various degrees of stress on copper surfaces. Vacuoles were damaged and disappeared and mitochondria suffered oxidative stress rapidly after contact with the copper surfaces (Fig. 5 to 7).
Identifying these target structures in yeasts now constitutes the first steps for a deeper understanding of the process of contact-mediated killing on copper and sets a framework for future studies. These studies will identify if certain membrane-bound proteins or membrane lipids themselves are the sensitive targets in cells. Damage to these specific targets is expected to lead to the rapid and efficient killing process on dry metallic copper surfaces.
Acknowledgments
This research was supported by a pilot grant from NIH (grant P20 RR-017675, from the National Center for Research Resources). We also acknowledge funds from the International Copper Association (ICA), the Copper Development Association (CDA), and a University of Nebraska—Lincoln Office of Research Faculty Seed grant to G.G. C.E.S. was supported by a Fundação para a Ciência e Tecnologia, Portugal, graduate fellowship. Equipment for this project was purchased with Nebraska Tobacco Settlement Biomedical Research Development Funds and start-up funds from the School of Biological Sciences (University of Nebraska—Lincoln).
We thank You (Joe) Zhou and Terri Fangman (University of Nebraska—Lincoln) for skillful technical assistance with microscopy, Charles A. Kuszynski (University of Nebraska Medical Center, Omaha, NE) for introduction to flow cytometry, and Javier Seravalli (University of Nebraska—Lincoln) for performing ICP-MS analysis. Thanks are due to Daniel Kornitzer (Israel Institute of Technology and the Rappaport Institute, Haifa, Israel) and Audrey L. Atkin and Jaekwon Lee (University of Nebraska—Lincoln) for the generous gifts of strains and Kenneth Nickerson (University of Nebraska—Lincoln) for helpful suggestions and critical reading of the manuscript.
The contents of this article are solely the responsibility of the authors and do not represent the official views of the NIH.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Adle, D. J., D. Sinani, H. Kim, and J. Lee. 2007. A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J. Biol. Chem. 282:947-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., et al. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Bucana, C., I. Saiki, and R. Nayar. 1986. Uptake and accumulation of the vital dye hydroethidine in neoplastic cells. J. Histochem. Cytochem. 34:1109-1115. [DOI] [PubMed] [Google Scholar]
- 6.Casey, A. L., et al. 2010. Role of copper in reducing hospital environment contamination. J. Hosp. Infect. 74:72-77. [DOI] [PubMed] [Google Scholar]
- 7.Chanet, R., and R. C. von Borstel. 1979. Genetic effects of formaldehyde in yeast. III. Nuclear and cytoplasmic mutagenic effects. Mutat. Res. 62:239-253. [DOI] [PubMed] [Google Scholar]
- 8.Cox, M. M., J. L. Keck, and J. R. Battista. 2010. Rising from the ashes: DNA repair in Deinococcus radiodurans. PLoS Genet. 6:e1000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dancis, A., D. Haile, D. S. Yuan, and R. D. Klausner. 1994. The Saccharomyces cerevisiae copper transport protein (CTR1p)—biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269:25660-25667. [PubMed] [Google Scholar]
- 10.Elguindi, J., J. Wagner, and C. Rensing. 2009. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J. Appl. Microbiol. 106:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espirito Santo, C., P. V. Morais, and G. Grass. 2010. Isolation and characterization of bacteria resistant to metallic copper surfaces. Appl. Environ. Microbiol. 76:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Espirito Santo, C., E. W. Lam, C. G. Elowsky, D. Quaranta, D. W. Domaille, C. J. Chang, and G. Grass. Bacterial killing by dry metallic copper surfaces. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 12.Espirito Santo, C., N. Taudte, D. H. Nies, and G. Grass. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl. Environ. Microbiol. 74:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillum, A. M., E. Y. Tsay, and D. R. Kirsch. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198:179-182. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell, B., and J. M. Gutteridge. 1984. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halliwell, B., and J. M. Gutteridge. 1990. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 186:1-85. [DOI] [PubMed] [Google Scholar]
- 17.Huibregtse, J. M., D. R. Engelke, and D. J. Thiele. 1989. Copper-induced binding of cellular factors to yeast metallothionein upstream activation sequences. Proc. Natl. Acad. Sci. U. S. A. 86:65-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imlay, J. A., and S. Linn. 1988. DNA damage and oxygen radical toxicity. Science 240:1302-1309. [DOI] [PubMed] [Google Scholar]
