Abstract
The isolation and results of genomic and functional analyses of Rhodococcus equi phages ReqiPepy6, ReqiDocB7, ReqiPine5, and ReqiPoco6 (hereafter referred to as Pepy6, DocB7, Pine5, and Poco6, respectively) are reported. Two phages, Pepy6 and Poco6, more than 75% identical, exhibited genome organization and protein sequence likeness to Lactococcus lactis phage 1706 and clostridial prophage elements. An unusually high fraction, 27%, of Pepy6 and Poco6 proteins were predicted to possess at least one transmembrane domain, a value much higher than the average of 8.5% transmembrane domain-containing proteins determined from a data set of 36,324 phage protein entries. Genome organization and protein sequence comparisons place phage Pine5 as the first nonmycobacteriophage member of the large Rosebush cluster. DocB7, which had the broadest host range among the four isolates, was not closely related to any phage or prophage in the database, and only 23 of 105 predicted encoded proteins could be assigned a functional annotation. Because of the relationship of Rhodococcus to Mycobacterium, it was anticipated that these phages should exhibit some of the features characteristic of mycobacteriophages. Traits that were identified as shared by the Rhodococcus phages and mycobacteriophages include the prevalent long-tailed morphology and the presence of genes encoding LysB-like mycolate-hydrolyzing lysis proteins. Application of DocB7 lysates to soils amended with a host strain of R. equi reduced recoverable bacterial CFU, suggesting that phage may be useful in limiting R. equi load in the environment while foals are susceptible to infection.
Although the “tailed phage,” or members of the virus order Caudovirales, constitute the majority of DNA diversity on the planet, the genomes of only a few hundred are known. Moreover, the genomic data that are available for bacteriophages are extremely unevenly distributed in terms of host species, with two-thirds derived from phages of only eight host genera or closely related bacterial species, including those belonging to the Staphylococcus, Mycobacterium, Pseudomonas, Lactococcus, and Burkholderia genera and the Enterobacteriaceae family (28). Nevertheless, some important themes have emerged from analyzing multiple phages that infect closely related host genera. One theme emerging is that despite rampant lateral gene transfer, there are recognizable “phage types” spanning geography and host taxa. Casjens (11a) was able to group 73 tailed-enterophage genomes into 13 phage types, the members of which were more closely related to each other by a number of criteria than they were to any member of another type. It should be noted that the classic enterophages do not include representatives of all phage types. When groups of phages from other host clades are sequenced, new phages and new phage types are identified. For example, among the sequenced phages of Burkholderia, there are members of the established λ, Mu, and P2 temperate phage types, as well as entirely new virulent phage types, such as Bcep781 and Bcep22 (42, 43, 67-69). Conversely, there are “host-specific” phage characteristics and genes that a phage is more likely to share with other phages that infect the same host, regardless of phage type. Ultimately, though, there do not appear to be phage type or host range restrictions on recombination between different phages, although rates of recombination are affected by both considerations. This has led phage genomes to be profoundly mosaic, a characteristic that confounds attempts at establishing traditional evolutionary relationships between phages. However, mosaicism can also be a valuable tool in functional analysis in that patterns of mosaic exchange tend to identify functional groups of cosegregating genes or interacting protein domains. Finally, although several thousand prophage elements have been identified in bacterial and archaeal genomes, the usefulness of these sequences is limited because only a few have been shown to be functional viruses. Because a prophage element may not be functional, the assumption cannot be made that the genes carried on a prophage element encode intact, functional proteins. This last consideration is particularly relevant when using sequence comparisons to make predictions about protein functions.
This perspective has led us to generate genomic data for new phages of bacterial genera currently underrepresented in the phage database. The approach of sequencing multiple genomes of phages selected for propagation on a particular host not only identifies new phage types and establishes sequences of many genes known to be functional but also contributes directly to the genomics of the host bacterium. Phages are ultimately part of the bacterial “pan-genome” (74, 75). Identifying sequences within a functional phage allows for the unambiguous categorization of homologous bacterial open reading frames that encode novel, hypothetical proteins of unknown function termed phage-associated proteins (16, 62).
A bacterial genus for which phage genomic data is currently lacking is Rhodococcus, a member of the class Actinobacteria. Most Rhodococcus species identified are versatile soil saprophytes capable of catabolizing chemically diverse products of plant secondary metabolism, including high-profile xenobiotics, such as dioxins, polychlorinated biphenyls (PCBs), and long-chain n-alkanes found in soils contaminated with crude oil (4, 18, 19, 26, 27, 40, 41). The genome of Rhodococcus RH1 is >60% GC and spans ∼9.7 Mbp, making it one of the largest in the microbial world (47). Phages of Rhodococcus have been propagated, although no genomic information is available. These include prophage induced from human and equine clinical Rhodococcus isolates (32, 52). Phages against Rhodococcus and other filamentous bacteria were also isolated from activated sludge samples taken from wastewater treatment plants (76). Many of the phages have particularly broad host ranges, covering several genera (76). To date, all Rhodococcus phages identified are of the Siphoviridae morphology (1). The prevalence of siphophages among phages of Actinobacteria, including 61 of the 70 phages identified for Mycobacterium (30), suggests that a flexible tail may be advantageous in penetrating the thick, mycolic acid-containing capsule characteristic of the mycolata (1). Beyond morphology and host range, however, little is known about the biology and diversity of Rhodococcus phages.
One rhodococcal species, R. equi, poses an important challenge to the equine industry, causing severe pneumonia in foals (14, 33, 48, 57). Locations where Rhodococcus infections are prevalent frequently contain high levels of Rhodococcus in the environment (13, 15, 49, 72). There is an age-dependent susceptibility of horses to R. equi (48, 57). The discrete window during which exposure results in clinical manifestation of illness in foals suggests that the R. equi infection occurs in the first few weeks of life (13, 14, 34). Therefore, it is possible that controlling environmental exposure to R. equi using phage during this brief window of time might be an effective and practical means for reducing the incidence of this disease. Thus, isolation and genomic characterization of phages against R. equi would not only be instructive in terms of phage and bacterial evolution but also could provide tools for phage-based prophylaxis or therapeutics. Here, we report the isolation and results of genomic analysis of four new phages of R. equi from soil samples as well as report on experiments designed to test whether exogenously applied phage can reduce bacterial loads in a soil matrix.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Rhodococcus strains used were provided by the Equine Infectious Disease Laboratory, Texas A&M University. The strains included soil and clinical isolates, as well as ATCC strains (see Table 2). Soil isolates include MillB, HDP5B, and HDP1C (13, 25). A total of 29 clinical isolates collected from various sources, including trans-tracheal wash, lung, lymph node, and the feces of infected foals, were used in this study (S1, S2, S3, L1, L1p-, 98-099, 99-110, 99-158, 99-171, 99-180, 99-228, 99-232, 99-120, 99-134, 00-051, 00-050, 01-116, 02-125, 04-172, 04-181, 04-195, 04-200, 04-239, 05-300, 05-305, 05-306, 05-338, 05-373, and 06-383). Additionally, phage host range was tested on ATCC strains 33701, a Rhodococcus equi type strain containing the virulence-associated plasmid, and plasmid-cured strains 33701-, 33703, 33704, and 33705 (58, 72). R. equi was cultured in brain heart infusion (BHI) broth and agar plates at 30°C or 37°C.
Phage isolation.
Rhodococcus equi phages were isolated from soil samples using an enrichment technique. Soil samples (10 g) were combined with 50 ml of tryptone nutrient broth (0.5% tryptone, 0.25% yeast extract, 0.01% glucose, 0.85% NaCl) and 0.5 ml chloroform and incubated with gentle shaking for 4 h at 22°C. Solid material was removed by centrifugation at 10,000 × g, passing the supernatant through miracloth (EMD Chemicals), and filtration through a sterile 0.22-μm filter unit to produce a sterile rinsate. Early-logarithmic-phase host cultures (0.1 ml for a 50-ml enrichment) were mixed with 25 ml of the rinsate and 25 ml of fresh BHI and incubated overnight at 30°C with shaking at 200 rpm. The enrichment cultures were treated with 0.1% chloroform, and bacterial debris was cleared by centrifugation at 10,000 × g and filtration through a sterile 0.22-μm filter unit. The presence of phage was determined by spotting dilution series onto agar lawns of the enrichment host. Lawns were prepared by mixing 100 μl of bacterial suspension (prepared by adjusting a fresh overnight culture to an A600 of 1) with 5 ml of soft agar (0.35% agar prepared in 1% tryptone, 0.5% NaCl, 3 mM MgCl2, 3 mM CaCl2, and 0.04% [wt/vol] glucose), and spreading on BHI agar plates. Clonal phage stocks were prepared following plaque purification. For this, samples were plated at a dilution that produced well-separated plaques, and individual plaques were excised and resuspended in 0.5-ml portions of phage buffer (10 mM Tris [pH 7.6], 5 mM MgSO4·7H2O, 0.01% gelatin) to make individual “pickates.” This process was repeated at least twice to produce clonally pure phage stocks. Finally, high-titer lysates (109 to 1010 PFU/ml) were made from nearly confluent plate lysates by standard methods. The phage titer was determined by mixing 100 μl of host cells (A600 of 1) and 100 μl of each phage dilution with 55°C melted top agar (0.1% agar prepared in BHI) and pouring the mixture over BHI plates. Plaques were scored following 24- to 48-h incubations.
Host range determination.
The host range of each phage was analyzed by spotting 10 μl of each phage lysate, diluted to a predetermined routine test dilution (RTD) concentration, onto overlays of each host. The RTD of each phage lysate was defined as the last 10-fold dilution that developed confluent clearing when spotted onto a lawn of the original enrichment host. The plates were incubated at 30°C for 24 h before they were scored for clearing.
TEM.
Transmission electron microscopy (TEM) of virions was performed by diluting lysates 1:1 with phage buffer and applying 5 μl onto a freshly glow-discharged, Formvar carbon-coated grid for 1 min. The grids were then washed briefly with deionized water drops and stained with 2% (wt/vol) aqueous uranyl acetate. Specimens were observed on a JEOL 1200EX transmission electron microscope operating at an acceleration voltage of 100 kV. Images were recorded at calibrated magnifications on Kodak 4489 film.
Phage DNA isolation and genome sequencing.
Phage DNA was isolated from high-titer lysates as described previously (64). The size of the genome was estimated by pulsed-field gel electrophoresis using a CHEF Mapper apparatus (Bio-Rad). The genomic sequences were determined using a combination of 454 pyrosequencing, random sequencing from a shotgun clone library, and primer walking. Genomic DNAs from phages ReqiPoco6 (Poco6), ReqiDocB7 (DocB7), ReqiPine5 (Pine5), and ReqiPepy6 (Pepy6), along with genomic DNAs from 12 additional phages, were combined at equal molar levels and sequenced by pyrosequencing (454 Life Technologies). Additionally, genomic DNA libraries were prepared in the pSmart-LCKan vector (Lucigen), and plasmids were prepared and sequenced to low coverage (64). The integrity of the assemblies was confirmed by long-range PCR (DyNAzyme EXT; New England Biolabs) using primers staggered approximately 9 kb from each other (data not shown). Taken together, these sequencing efforts resulted in 13-fold coverage of Pepy6 and Pine5, 29-fold coverage of Poco6, and 23-fold coverage of DocB7. The genomic terminal structure and end sequences were determined using a previously described strategy (64). The primers used to determine the 3′ cos overhangs of Pepy6 and Poco6 were as follows: for Pepy6, primers Pepy6CM.ENDA (5′-GGAATTGAAACACGGCATCT) and Pepy6CM.ENDB (TTTGAAGCTCGTCTGTTCTTGA); for Poco6, primers PocoKL.NN (5′-ACCAGAGATGACCCAGTTGC) and PocoKL.MM (5′-CACACAGTGGGAACAGTGGT). The circular genetic maps of DocB7 and Pine5 were confirmed by sequencing the PCR product generated by amplification of genomic DNA with primers extending out from either end of the assembly.
Genome annotation.
Sequence assembly and analyses were performed essentially as described previously (66, 68, 69). Sequencher (Gene Codes Corporation) was used for sequence assembly and editing. Protein-coding regions were predicted using Genemark (http://opal.biology.gatech.edu/GeneMark/gmhmm2_prok.cgi) and manually edited in Artemis (http://www.sanger.ac.uk/resources/software/artemis/) using the phage genome annotation tool ArtAnnoPipe (http://athena.bioc.uvic.ca/node/541) (8, 60). Dot plots were generated using JDotter (9). Predicted proteins were compared to proteins in the GenBank database using BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Transmembrane domains were identified with TMHMM (http://www.cbs.dtu.dk/services/TMHMM/). Conserved domains in proteins were identified through searches with InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) and the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (46).
Phage efficacy experiments. (i) Preparation of soil substrate.
Soil samples were taken from the top 3 in. in a wooded area in College Station, Texas. These samples were passed through a 2-cm sieve and dried for 24 h at 105°C. The soil matrix was a sandy loam (64% sand, 25% silt, 11% clay, 0% inorganic carbon, 2.7% organic carbon) (pH 5.8). Analysis of soil composition was kindly provided by Christine Morgan (Department of Soil and Crops, Texas A&M University). To sterilize soil and eradicate spores, the moisture level of the soil was first adjusted to 20% (0.2 ml water per gram [dry weight] of soil), and the soil was spread in a thin layer (1 to 2 cm thick) and autoclaved with wet heat (121°C, 1 h). The autoclaved soil was then incubated at 30°C for 1 to 2 days. After incubation, the soil was reautoclaved and dried for 2 days at room temperature.
(ii) R. equi and phage application in soil.
Phage efficacy experiments were conducted with 20 g of sterile soil aliquoted into deep petri dishes (20 mm deep; 100-mm diameter). Two independent experiments were performed, and each experiment contained 7 treatment groups. In each treatment group, 3 subsamples (3 soil matrixes contained in 3 separate petri dishes subjected to R. equi inoculation and further treatment) were set up. All plates received 3 ml of an R. equi ATCC 33701 suspension which was prepared by adjusting an overnight culture to an A600 of 1 and further diluting the culture 1,000-fold in phosphate-buffered saline (PBS). The R. equi suspension was applied to the soil matrix using a sterile spray bottle, thoroughly mixed using a sterile spatula, and subjected to a 2.5-h preincubation at 30°C. One group of three plates was used to determine the initial bacterial CFU measurements. For the remaining 6 groups of 3 plates, 3 ml of either phage DocB7 suspensions (105, 106, 107, 108, or 109 PFU/ml) or phage buffer (negative control) were sprayed on the soil surface. The petri dishes were placed in a sealed plastic container along with a layer of moist paper towels (to maintain humidity) and incubated at 30°C for 48 h. To determine the number of R. equi CFU for each treatment, the contents of each petri dish were analyzed independently by transferring the petri dish contents to a 50-ml sterile screw-cap tube containing 20 ml of sterile PBS and mixing the contents of each tube. The soil samples were pelleted by centrifugation for 10 min at 8,000 × g, and the supernatant was removed and replaced with an equal volume of fresh PBS. After thorough mixing, dilutions were plated in duplicate on BHI agar and incubated for 48 h at 37°C.
To determine the recovery of phage from soil samples, two independent experiments were set up in which phage DocB7 was applied to 20 g of sterile soil to a final concentration of 1.5 × 106 PFU/g. The contents of each petri dish were analyzed independently by transferring the petri dish contents to a sterile, 50-ml screw-cap tube, and the soil samples were resuspended in 20 ml of phage buffer. After the soil samples were pelleted (10 min at 10,000 × g), the supernatants were filter sterilized, and the titer of recovered phage was determined in duplicate.
Nucleotide sequence accession numbers.
The genome sequences of Rhodococcus equi phages were deposited in GenBank under accession numbers GU580940 (ReqiDocB7), GU580943 (ReqiPine5), GU580941 (ReqiPepy6), and GU580942 (ReqiPoco6).
RESULTS
Isolation of novel R. equi phages.
Soil samples were obtained from various locations around equine breeding farms where R. equi infections of foals were known to occur. Four new phages, capable of forming plaques on overlays of R. equi, were isolated from soil samples using an enrichment method. Phages ReqiPoco6 (Poco6) and ReqiDocB7 (DocB7) were isolated using R. equi host strains that had originated from the same environmental samples (MillB and HDP1C, respectively). Phages ReqiPine5 (Pine5) and ReqiPepy6 (Pepy6) were isolated using R. equi host strains 05-305 and 05-306, respectively, which were initially cultured from tracheobronchial aspirate fluid samples from pneumonic foals.
Transmission electron microscopy revealed that Poco6, DocB7, Pine5, and Pepy6 had characteristic siphophage morphologies, with icosahedral heads (average diameters of 82 nm, 75 nm, 66 nm, and 80 nm, respectively) and long flexible, noncontractile tails (281 nm, 489 nm, 236 nm, and 285 nm, respectively) (Fig. 1 and Table 1). The International Committee on Taxonomy of Viruses (ICTV) taxonomic prefix of these phages would be vB_ReqS (39). The host range of these phages was assessed by spotting dilutions of lysates onto lawns of 37 R. equi strains, including clinical and environmental isolates collected from different epidemiological regions across the world (Table 2). These hosts include representative VapA+ and VapA− strains (73). Each of the phages was able to generate clearing spots on multiple hosts, including representatives from clinical strains. DocB7 exhibited the broadest host range, forming clearing spots on 35 isolates. Pine5 showed clearing on only 3 isolates, while Poco6 and Pepy6 showed clearing on 8 and 19 isolates, respectively. There was not a correlation between phage sensitivity and the presence or absence of the virulence plasmid.
FIG. 1.
Negative-stained images of new rhodococcal siphophages. ReqiDocB7 (A), ReqiPine5 (B), ReqiPoco6 (C), and ReqiPepy6 (D) negatively stained with 2% (wt/vol) aqueous uranyl acetate. Bars, 200 nm.
TABLE 1.
Rhodococcus equi phage characteristics and features
| Phage | Virion head diam (nm) | Tail length (nm) | TMPa |
Genome length (bp)b | Genomic terminic | No. of CDSd | No. of CDS with TMD (%)e | Pre-tmp frameshift/slip. seq.f | tRNA | Notable feature(s) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | % α-helix | Pred. tail length (nm) | ||||||||||
| ReqiDocB7 | 75 | 489 | 3,101 | 58 | 465 | 75,772 | pac | 105 | 9 | Yes (GGAAAAA) | No | Unique; broad host range |
| ReqiPepy6 | 80 | 285 | 1,950 | 67 | 292 | 76,806 | 3′ cos | 107 | 29 (29) | No | Yes | Phage 1706 like; many TMD proteins |
| ReqiPoco6 | 82 | 281 | 1,951 | 72 | 293 | 78,064 | 3′ cos | 107 | 26 (24) | No | Yes | Phage 1706 like; many TMD proteins |
| ReqiPine5 | 66 | 236 | 1,732 | 68 | 259 | 59,231 | pac | 85 | 13 (15) | No | No | Rosebush like |
TMP, tape measure protein. The measured length (in amino acids [aa]), % alpha-helical content (% α-helix), and predicted tail length (Pred. tail length) (in nanometers) of the tape measure protein is shown.
Genome length, genome nucleotide base pair length.
The structure of the packaged genomic termini is shown. pac, headful packaging; cos, cohesive ends.
The number of protein-coding genes (CDS) is shown.
The number and percentage of protein-coding genes with predicted transmembrane domains (CDS with TMD) is shown.
The presence or absence of a pre-tape measure protein ribosomal frameshift slippery sequence (Pre-tmp frameshift/slip. seq.) is shown.
TABLE 2.
Host ranges of phages Poco6, DocB7, Pine5, and Pepy6a
| Host strain or parameter | Host range of phageb: |
|||
|---|---|---|---|---|
| Poco6 | DocB7 | Pine5 | Pepy6 | |
| ATCC 33701 | X | X | X | |
| ATCC 33701- | X | X | ||
| ATCC 33704 | X | |||
| MillB | X | X | ||
| HDP5B | X | |||
| HDP1C | X | X | ||
| S1 | X | X | ||
| S2 | X | X | X | |
| S3 | X | |||
| L1 | X | |||
| L1p- | X | X | ||
| ATCC 33703 | X | X | ||
| ATCC 33705 | X | |||
| 98-099 | X | X | ||
| 99-110 | X | X | X | |
| 99-171 | X | X | X | |
| 99-180 | X | X | ||
| 99-228 | X | X | X | |
| 99-232 | X | X | ||
| 99-120 | X | |||
| 99-134 | X | |||
| 99-158 | ||||
| 00-051 | X | |||
| 00-050 | X | X | X | |
| 01-116 | X | X | X | |
| 02-125 | X | X | X | |
| 04-172 | X | |||
| 04-181 | X | |||
| 04-195 | X | X | ||
| 04-200 | X | |||
| 04-239 | X | |||
| 05-300 | X | X | ||
| 05-305 | X | X | ||
| 05-306 | X | X | ||
| 05-338 | X | |||
| 05-373 | X | |||
| 06-383 | X | |||
| Total no. of strains with clearing | 8 | 35 | 3 | 19 |
The host ranges of phages Poco6, DocB7, Pine5, and Pepy6 were determined by spotting a routine test dilution onto overlays of the indicated host strains.