- 19.Johnson-Cadwell, L. I., M. B. Jekabsons, A. Wang, B. M. Polster, and D. G. Nicholls. 2007. ‘Mild uncoupling’ does not decrease mitochondrial superoxide levels in cultured cerebellar granule neurons but decreases spare respiratory capacity and increases toxicity to glutamate and oxidative stress. J. Neurochem. 101:1619-1631. [DOI] [PubMed] [Google Scholar]
- 20.Kachur, A. V., C. J. Koch, and J. E. Biaglow. 1998. Mechanism of copper-catalyzed oxidation of glutathione. Free Radic. Res. 28:259-269. [DOI] [PubMed] [Google Scholar]
- 21.Macomber, L., and J. A. Imlay. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macomber, L., C. Rensing, and J. A. Imlay. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda, H., and N. Ishida. 1967. Specificity of binding of hexopyranosyl polysaccharides with fluorescent brightener. J. Biochem. 62:276-278. [DOI] [PubMed] [Google Scholar]
- 24.Marais, F., S. Mehtar, and L. Chalkley. 2010. Antimicrobial efficacy of copper touch surfaces in reducing environmental bioburden in a South African community healthcare facility. J. Hosp. Infect. 74:80-82. [DOI] [PubMed] [Google Scholar]
- 25.Mehtar, S., I. Wiid, and S. D. Todorov. 2008. The antimicrobial activity of copper and copper alloys against nosocomial pathogens and Mycobacterium tuberculosis isolated from healthcare facilities in the Western Cape: an in-vitro study. J. Hosp. Infect. 68:45-51. [DOI] [PubMed] [Google Scholar]
- 26.Mikolay, A., et al. 2010. Survival of bacteria on metallic copper surfaces in a hospital trial. Appl. Microbiol. Biotechnol. 87:1875-1879. [DOI] [PubMed] [Google Scholar]
- 27.Miller, E. W., L. Zeng, D. W. Domaille, and C. J. Chang. 2006. Preparation and use of Coppersensor-1, a synthetic fluorophore for live-cell copper imaging. Nat. Protoc. 1:824-827. [DOI] [PubMed] [Google Scholar]
- 28.Molteni, C., H. K. Abicht, and M. Solioz. 2010. Killing of bacteria by copper surfaces involves dissolved copper. Appl. Environ. Microbiol. 76:4099-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortimer, R. K., and J. R. Johnston. 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics 113:35-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Infect. 63:289-297. [DOI] [PubMed] [Google Scholar]
- 31.Outten, F. W., D. L. Huffman, J. A. Hale, and T. V. O'Halloran. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J. Biol. Chem. 276:30670-30677. [DOI] [PubMed] [Google Scholar]
- 32.Pennsylvania Health Care Cost Containment Council (PHC4). 2008. Hospital-acquired infections in Pennsylvania: calendar year 2006. http://www.phc4.org/reports/hai/06/docs/hai2006report.pdf.
- 33.Rex, J. H., M. G. Rinaldi, and M. A. Pfaller. 1995. Resistance of Candida species to fluconazole. Antimicrob. Agents Chemother. 39:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruissen, A. L., et al. 2001. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 356:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadtman, E. R. 1992. Protein oxidation and aging. Science 257:1220-1224. [DOI] [PubMed] [Google Scholar]
- 36.Tkeshelashvili, L. K., T. McBride, K. Spence, and L. A. Loeb. 1991. Mutation spectrum of copper-induced DNA damage. J. Biol. Chem. 266:6401-6406. [PubMed] [Google Scholar]
- 37.Warnes, S. L., S. M. Green, H. T. Michels, and C. W. Keevil. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl. Environ. Microbiol. 76:5390-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weaver, L., H. T. Michels, and C. W. Keevil. 2010. Potential for preventing spread of fungi in air-conditioning systems constructed using copper instead of aluminium. Lett. Appl. Microbiol. 50:18-23. [DOI] [PubMed] [Google Scholar]
- 39.Weaver, L., H. T. Michels, and C. W. Keevil. 2008. Survival of Clostridium difficile on copper and steel: Futuristic options for hospital hygiene. J. Hosp. Infect. 68:145-151. [DOI] [PubMed] [Google Scholar]
- 40.Weissman, Z., I. Berdicevsky, B. Z. Cavari, and D. Kornitzer. 2000. The high copper tolerance of Candida albicans is mediated by a P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 97:3520-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, X., D. Sinani, H. Kim, and J. Lee. 2009. Copper transport activity of yeast Ctr1 is down-regulated via its C terminus in response to excess copper. J. Biol. Chem. 284:4112-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng, L., E. W. Miller, A. Pralle, E. Y. Isacoff, and C. J. Chang. 2006. A selective turn-on fluorescent sensor for imaging copper in living cells. J. Am. Chem. Soc. 128:10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]