X indicates clearing on the indicated host strain. A boldface letter X corresponds to the original enrichment host for that phage.
Genome organization of ReqiPepy6 and ReqiPoco6 and relationship to phage 1706.
The genomic sequences of Pepy6 (76,806 bp) and Poco6 (78,064 bp) were closely related and colinear with isolated blocks of nonidentity (Fig. 2). The majority of the Pepy6 and Poco6 genomes could be aligned, with multiple segments larger than 5 kb with up to 92% identity at the DNA level. Pepy6 and Poco6 were predicted to encode 107 proteins and a single tRNA of unknown specificity, all on the same coding strand (Fig. 3). Both phages possessed 3′ extended cos ends, with the sequence 5′-CGCCGCCCT. This is related to the 3′ extended cos termini (GCCCTGTCT [variant bases underlined]) of Lactococcus phage 1706, whose terminase large subunit (TerL) (LaP1706_gp8) was 60% identical to the predicted TerL subunits of Pepy6 and Poco6 (gp14 and gp16, respectively). The Pepy6 and Poco6 cos sites were located approximately 13 kb upstream of TerL (Fig. 3).
FIG. 2.
Dot plot alignment of the nucleotide sequences of Pepy6 and Poco6. Pepy6 and Poco6 are closely related, colinear phages with isolated blocks of nonidentity. The percentages of identity of the longer aligned blocks are indicated.
FIG. 3.
Genome maps of Pepy6 and Poco6 and related phages and prophages. (A) Genome maps of phages ReqiPepy6 and ReqiPoco6. Purple shading between the genome maps indicates segments of DNA that align with greater than 75% identity. The colors of the genes or proteins in the gene maps indicate the relationship of the encoded protein with other proteins as follows. Yellow indicates a homologue is encoded by phage 1706. Yellow genes with green outlines encode homologues of phage 1706 virion-associated proteins. The 1706 gene is indicated below the Rhodococcus phage gene. Turquoise indicates that there was no homologue in 1706, nor was a functional annotation possible. Dark blue indicates proteins for which a functional annotation could be made but lacked homologues in 1706. Green proteins possess 56 invariant, N-terminal amino acid residues, indicated by peach bars. Genes outlined in red encode proteins with predicted transmembrane helices. Regions shaded in peach indicate locations of gene insertions relative to phage 1706. (B) Genome maps of Lactococcus lactis phage 1706 and prophage elements BRYFOR and CLOSS21, present in the genomic sequences of Bryantella formatexigens DSM 14469 and Clostridium sp. strain SS2/1 genome, respectively. The phage 1706 genome map was derived from the GenBank entry with accession no. EU081845. The BRYFOR prophage encompasses the region encoding locus tags BRYFOR_08504 to BRYFOR_08569. The CLOSS21 prophage encompasses the region encoding locus tags CLOSS21_01492 to CLOSS21_01580. In order to align with phage 1706, the BRYFOR and CLOSS21 maps are drawn with the order of prophages changed. The locations of the prophage genomic termini, between the coding region of BRYFOR gene73 and gene1 and CLOSS21 gene89 and gene1, are indicated on the genome maps by turquoise shading. The colors of the genes or proteins indicate the relationship of the encoded protein with other proteins as follows: yellow, homologue encoded by Pepy6 and Poco6; gray, no homologue encoded by Pepy6 or Poco6. A green outline indicates that the protein encoded by the gene is a homologue of phage 1706 virion-associated protein. A red outline indicates that the protein encoded by the gene has predicted transmembrane helices. The gene number of the corresponding protein encoded by Pepy6 and 1706 is indicated below each map (-, no protein). Regions shaded in peach indicate locations of gene insertions in the map relative to phage 1706. Select functional annotations are shown above genes by the following abbreviations: e15, Salmonella phage epsilon15 tail fiber protein; lip, lipase; hol, holin; tmp, tape measure protein; Prim, primase; pol, DNA polymerase; HNH, homing endonuclease; Int, integrase; endo, endonuclease; ligA, ligase A domain; CMP deaminase, deoxycytidinylate deaminase. Homologous gene products in Pepy6 and 1706 (Pepy6yp and 1706yp, respectively) are indicated below each map.
Based on the total number and amino acid similarity of shared proteins, the phage in the public database identified as being most closely related to Pepy6 and Poco6 was Lactococcus lactis phage 1706, a virulent siphophage initially isolated from an industrial cheese production site (Fig. 3 and Table 3) (17, 20). Homologues of 23 proteins encoded by genes carried on phages Pepy6 and Poco6 could be identified among the 76 proteins encoded by phage 1706, with individual amino acid identities from 47% to 75% (Fig. 3 and data not shown). These homologues included nine out of the 15 phage 1706 proteins demonstrated to be virion associated (20). Many of the proteins encoded by genes carried on phage 1706 predicted to be involved in virion morphogenesis and DNA packaging had homologues in Pepy6 and Poco6, include LaP1706_gp13 (major capsid subunit, corresponding to Pepy6 gp28 and gp30 and Poco6 gp29 and gp31), LaP1706_gp10 (prohead protease, corresponding to Pepy6 gp25 and Poco6 gp26), LaP1706_gp18 (tape measure protein, corresponding to Pepy6 gp35 and Poco6 gp36), and LaP1706_gp9 (portal protein, corresponding to Pepy6 gp22 and Poco6 gp24) as well as LaP1706_gp8 (TerL).
TABLE 3.
Phages and prophage elements related to Rhodococcus phage Pepy6, Poco6, Pine5, or DocB7
| Phage or prophage (host)a | Genome length (kb)b | Genomic terminic | No. of CDSd | No. of CDS with TMD (%)e | Relationship to Rhodococcus phage (no. of shared proteins)f |
|---|---|---|---|---|---|
| 1706 (Lactococcus lactis) | 55 | 3′ cos GCCCTGTCT | 76 | 12 (16) | 21 proteins shared with Pepy6 |
| BRYFOR.phiA (Bryantella formatexigens DSM 14469) | 57 | 3′ cos (based on TerL similarity to Pine5 gp14) | 73 | 17 (23) | 23 proteins shared with Pepy6; 21 proteins shared with 1706 |
| CLOSS21 (Clostridium sp. strain SS2/1) | 66 | 3′ cos (based on TerL similarity to Pine5 gp14) | 89 | 18 (20) | 24 proteins shared with Pepy6; 22 proteins shared with 1706 |
| CLOLEP (Clostridium leptum DSM 753) | 52 | 3′ cos (based on TerL similarity to Pine5 gp14) | 78 | 11 (14) | 34 proteins shared with Pepy6; 21 proteins shared with 1706 |
| Mycobacteriophage Rosebush-like phages | 67-71 | pac (based on TerL related to Rosebush gp7) | 81-104 | (∼10) | 31 proteins shared with Pine5 |
1706, Lactococcus lactis phage 1706 (NC_010576); BRYFOR, Bryantella formatexigens DSM 14469 prophage (derived from GenBank entry NZ_ACCL02000018.1 and includes coding region for locus tags BRYFOR_08569 to BRYFOR_08504); CLOSS21, Clostridium sp. strain SS2/1 prophage (derived from GenBank entry NZ_ABGC03000034 and includes locus tags CLOSS21_01580 to CLOSS21_01492); CLOLEP, Clostridium leptum DSM 753 (derived from GenBank entry NZ_ABCB02000017 and includes locus tags CLOLEP_01347 to CLOLEP_01425). Mycobacteriophage Rosebush-like phages (with GenBank entries in parentheses) included Rosebush (NC_004684), Qyrzula (NC_008204), Cooper (NC_008195), PG1 (NC_005259), Orion (NC_008197), Pipefish (NC_008199), Chah (NC_011284), Colbert (GQ303259.1), Nigel (NC_011044), Puhltonio (GQ303264.1), Phaedrus (NC_011057), Phlyer (NC_012027), and UncleHowie (GQ303266.1).
Genome length, genome nucleotide kilobase pair length.
The structure of the packaged genomic termini is shown. pac, headful packaging.
The number of protein-coding genes (CDS) is shown.
The numbers and percentages of the proteins encoded by the element that are predicted to have transmembrane domains (CDS with TMD) are shown.
Maximum E values of 1e−10 and scores over 50 were used to define shared proteins.
Domain shuffling in DNA metabolism genes.
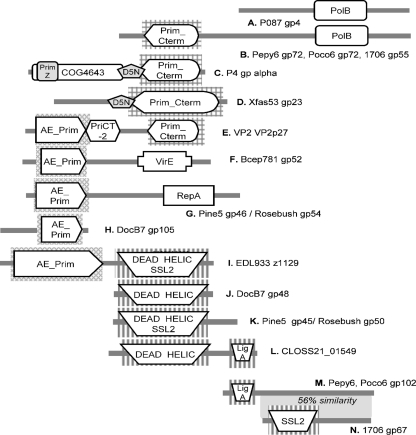
Two of the additional Pepy6 and Poco6 predicted proteins with identifiable homologues in phage 1706 had conserved domains suggestive of functions in DNA replication, but the novel combination of domains meant that specific functions could not be robustly assigned. The first of these were the Pepy6 and Poco6 gene72 products, which were similar over their entire length to phage 1706 gp55. Two distinct conserved domains were identified in these proteins: a COG3378 domain at the N terminus and a COG0417 domain at the C terminus. The COG3378 domain, originally identified as a C-terminal conserved domain in DNA primase, is one of a number of domains that have undergone extensive domain shuffling and thus can be found either by themselves or in combination with several other types of conserved domains (36). Likewise, Pepy6 gp102 and Poco6 gp102, which are homologues of phage 1706 gp67, had an atypical association of conserved domains likely to function in DNA replication (Fig. 4). The Rhodococcus phage proteins were significantly longer (542 residues compared to 412 residues) than phage 1706 gp67, and the amino-terminal extension appeared to be derived from LigA, a DNA ligase domain-containing protein. This fusion may have occurred during the acquisition of an 11-kb insertion between Pepy6 gene77 and gene102, encoding homologues of phage 1706 gene66 and gene67, respectively (Fig. 4 and 5 and see below). The phage 1706 protein contained an SSL2 domain commonly present in DEAD box helicase (Fig. 4). The region of Pepy6 gp102 that aligned with 1706 gp67 included the SSL2 domain-containing region but was different enough such that a conserved domain was not identified in the Rhodococcus proteins. SSL2 and related DEAD box helicase domains have been found associated with other domains. For example, one group of bacterial proteins has carboxy-terminal DEAD box helicase domains and amino-terminal AE-Prim_S domains. A permuted arrangement of these domains was observed in hypothetical protein CLOSS21_01549, encoded by the CLOSS21 prophage element related to Pepy6 and Poco6 (Fig. 4 and see below). CLOSS21_1549 had an amino-terminal DEAD box helicase domain (that aligned with Pepy6 gp102 residues 137 to 540, with 47% identity) and a carboxy-terminal LigC domain (that aligned with Pepy6 gp102 LigC domain-containing residues 15 to 86, with 38% identity).
FIG. 4.
Novel combinations and permutations of DNA metabolism-associated conserved domains in phages and proteins encoded by bacterial genes. (A) Lactococcus phage P087 (gp4); (B) Pepy6 (gp72), Poco6 (gp72), and Lactococcus phage 1706 (gp55); (C) satellite phage P4 (RPBPP4); (D) Xylella phage Xfas53 (gp23); (E) Vibrio phage Vp2 (VP2p2); (F) Burkholderia phage Bcep781 (gp53); (G) Pine5 (gp46) and Rosebush (gp54); (H) DocB7 (gp105); (I) Escherichia coli O157:H7 EDL933 (z1129); (J) DocB7 (gp48); (K) Pine5 (gp45) and mycobacteriophage Rosebush (gp50); (L) CLOS21 (CLOSS21_01549); (M) Pepy6 (gp102) and Poco6 (gp102); (N) Lactococcus phage 1706 (gp67). Related domains are shaped and shaded similarly. Domain abbreviations are based on the Conserved Domain Database entries as follows: PolB, COG0417; Prim_Cterm, COG3378; AE_Prim, cl01287; DEAD HELIC, CD00046; SSL2, COG1061; LigA, CL03295; PriCT-2D5N, pfam08707; D5_N superfamily, cl07360; Prim Z, cl07038; VirE, COG5545; RepA, COG3598; COG4643, COG4643.
FIG. 5.
Complex relationships between potential tail fibers encoded by Pepy6 and Poco6. (A) Alignment of the first 56 residues of Pepy6 gp1, gp2, gp4, gp5, and gp7 with Poco6 gp1, gp2, gp3, gp5, gp6, and gp8 and the amino-terminal residues of PSSM4_087 (encoded by Prochlorococcus phage P-SSM4 genes), XF_2114 (encoded by Xylella fastidiosa prophage Xfp6 genes), and 3396_48 (encoded by Streptococcus phage phi3396 genes). Invariant residues are shaded. (B) Schematic alignment of Pepy6 gp1, gp2, gp4, gp5, and gp7 with Poco6 gp1, gp2, gp3, gp5, gp6, and gp8 and also to e15 (tail fiber, encoded by Salmonella phage epsilon15 gene), PSSM4_087 (encoded by Prochlorococcus phage P-SSM4 gene), 3396_48 (encoded by Streptococcus phage phi3396 gene) and (FRAAL2677 encoded by Frankia alni ACN14a gene). Alignment scores are color coded as indicated. Conserved domains are indicated by shaded regions.
Prophage elements related to Pepy6, Poco6, and 1706.
Four prophage elements related to Pepy6, Poco6, and Lactococcus phage 1706 were identified in the genome assemblies from four members of the Firmicutes order Clostridiales: Bryantella formatexigens DSM 14469, Clostridium leptum DSM 753, Ruminococcus torques ATCC 27756, and Clostridium sp. strain SS2/1 (Fig. 3 and Table 3). The Bryantella formatexigens DSM 14469, Clostridium leptum DSM 753, Ruminococcus torques ATCC 27756, and Clostridium sp. strain SS2/1 prophage elements are referred to as BRYFOR, CLOLEP, RUMTOR, and CLOSS21 prophage elements, respectively. The relationship of phage 1706 to the RUMTOR and CLOLEP prophage elements was described previously (20). Each of these phage and prophage elements encoded a common set of 22 proteins that included most of the virion-associated and structural proteins, TerL, and proteins containing various DNA metabolism-related conserved domains (Fig. 3). Integrase genes could be identified at the beginning of each prophage element, which would correspond to a genome location between the Pepy6 gp35 and gp72 equivalents (Fig. 4). As in phage 1706, no integrase gene candidate was identified in Poco6 or Pepy6 (20).
Tail fiber gene complexity.
Pepy6 and Poco6 are >20 kb larger than phage 1706 and the clostridial prophage, including a segment of >8 kb of novel sequence inserted between TerL and cosL (Fig. 3 and 5). This region carried genes encoding a novel set of proteins, 5 in Pepy6 and 6 in Poco6, that although widely variant in size (278 to 856 amino acids [aa]), have an unusual, modular relationship to each other. The first 53 residues of each of these proteins were 56% to 94% identical, with 21 invariant residues present in all 11 proteins (Fig. 5). Numerous other proteins in the public database with related amino-terminal domains were identified by BLAST, and alignments reveled 11 invariant residues in proteins from phages and prophages from taxonomically diverse hosts such as cyanobacteria (e.g., Prochlorococcus phage P-SSM4 gp87), gammaproteobacteria (e.g., XF_2114 in Xylella prophage Xfp6), and Firmicutes (e.g., Streptococcus phage phi3396 gp48). However, except for Pepy6 gp2, Poco6 gp3, and Poco6 gp6, these proteins were not recognizably related to proteins in the public database beyond the conserved amino-terminal domain. Poco6 gp3 and Pepy6 gp2 C-terminal domains were 31% identical to the pectate lyase conserved domain of gp20, the tail fiber protein Salmonella phage ɛ15 (38). Several other Poco6 and Pepy6 proteins in these clusters also contained pectin-lyase domains and other carbohydrate-interaction domains as well, including SGNH hydrolase and concanavalin A-like lectins/glucanases. Likewise, the C terminus of Poco6 gp6 was related to lipases from a number of Actinomycetes. GDSL/SGNH hydrolase domain proteins are involved in the hydrolysis of wide variety of substrates, including fatty acids, aromatic esters, and amino acid derivatives, and are particularly abundant in Actinomycetes (3, 6). These relationships suggested that this gene clusters might encode proteins that interact with the cell envelope in Rhodococcus.
A second large insertion in the genomes of Pepy6 and Poco6 relative to phage 1706 was identified. This 11-kb insertion, encompassing Pepy6 genes gene77 to gene102, corresponds to an insertion between gene66 and gene67 in phage 1706 (Fig. 4). The BRYFOR and CLOSS21 prophage elements contained four genes and one gene, respectively, between their 1706 gene66 and gene67 homologues (Fig. 4). As previously discussed, rearrangements associated with this region may have been the source of the novel domain structure of Pepy6 and Poco6 gp102. The genes present in the Pepy6 and Poco6 11-kb insertion were predicted to encode primarily small, hypothetical novel proteins, although one gene encoding a conserved thymidylate synthase was identified.
Poco6 and Pepy6 lysis genes.
In addition to the cytoplasmic membrane and the peptidoglycan cell wall, the envelope of the Rhodococcus and the other genera of the mycolata, a suprageneric taxon including Nocardia, Corynebacterium, and Mycobacterium, has a third layer comprised of an arabinogalactan polymer linked to an “outer membrane” of mycolic acid chains (70). Extrapolating from the lysis systems of mycobacteriophages, rhodococcal phages can be expected to encode not only the canonical holin and endolysin (LysA) lysis proteins but also an enzyme, LysB, that attacks this outer layer (21, 22, 54). The LysA and holin genes were readily identified as adjacent reading frames, gene10 and gene11 in Pepy6, and as a nearly identical pair of genes gene12 and gene13 (>90% identity with the Pepy6 proteins) found in the same position in Poco6. The LysA annotation was made particularly robust by the presence of a conserved N-terminal peptidoglycan-binding domain and N-acetylmuramoyl-l-alanine amidase catalytic residues and by similarity to actinobacterial amidases (data not shown). The holin annotation was based on gene location, prediction of a transmembrane domain, and the weak but significant primary sequence similarity (46%) to the experimentally confirmed holin encoded by Brevibacterium flavum phage BFK20 (10). The predicted length and topology of the two transmembrane domains (TMDs) and the distribution of charged residues are remarkably conserved between the Pepy6/Poco6 proteins and the BFK20 holin (not shown). The presence of two unambiguous TMDs allowed these proteins to be assigned as class II holins. Moreover, the absence of a secretion signal in the LysA sequence indicated that these holins must form large holes, unlike many other members of class II, which have pinholin character (i.e., make only small holes in the membrane) and serve only to effect a temporally scheduled depolarization of the host membrane (53).
In mycobacteriophage genomes, LysB genes are invariably closely linked, or adjacent to, the holin-endolysin loci (54). However, the only candidates for LysB in Pepy6 and Poco6, gp33 and gp34, respectively, were located in the virion structural protein cluster. The protein most closely related (49% similar) to these candidates was ROP_59440, a PE-PPE domain-containing protein encoded by Rhodococcus opacus B4. ROP_59440 was a member of a small cluster of related proteins encoded by several Nocardia and Rhodococcus species (data not shown). However, psi-BLAST analysis detected a significant relationship between these proteins and mycobacteriophage LysB proteins, primarily Cjw1 gp35 (score of 58.5) but also to D29 gp12 (54). As was found for the mycobacteriophage proteins, the Pepy6 and Poco6 putative LysB equivalents were predicted to be cytoplasmic and thus may be released by holin function along with LysA.
Abundance of predicted membrane-associated proteins in Pepy6, Poco6, and related prophages.
The Pepy6 and Poco6 genomes carried an unusually high fraction of genes predicted to contain at least one TMD (Tables 3 and 4). For Pepy6, 29 out of 107 proteins (29%) and for Poco6, 26 out of 107 proteins (24%) were predicted to have TMDs. In order to determine how these values compare to other phages, the predicted proteins encoded by 406 Caudovirales genomes were screened for TMDs. Out of a total of 36,324 proteins encoded by phage genes, 3,081 (8.5%) were predicted to have at least one TMD (Table 4). When the data set was broken down by host clade, there was very little difference in the percentage of TMD proteins among the Proteobacteria (8.3%), Actinobacteria (8.0%), Firmicutes (9.5%) and “other” (which included Cyanobacteria and Flavobacterium) (7.0%). All phages encoded at least one potential TMD protein. Vibrio phage VP5 encoded the lowest percentage of TMD proteins (2.1%), and when Pepy6 and Poco6 were excluded, Bacillus phage SPP1 encoded the highest percentage of TMD proteins (17.8%). The clostridial prophages related to Pepy6 and Poco6 as well as phage 1706 were enriched in TMD protein-coding regions (14% to 23%) (Table 3).
TABLE 4.
Analysis of the presence of transmembrane domains (TMD) in proteins encoded by phage genes broken down by host taxonomy
| Host clade | No. of phage | Total no. of proteinsa | No. of proteins with TMDb | % proteins with TMD | Phage encoding the lowest or highest % of TMD proteins (% TMD proteins)c |
|
|---|---|---|---|---|---|---|
| Lowest | Highest | |||||
| Proteobacteria | 178 | 16,407 | 1,360 | 8.3 | Vibrio VP5 (2.1) | Klebsiella K11 (19.6) |
| Actinobacteriad | 69 | 7,478 | 603 | 8.0 | Mycobacterium Porky (5.4) | Corynebacterium P1201 (12.4) |
| Firmicutes | 141 | 9,958 | 943 | 9.5 | Bacillus BCJA1c (2.1) | Bacillus SPP1 (17.8) |
| Othere | 18 | 2,481 | 175 | 7.0 | Cyanophage P60 (2.5) | Flavobacterium 11b (12.3) |
| Total | 406 | 36,324 | 3,081 | 8.5 | VP5 BCJA1c (2.1) | Bacillus SPP1 (17.8) |
Total number of proteins encoded by phage genes.
Number of proteins encoded by phage genes and predicted to have at least one TMD.
The phage with genes encoding the lowest or highest percentage of proteins predicted to have at least one TMD. The percentage of proteins predicted to have at least one TMD is shown in parentheses.
Actinobacteria does not include Rhodococcus phages Pepy6, Poco6, Pine5, and DocB7.
Includes Cyanobacteria and Flavobacterium.
Pine5 genome organization and relationship to mycobacteriophage Rosebush.
At 59 kb, Pine5 was the smallest of the Rhodococcus phages analyzed. Pine5 genes were predicted to encode 85 proteins (Fig. 6), of which 41 (52%) were small, hypothetical novel proteins with no recognizable homologues in the public database. Thirty-one proteins encoded by genes carried on Pine5 (39%) were related in sequence and gene order to proteins encoded by a cluster of 13 closely related mycobacteriophages that included Rosebush, Qyrzula, Phlyer, Phaedrus, Pipefish, Nigel, Cooper, Chah, PG1, Orion, Colbert, Puhltonio, and UncleHowie (29-31). Rosebush was the first member of this group to be described (56). The 13 Rosebush-like phages exhibited an average of 62 to 99% DNA sequence identity to each other across the majority of the length of their genome (30, 31). The regions of Pine5 genomic DNA that aligned with Rosebush genomic DNA were located primarily in nine blocks, ranging from 0.4 to 1.1 kb, with up to 73% identity, which were predicted to encode proteins involved in virion morphogenesis and DNA metabolism (Fig. 6). While significant, this level of similarity falls below the threshold used to define the clusters within the mycobacteriophages (30). However, Pine5 is the first nonmycobacteriophage that exhibits shared synteny with this group of Mycobacterium phage.
FIG. 6.
Genome maps of Pine5 and DocB7. Phage genome maps are drawn to scale with the coding strand indicated by the position of the genes above (rightwards) and below (leftwards) the central ruler. (A) Pine5 aligned with mycobacteriophage Rosebush. The Rosebush map was generated from the GenBank entry with accession no. AY129334.1. Purple lines between Pine5 and Rosebush indicate regions of greater than 60% DNA identity between the two phages. Genes are color coded to indicate similarity with other proteins in the public database as follows: yellow, homologues are found in the mycobacteriophages Rosebush, Qyrzula, Phlyer, Phaedrus, Pipefish, Nigel, Cooper, Chah, PG1, Orion, Colbert, Puhltonio, and UncleHowie; green, homologues in the public database but not in Rosebush; blue, no homologue in the public database; gray, Rosebush genes without homologues in Pine5. Genes with red outlines encode proteins with predicted transmembrane domains. Annotations of selected genes are abbreviated as follows: PAPSr, phosphoadenosine phosphosulfate reductase; sub, subunit; RuvC, Holliday junction resolvases. (B) DocB7.Genes are color coded to indicate similarity with other proteins in the public database as follows: yellow genes encode proteins with at least one homologue in the public database; blue genes, no homologue in the public database. Genes with red outlines encode proteins with predicted transmembrane domains. The 2,483-bp region highlighted in peach lacks recognizable protein-coding genes (no cds, no protein-coding sequences) and contains 13 perfect inverted repeats and one direct repeat.
Morphogenesis protein homologues in Pine5 and the Rosebush-like mycobacteriophage included the tape measure protein, portal protein, capsid subunit, and major tail subunit. Candidate genes encoding the frameshift tail chaperone, prohead protease, and scaffolding subunit were not identified in Pine5. Like Rosebush, the Pine5 genome assembled into a circular map, and no end sequences were identified, strongly suggesting that the phage utilizes a pac-type headful packaging mechanism (56). Additional TerL proteins significantly related to the TerL protein from Pine5 and Rosebush encoded by other phages were demonstrated to have circularly permuted genomes, included Bcep781, Aaphi23, and PY100 (59, 61, 68).
Pine5 and Rosebush carried genes encoding a related set of proteins with conserved domains, suggesting that they play various roles in DNA metabolism. These proteins included Pine5 gp45 and Rosebush gp54, which possessed SSL2/helicase domains (Fig. 4). Pine5 gp46 and Rosebush gp54 proteins had N-terminal cd04859 primase-polymerase (Prim_Pol) domain and a carboxy-terminal region that includes Walker A and Walker B nucleotide-binding sites as part of a RepA helicase domain. Proteins identified by BLAST searches that could be aligned to Pine5 gp46 across their entire length included only the mycobacteriophage homologues and gp43 of Corynebacterium phage BFK20. However, proteins unrelated to Pine5 gp46 at a primary structural level but with conserved domain architecture are widespread among prokaryotes (36).
The only readily identifiable component of lysis predicted to be encoded by Pine5 was the LysA endolysin candidate, Pine5 gp40. Pine5 gp40 has two domains that support this annotation, an N-terminal peptidase domain and a muramidase/glycosyl hydrolase domain. Some proteins encoded by genes carried on mycobacteriophages, including LysA, were grouped into protein families characterized by members having complex chimeric interrelationships (31). The relationship of Pine5 LysA to the mycobacteriophage LysA proteins was also chimeric. No holin gene candidate could be identified for Pine5. In Rosebush, the holin is encoded by the gene immediately downstream of the LysA gene, but there is no downstream gene in Pine5 at this location (Fig. 6, ReqiPine5 genome map). Thirteen Pine5-encoded proteins were predicted to have transmembrane domains, but most of these proteins lacked homologues in the public database and none possessed sequence similarity to a holin. However, the predicted topology of one protein, Pine5 gp17, was markedly similar to that of the BKF20, Pepy6, and Poco6 holin proteins and is thus the best candidate holin (data not shown). No LysB equivalent was identified for Pine5. This was consistent with the results of a global search of LysB homologues in 60 mycobacteriophage genomes in which only four phages, Rosebush, Qyrzula, Myrna, and Che12 lacked identifiable LysB equivalents (54). While not related to any protein encoded by genes carried on a phage, Pine5 gp21 possessed a SGNH-hydrolase domain, found in numerous lipases and esterases, and thus may play a role in lysis.
DocB7, a new phage type.
The DocB7 genome was 75,772 bp and predicted to carry genes encoding 105 proteins (Fig. 6). DocB7 represents the first isolate of a new phage type with a distinct genome organization unrelated to any entries in the public database. Moreover, only 31% of DocB7 proteins had significant similarity to known proteins, and no genetic element that shared significant gene content with DocB7 could be identified in bacterial genomes. Functional assignments could be made for only 23 DocB7 proteins. Even with so few functional annotations, it was apparent that the genome was organized into two convergent transcriptional arrays, with virion morphogenesis loci located on the left arm and DNA metabolism and regulatory genes largely encoded on the right arm. The left arm of DocB7, from gene1 to gene30, encoded proteins with functions in DNA packaging and virion morphogenesis. The DocB7 TerL homologue, encoded by gene3, showed only distant relationships to other TerL proteins. Only two significantly related proteins were identified in the public database—the TerL homologues from Thermus phage phiYS40, and Microcystis phage Ma-LMM01, both large myophages (51, 81). Defined termini were not identified for DocB7, indicating that DocB7 utilizes a headful packaging strategy, as does Ma-LMM01 (81). DocB7 gp4 had some similarity to proteins of unknown function encoded by genes in a number of bacteria present in the human gut microbial community, including Eubacterium rectale, Clostridium methylpentosum, and Ruminococcus (45). This protein contained a conserved domain (DUF935) also present in many proteins encoded by phage genes, notably Mu gp29, and probably functions in portal morphogenesis. DocB7 gp5 was significantly related to the phage T7 gene 17 tail fiber (63). While lacking identifiable conserved domains, DocB7 gp7 was 47% identical to Gordonia terrae phage GTE5 gp1, predicted to have an endopolygalacturonase (PGU1) domain (7). The amino-terminal 80 residues of DocB7 gp7 were significantly related to the amino termini of hundreds of bacterial and fungal glycoside hydrolase family proteins, such as Lam55a from Phanerochaete chrysosporium (35).
DocB7 gene20 and gene21 were annotated as the λ G/G-T tail chaperone equivalents due to their location upstream of the tape measure protein gene (gene22) and presence of a predicted ribosomal frameshift sequence (80). Ribosomal slippage at the sequence GGAAAAA (nucleotide [nt] 17359) would direct a +1 translational reading frameshift, leading to the DocB7 gp20-21 frameshift protein. The tape measure protein candidate gp22 had a mosaic relationship with other proteins in the public database, as was reported for mycobacteriophage tape measure proteins (31). At 3,101 residues, the unusual length of DocB7 gp22 is consistent with the unusually long tail (489 nm) of the virion (Fig. 1). Three conserved domains were identified in gp22: (i) a tape_meas_TP901 domain near the amino terminus; (ii) a region that was recognized by multiple conserved domain clusters, including PRK01156, chromosome segregation protein; and finally, (iii) near the carboxy terminus, a “COG5412, Phage-related protein of unknown function” domain.
The right arm of DocB7 carried genes encoding proteins implicated in regulation and DNA replication. The protein encoded by gene31 was identified as a possible WhiB transcription factor. WhiB transcription factors were first identified in Streptomyces aureofaciens as factors involved in the development of the mycelium (37). WhiB-related proteins are common among the mycobacteriophages (56). Two DocB7 genes were predicted to encode proteins with some similarity to subunits of DNA polymerase III, including the beta clamp domain subunit (gp44) and the epsilon subunit (gp45). While T4 phages frequently encode functional equivalents of DNA polymerase III subunits, the DocB7 proteins were more closely related to bacterial homologues than phage homologues. However, the low similarity scores suggested that these genes were not the results of a recent acquisition.
Like the other Rhodococcus phages, a gene on DocB7 encoded a protein with conserved helicase and primase domains (Fig. 4). The helicase gp105 has a typical DEAD box helicase domain. gp105, has a C-terminal AE_Prim_S domain and exhibits only weak similarity to three other proteins in the public database. Two additional proteins have notable conserved domains. gp68 is a large protein (644 amino acids) with a C-terminal Von Willebrand factor type A (vWA) domain. vWA domains have a metal ion-dependent adhesion site that promotes oligomerization (79). gp69 (535 aa) has a C-terminal AAA domain. AAA domains are ATP-hydrolyzing domains. Both vWA and AAA domains are found in numerous, otherwise functionally unrelated, and widely diverse proteins. The lack of other significant relationships in the large N-terminal domains of gp68 and gp69 to other proteins precluded any functional assignment. Additional proteins predicted to be encoded by DocB7 genes include a homing endonuclease, gp54, and a tyrosine recombinase/possible integrase, gp59. The presence of a putative integrase and a WhiB transcription factor homologue raised the possibility that DocB7 might be a temperate phage, although DocB7 plaques were not detectably turbid.
The region between DocB7 gene30 and gene31, separating the two coding arms of DocB7, contained a putative convergent transcription terminator domain. The 46 bp separating the stop codons of these two genes, were completely symmetric, which if transcribed, would form an extremely large stem-loop structure (34519-TAAGAAAACCCCCGGTACCTAATGAAAGGTACCGGGGGTTTTCTTA-34564 [potential stem formation residues are underlined]), likely promoting bidirectional, rho-independent transcription termination.
DocB7 lysis genes.
Compared to the other Rhodococcus phages, DocB7 possessed the most complete lysis cassette. DocB7 lysis genes encoding LysA (gp27), LysB (gp38), and three holin candidates (gp26, gp28, and gp30) were identified. Like Pepy6 and Poco6, the DocB7 gp27 LysA had an N-terminal peptidoglycan recognition protein conserved domain (cd06583) and conserved catalytic residues required for amidase activity. Moreover, gp27 is related to the mycobacteriophage LysA equivalents, including PG1 gp49, as well as the BFK20 gp24 lysin. Like these proteins, DocB7 gp27 was predicted to be a cytoplasmic protein and would thus require holin activity to gain access to the cell wall. Proteins encoded by gene26, gene28, and gene30 were predicted to possess two, one, and four transmembrane domains, respectively. The predicted membrane orientation and charge distribution of DocB7 gp28 were compared to those of the BFK20 gp26 holin (data not shown). Of these, only gp26 exhibited significant sequence similarity to proteins in the public database and were related to members of mycobacteriophage Pham107 members, like gp27 of phage Bxb1 (31). Some lysis systems in phages of Gram-negative hosts are fairly well characterized, allowing definitive identification and functional characterizations not only of holins of several different topologies but also of antiholins, which are holin-specific negative regulators. However, holin function in Gram-positive bacteria in general, and especially in the Actinomycetes, is poorly understood. It is thus not possible to discern which of these proteins might be a holin or which might serve as an antiholin regulator.
DocB7 gene38 encodes a protein with a PE-PPE domain, believed to be associated with cell surface features (2). This protein was weakly similar to cutinases and hypothetical proteins from numerous Mycobacterium and other Corynebacterium species, for example exhibiting 40% similarity to Mycobacterium smegmatis cutinase GI:302566037. gp38 lacks the secretory signals common to most mycobacterial cutinases (78). When gp38 was queried against the database of Caudovirales proteins, similarity to mycobacteriophage LysB proteins, especially Bethlehem gp9, was detected. Thus, gp38 may be a LysB protein dependent on holin permeabilization of the membrane for release.
Gene-free region in DocB7.
A notable feature of the DocB7 genome is an unusually long region, 2,484 bp (from 68369 to 70852; Fig. 6B) lacking any predicted protein- or RNA-encoding genes. There were no homologues to this region in the database at a DNA level, and a translation BLAST algorithm did not detect any significant amino acid similarity with proteins in the public database. Manual inspection identified a number of small open reading frames with potential start codons, but none had a reasonable Shine-Dalgarno motif. This region was contained 14 inverted repeats, 13 perfect and one with a single mismatch, and one direct repeat (data not shown). The dinucleotide composition of this region is significantly different from the rest of the genome, implying that it might be recently acquired (data not shown).
Phage efficacy at reducing R. equi in soil matrix.
Phage DocB7 was chosen to determine whether application of phage to soil samples containing R. equi would result in a reduction in bacterial CFU. The soil matrix used in these assays was a moist sandy loam. Preliminary experiments were conducted to determine the stability and recoverability of DocB7 from soils indicated that approximately 10% of the applied phage could be recovered following 48 h of incubation in soil at 30°C in the absence of host cells (data not shown). In two independent experiments, an R. equi suspension was applied to sterile soil samples and incubated at 30°C prior to the application of phage. Phage was applied by spraying, and then the number of surviving CFU in the soil was assessed. Beginning at an initial R. equi concentration at 1.0 × 105 CFU/g, viable counts increased by nearly 3 orders of magnitude (to 7.4 × 107 CFU/g) in 48 h at 30°C (Fig. 7). In contrast, all phage-treated samples exhibited a reduction in CFU after incubation at all levels of initial input phage. The reduction was not colinear with increasing concentrations of phage. Application of phage at an approximate multiplicity of infection (MOI) of 10 resulted in the lowest recoverable CFU (6.0 × 104 CFU/g) after the 48-h incubation, a level even lower than the initial R. equi inoculums (1.0 × 105 CFU/g).
FIG. 7.
Effect of phage application on recovery of R. equi from soil. Phage effect on R. equi was evaluated via 7 treatment groups. Each treatment group contained 3 parallel subsamples, where 3 sterile soil matrixes were inoculated independently with R. equi strain ATCC 33701 and were subjected to further treatment. R. equi was inoculated at an initial concentration of 1.0 × 105 CFU/g soil. The R. equi levels were determined before (0-h) and after (48-h) incubation in the absence of phage treatment (No phage). The R. equi CFU levels were also determined following 48 h of incubation in the presence of phage DocB7. Phage DocB7 was applied at initial multiplicity of infections (MOIs) of 1,000, 100, 10, 1, and 0.1 PFU/CFU. After each subsample was analyzed in two replicate samples, the mean values± standard deviations of the subsamples (error bars) were plotted. Two independent experiments were carried out, and the results of one representative experiment are shown.
DISCUSSION
Rhodococcus phage diversity.
The sequences of Pepy6, Poco6, DocB7, and Pine5 represent the first complete genomic data available for phages that infect Rhodococcus. It is relevant, therefore, to discuss the relationship of these Rhodococcus phages to the >450 complete phage genomes, and even more prophage elements located in bacterial genome entries, present in the public database. The relationships of these phages to each other and to phages that infect other host clades follow several of the patterns noted when genomic data from other host-defined groups of phages have been analyzed. One of the four phages, DocB7, fit the criteria of being a new phage type. DocB7 not only has a distinct genome organization but encodes primarily novel proteins of unknown function and has only a few proteins in common with any other phages or prophage sequences available at the time of analysis. This is expected, as it is still common to isolate completely new phage types, especially for a host for which there is only limited phage genomic data available. It is anticipated, however, that as more Rhodococcus and other phages are analyzed, DocB7 will ultimately become part of a cluster of related phages whose host range spans multiple genera. This is the case for Pine5, for which over 30% of encoded proteins are present in members of a cluster of at least 13 mycobacteriophages, including Rosebush. While the mycobacteriophage members of this cluster share significant DNA similarity to each other, Pine5 has only limited DNA sequence similarity, well below the threshold used to define the mycobacteriophage clusters (30). Considering the taxonomic proximity of Rhodococcus to Mycobacterium and the relative abundance of mycobacteriophage genomic data, it was expected that at least some Rhodococcus phages would be similar to mycobacteriophages. Finally, over 20% of the proteins encoded by Pepy6 and Poco6 shared similarity to proteins from phages that infect Firmicutes hosts, including Lactococcus phage 1706 and Clostridium prophage elements sequenced as part of the Human Gut Microbiome Initiative (24). The somewhat atypical genome organization of these phages might contribute to the maintenance of genome identity. Unusual genome features identified in 1706, Poco6, and Pepy6 include the atypical location of the cos termini (i.e., not adjacent to the terminase genes) and the lack of clustering of lysis genes. There also appear to be two preferred spots for genome expansion, which accounted for the majority of the broad genome size range observed in this cluster of phages (from 52 kb to 78 kb). This relationship in which otherwise closely related phages differ considerably in gene content due to the insertion of large clusters of small genes encoding mostly hypothetical novel proteins is seen in some other phage families. For example, Burkholderia phage BcepF1 has an ∼20-kb region that encodes 62 small hypothetical proteins, representing the majority of gene content differences between BcepF1 and Pseudomonas phage F8 (42).
While there are phage types whose hosts span broad taxonomic lineages, it is also expected that regardless of phage type, phages that infect a specific host clade will have specialized, host-specific, adaptations. One of the features that Rhodococcus shares with Mycobacterium is the presence of an outer layer composed of mycolic acids covalently linked to the peptidoglycan (71). This layer presents an additional challenge to phages of the mycolata, both in terms of adsorption and DNA injection process as well as host cell lysis. Evidence has been provided recently that efficient lysis by mycobacteriophage requires, in addition to the holin and endolysin, a mycolylarabinogalactan esterase (LysB) in order to disrupt this outer membrane (21, 22, 54). Analogies can be made between the function of LysB in disruption of the outer membrane with the Rz-Rz1 equivalents of Gram-negative hosts, which function in outer membrane disruption (54, 65). Most mycobacteriophages encode LysB equivalents, so it was anticipated that phages that infect other members of the Corynebacterineae, including Rhodococcus, would also carry genes encoding LysB equivalents (31, 54). DocB7, Pepy6, and Poco6 carry genes encoding potential LysB homologues, despite otherwise insignificant similarity to mycobacteriophages. However, the genetic structure of the lysis genes of the Rhodococcus phages differed significantly from that of the mycobacteriophage in that they lacked well-defined lysis gene cassettes. Rosebush was among the few mycobacteriophages in which LysB genes were not identified, and the lack of recognizable LysB homologues is one of the many genome features shared by Pine5 and Rosebush. It is possible that these phages have a functionally equivalent but mechanistically unrelated strategy for disruption of the mycolate layer.
A profusion of membrane proteins.
Perhaps the most remarkable feature identified in the Rhodococcus phages was the elevated percentage of proteins encoded by Pepy6 and Poco6 that contained predicted transmembrane domains. Pepy6 and Poco6 encode 29% and 24% proteins with predicted TMDs, respectively, which is significantly higher than the average (8.5%) for all tailed phages. The phage with the next highest percentage of TMD-containing proteins was identified as Klebsiella phage K11, at 19.6% TMD-containing proteins. This was not found to be a general feature of phages of the mycolata, nor is it something observed among phages of the closely related Mycobacterium. The majority of phages of Actinomycetes analyzed were from Mycobacterium hosts (63 out of 69), and there was no general enrichment in TMD domains among these phages. When the data on phage gene-encoded transmembrane domains were analyzed based on phage type, there was not a consistent correlation between phage type and percentage of TMD (data not shown). For example, Klebsiella phage K11 is a T7-like phage, but only 79 out of 829 (9.5%) proteins encoded by 17 T7-like phages were predicted to have at least one TMD, suggesting that as a whole, the T7-like phages are not enriched in TMD-containing proteins. In contrast, phage 1706 and the prophage related to Pepy6 and Poco6 encode a higher than average percentage of TMD-containing proteins (14% to 23%), suggesting that this might be a general characteristic of the “1706-like” cluster of phages. The TMD-containing proteins encoded by Pepy6, Poco6, and the related Firmicute phages were relatively small proteins of unknown function and were poorly conserved, even among this group of phages. It should be noted that all tailed phages present as whole-genome entries in the public database carry genes encoding at least one TMD-containing protein. This was expected, because of the key role in lysis played by holins, which always have at least one TMD (82). Additionally, nearly all phages of Gram-negative hosts encode a spanin like λ Rz, which always has a single N-terminal TMD (65). Another class of TMD-containing proteins encoded by phage genes includes the small, poorly conserved inner membrane components of superinfection exclusion (44). In bacteriophage λ, 5 of 73 proteins are predicted to be integral membrane proteins possessing at least one TMD. These include three lysis proteins (S107 antiholin, S105 holin, and Rz) and two superinfection exclusion proteins. It is likely that at least some of the TMD-containing proteins encoded by Pepy6, Poco6, 1706, and the clostridial prophage function in lysis or superinfection exclusion. Ultimately, the shear abundance, and topological distribution, of membrane proteins predicted to be encoded by phage genes suggests a variety of functional roles that awaits further elucidation.
Rhodococcus phage as a prophylactic.
The efficacy of phage against R. equi in soil demonstrated here suggests the potential for phage-based environmental prophylaxis against this pathogen. Although many aspects of the epidemiology of R. equi infections remain to be determined, there has been an observed association between the prevalence of virulent R. equi strains in the environment—particularly in the soil—with the occurrence of disease in foals (15, 50, 72). The highest indigenous level of R. equi in surface soil has been reported as approximately 104 CFU/g, although this number is highly variable, depending on the chemical characteristics and structure of the soil, as well as the sampling seasons (72). In the sterile soil system presented here, R. equi was inoculated at a final density of 1 × 105 CFU/g, and this initial population increased further to 7.4 × 107 CFU/g after 48 h of incubation at 30°C in the absence of phage. This rapid growth to such high bacterial density is likely a reflection of the nutrient content of the soil, the growth state of the inoculated R. equi culture, and the lack of any indigenous soil microflora which would normally compete against R. equi. Due to the high density (108 to 109 bacteria per gram of soil) and genetic diversity of bacterial populations in soil, competition among the mixed populations of bacteria in situ is inevitable (77). The relatively rapid growth of R. equi observed probably contributed to the efficient bacterial killing exhibited by phage ReqiDocB7, but it should be noted that the physiology and density of natural R. equi populations will likely vary significantly from the model system reported here.
In the soil model, sterile soil was inoculated with a known virulent R. equi strain and subsequently challenged with various concentrations of phage DocB7. As might be expected, increasing levels of phage resulted in decreasing levels of recoverable bacteria, up to a >3-log-unit order reduction observed at an MOI of 10. However, efficacy appeared to decrease at MOIs above 10. The existence of an apparently optimal phage dose in this system is predicted by theoretical models of phage therapy, which suggest that very high initial loads of phage can result in reduced treatment efficacy, as the initial round of bacterial killing results in too few surviving cells to allow continued phage replication (23, 55). Optimal phage dosing, in which reduced treatment efficacy is observed when the applied phage concentration is either too high or too low, is predicted to be observed in systems where the phage population exhibits a net decline over time (23). The poor recoverability of phage DocB7 from soils in the absence of host indicates that the soil system used here exhibited a rapid decline of phage. Phages have been shown to adsorb to, or be inactivated by, charged particles in soils such as clays, with removal rates affected by factors, including ion-exchange capacity, surface area, organic content, and pH (5, 11, 12). A better understanding of the ecological interactions between R. equi and its phages in the soil environment would greatly benefit the development of phage-based strategies for the biological control of this important pathogen in the foaling environment.
Acknowledgments
This work was supported by grants EF 0523951 and EF 0949351 from the National Science Foundation and funding from Texas AgriLife (to R.Y.). Support for Mei Liu, acquisition of Rhodococcus equi isolates, and application of phage to specimens containing R. equi was supported by the Link Equine Research Endowment, Texas A&M University.
We thank Christine Morgan for her insight into choosing and preparing the soil used in the efficacy study. We thank Nathan Slovis and Jacqueline Smith for providing soil samples from which R. equi was isolated.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Ackermann, H. W. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227-243. [DOI] [PubMed] [Google Scholar]
- 2.Adindla, S., and L. Guruprasad. 2003. Sequence analysis corresponding to the PPE and PE proteins in Mycobacterium tuberculosis and other genomes. J. Biosci. 28:169-179. [DOI] [PubMed] [Google Scholar]
- 3.Akoh, C. C., G. C. Lee, Y. C. Liaw, T. H. Huang, and J. F. Shaw. 2004. GDSL family of serine esterases/lipases. Prog. Lipid Res. 43:534-552. [DOI] [PubMed] [Google Scholar]
- 4.Alvarez, V. M., et al. 2008. Bioremediation potential of a tropical soil contaminated with a mixture of crude oil and production water. J. Microbiol. Biotechnol. 18:1966-1974. [PubMed] [Google Scholar]
- 5.Ashelford, K. E., M. J. Day, and J. C. Fry. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bielen, A., et al. 2009. The SGNH-hydrolase of Streptomyces coelicolor has (aryl)esterase and a true lipase activity. Biochimie 91:390-400. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, P., C. Sieiro, N. M. Reboredo, and T. G. Villa. 1998. Cloning, molecular characterization, and expression of an endo-polygalacturonase-encoding gene from Saccharomyces cerevisiae IM1-8b. FEMS Microbiol. Lett. 164:249-255. [DOI] [PubMed] [Google Scholar]
- 8.Borodovsky, M., R. Mills, J. Besemer, and A. Lomsadze. 2003. Prokaryotic gene prediction using GeneMark and GeneMark.hmm. Curr. Protoc. Bioinformatics Chapter 4, Unit 4.5. doi: 10.1002/0471250953.bi0405s01. [DOI] [PubMed]
- 9.Brodie, R., R. L. Roper, and C. Upton. 2004. JDotter: a Java interface to multiple dotplots generated by dotter. Bioinformatics 20:279-281. [DOI] [PubMed] [Google Scholar]
- 10.Bukovska, G., et al. 2006. Complete nucleotide sequence and genome analysis of bacteriophage BFK20-a lytic phage of the industrial producer Brevibacterium flavum. Virology 348:57-71. [DOI] [PubMed] [Google Scholar]
- 11.Burge, W. D., and N. K. Enkiri. 1978. Virus adsorption by five soils. J. Environ. Qual. 7:73-76. [Google Scholar]
- 11a.Casjens, S. R. 2008. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res. Microbiol. 159:340-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay, S., and R. W. Puls. 2000. Forces dictating colloidal interactions between viruses and soil. Chemosphere 41:1279-1286. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, N. D., et al. 2008. Association of soil concentrations of Rhodococcus equi and incidence of pneumonia attributable to Rhodococcus equi in foals on farms in central Kentucky. Am. J. Vet. Res. 69:385-395. [DOI] [PubMed] [Google Scholar]
- 14.Cohen, N. D., and R. J. Martens. 2007. Rhodococcus equi foal pneumonia, p. 355-366. In B. C. McGorum, P. M. Dixon, N. E. Robinson, and J. Schumacher (ed.), Equine respiratory medicine and surgery. Elsevier Limited, Philadelphia, PA.
- 15.Cohen, N. D., M. S. O'Conor, M. K. Chaffin, and R. J. Martens. 2005. Farm characteristics and management practices associated with development of Rhodococcus equi pneumonia in foals. J. Am. Vet. Med. Assoc. 226:404-413. [DOI] [PubMed] [Google Scholar]
- 16.Cortez, D., P. Forterre, and S. Gribaldo. 2009. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 10:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deveau, H., S. J. Labrie, M. C. Chopin, and S. Moineau. 2006. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72:4338-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field, J. A., and R. Sierra-Alvarez. 2008. Microbial degradation of chlorinated benzenes. Biodegradation 19:463-480. [DOI] [PubMed] [Google Scholar]
- 19.Field, J. A., and R. Sierra-Alvarez. 2008. Microbial degradation of chlorinated dioxins. Chemosphere 71:1005-1018. [DOI] [PubMed] [Google Scholar]
- 20.Garneau, J. E., D. M. Tremblay, and S. Moineau. 2008. Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 373:298-309. [DOI] [PubMed] [Google Scholar]
- 21.Gil, F., et al. 2008. The lytic cassette of mycobacteriophage Ms6 encodes an enzyme with lipolytic activity. Microbiology 154:1364-1371. [DOI] [PubMed] [Google Scholar]
- 22.Gil, F., et al. 2010. Mycobacteriophage Ms6 LysB specifically targets the outer membrane of Mycobacterium smegmatis. Microbiology 156:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill, J. J. 2008. Modeling bacteriophage therapy, p. 439-464. In S. Abedon (ed.), Bacteriophage ecology. Cambridge University Press, Cambridge, United Kingdom.
- 24.Gordon, J. I., et al. 2005, posting date. Extending our view of self: the human gut microbiome initiative (HGMI). National Human Genome Research Institute white paper. http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/HGMISeq.pdf.
- 25.Grimm, M. B., et al. 2007. Evaluation of fecal samples from mares as a source of Rhodococcus equi for their foals by use of quantitative bacteriologic culture and colony immunoblot analyses. Am. J. Vet. Res. 68:63-71. [DOI] [PubMed] [Google Scholar]
- 26.Hamamura, N., M. Fukui, D. M. Ward, and W. P. Inskeep. 2008. Assessing soil microbial populations responding to crude-oil amendment at different temperatures using phylogenetic, functional gene (alkB) and physiological analyses. Environ. Sci. Technol. 42:7580-7586. [DOI] [PubMed] [Google Scholar]
- 27.Hamamura, N., S. H. Olson, D. M. Ward, and W. P. Inskeep. 2006. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl. Environ. Microbiol. 72:6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hatfull, G. F. 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatfull, G. F., S. G. Cresawn, and R. W. Hendrix. 2008. Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res. Microbiol. 159:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatfull, G. F., et al. 2010. Comparative genomic analysis of sixty mycobacteriophage genomes: genome clustering, gene acquisition and gene size. J. Mol. Biol. 397:119-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatfull, G. F., et al. 2006. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiddema, R., M. D. Curran, N. P. Ferreira, J. N. Coetzee, and G. Lecatsas. 1985. Characterization of phages derived from strains of Rhodococcus australis and R. equii. Intervirology 23:109-111. [DOI] [PubMed] [Google Scholar]
- 33.Hondalus, M. K. 1997. Pathogenesis and virulence of Rhodococcus equi. Vet. Microbiol. 56:257-268. [DOI] [PubMed] [Google Scholar]
- 34.Horowitz, M. L., et al. 2001. Application of Sartwell's model (lognormal distribution of incubation periods) to age at onset and age at death of foals with Rhodococcus equi pneumonia as evidence of perinatal infection. J. Vet. Intern. Med. 15:171-175. [DOI] [PubMed] [Google Scholar]
- 35.Ishida, T., et al. 2009. Crystal structure of glycoside hydrolase family 55 β-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 284:10100-10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer, L. M., E. V. Koonin, D. D. Leipe, and L. Aravind. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33:3875-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kormanec, J., and D. Homerova. 1993. Streptomyces aureofaciens whiB gene encoding putative transcription factor essential for differentiation. Nucleic Acids Res. 21:2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kropinski, A. M., et al. 2007. The genome of epsilon15, a serotype-converting, group E1 Salmonella enterica-specific bacteriophage. Virology 369:234-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kropinski, A. M., D. Prangishvili, and R. Lavigne. 2009. Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ. Microbiol. 11:2775-2777. [DOI] [PubMed] [Google Scholar]
- 40.Larkin, M. J., L. A. Kulakov, and C. C. Allen. 2005. Biodegradation and Rhodococcus-masters of catabolic versatility. Curr. Opin. Biotechnol. 16:282-290. [DOI] [PubMed] [Google Scholar]
- 41.Larkin, M. J., L. A. Kulakov, and C. C. Allen. 2006. Biodegradation by members of the genus Rhodococcus: biochemistry, physiology, and genetic adaptation. Adv. Appl. Microbiol. 59:1-29. [DOI] [PubMed] [Google Scholar]
- 42.Lavigne, R., et al. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavigne, R., D. Seto, P. Mahadevan, H. W. Ackermann, and A. M. Kropinski. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406-414. [DOI] [PubMed] [Google Scholar]
- 44.Lu, M. J., and U. Henning. 1989. The immunity (imm) gene of Escherichia coli bacteriophage T4. J. Virol. 63:3472-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahowald, M. A., et al. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 106:5859-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchler-Bauer, A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLeod, M. P., et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meijer, W. G., and J. F. Prescott. 2004. Rhodococcus equi. Vet. Res. 35:383-396. [DOI] [PubMed] [Google Scholar]
- 49.Muscatello, G., G. A. Anderson, J. R. Gilkerson, and G. F. Browning. 2006. Associations between the ecology of virulent Rhodococcus equi and the epidemiology of R. equi pneumonia on Australian thoroughbred farms. Appl. Environ. Microbiol. 72:6152-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muscatello, G., et al. 2007. Rhodococcus equi infection in foals: the science of ‘rattles’. Equine Vet. J. 39:470-478. [DOI] [PubMed] [Google Scholar]
- 51.Naryshkina, T., et al. 2006. Thermus thermophilus bacteriophage phiYS40 genome and proteomic characterization of virions. J. Mol. Biol. 364:667-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nordmann, P., M. Keller, F. Espinasse, and E. Ronco. 1994. Correlation between antibiotic resistance, phage-like particle presence, and virulence in Rhodococcus equi human isolates. J. Clin. Microbiol. 32:377-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park, T., D. K. Struck, C. A. Dankenbring, and R. Young. 2007. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. J. Bacteriol. 189:9135-9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Payne, K., Q. Sun, J. Sacchettini, and G. F. Hatfull. 2009. Mycobacteriophage Lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 73:367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Payne, R. J., and V. A. Jansen. 2001. Understanding bacteriophage therapy as a density-dependent kinetic process. J. Theor. Biol. 208:37-48. [DOI] [PubMed] [Google Scholar]
- 56.Pedulla, M. L., et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 57.Prescott, J. F. 1991. Rhodococcus equi: an animal and human pathogen. Clin. Microbiol. Rev. 4:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahman, M. T., et al. 2003. Partial genome sequencing of Rhodococcus equi ATCC 33701. Vet. Microbiol. 94:143-158. [DOI] [PubMed] [Google Scholar]
- 59.Resch, G., E. M. Kulik, F. S. Dietrich, and J. Meyer. 2004. Complete genomic nucleotide sequence of the temperate bacteriophage Aa Phi 23 of Actinobacillus actinomycetemcomitans. J. Bacteriol. 186:5523-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rutherford, K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 61.Schwudke, D., et al. 2008. Broad-host-range Yersinia phage PY100: genome sequence, proteome analysis of virions, and DNA packaging strategy. J. Bacteriol. 190:332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siew, N., and D. Fischer. 2003. Twenty thousand ORFan microbial protein families for the biologist? Structure 11:7-9. [DOI] [PubMed] [Google Scholar]
- 63.Steven, A. C., et al. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 64.Summer, E. J. 2009. Preparation of a phage DNA fragment library for whole genome shotgun sequencing. Methods Mol. Biol. 502:27-46. [DOI] [PubMed] [Google Scholar]
- 65.Summer, E. J., et al. 2007. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 373:1098-1112. [DOI] [PubMed] [Google Scholar]
- 66.Summer, E. J., et al. 2010. Genomic and biological analysis of phage Xfas53 and related prophages of Xylella fastidiosa. J. Bacteriol. 192:179-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Summer, E. J., J. J. Gill, C. Upton, C. F. Gonzalez, and R. Young. 2007. Role of phages in the pathogenesis of Burkholderia, or ‘Where are the toxin genes in Burkholderia phages?’. Curr. Opin. Microbiol. 10:410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Summer, E. J., et al. 2006. Divergence and mosaicism among virulent soil phages of the Burkholderia cepacia complex. J. Bacteriol. 188:255-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Summer, E. J., et al. 2004. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J. Mol. Biol. 340:49-65. [DOI] [PubMed] [Google Scholar]
- 70.Sutcliffe, I. C. 1998. Cell envelope composition and organisation in the genus Rhodococcus. Antonie Van Leeuwenhoek 74:49-58. [DOI] [PubMed] [Google Scholar]
- 71.Sutcliffe, I. C. 1997. Macroamphiphilic cell envelope components of Rhodococcus equi and closely related bacteria. Vet. Microbiol. 56:287-299. [DOI] [PubMed] [Google Scholar]
- 72.Takai, S., et al. 2001. Prevalence of virulent Rhodococcus equi in soil from five R. equi-endemic horse-breeding farms and restriction fragment length polymorphisms of virulence plasmids in isolates from soil and infected foals in Texas. J. Vet. Diagn. Invest. 13:489-494. [DOI] [PubMed] [Google Scholar]
- 73.Takai, S., et al. 1995. Identification of virulent Rhodococcus equi by amplification of gene coding for 15- to 17-kilodalton antigens. J. Clin. Microbiol. 33:1624-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tettelin, H., et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950-13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tettelin, H., D. Riley, C. Cattuto, and D. Medini. 2008. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11:472-477. [DOI] [PubMed] [Google Scholar]
- 76.Thomas, J. A., J. A. Soddell, and D. I. Kurtboke. 2002. Fighting foam with phages? Water Sci. Technol 46:511-518. [PubMed] [Google Scholar]
- 77.Torsvik, V., and L. Ovreas. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240-245. [DOI] [PubMed] [Google Scholar]
- 78.West, N. P., et al. 2009. Cutinase-like proteins of Mycobacterium tuberculosis: characterization of their variable enzymatic functions and active site identification. FASEB J. 23:1694-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whittaker, C. A., and R. O. Hynes. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13:3369-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu, J., R. W. Hendrix, and R. L. Duda. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21. [DOI] [PubMed] [Google Scholar]
- 81.Yoshida, T., et al. 2008. Ma-LMM01 infecting toxic Microcystis aeruginosa illuminates diverse cyanophage genome strategies. J. Bacteriol. 190:1762-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Young, R., and I. N. Wang. 2006. Phage lysis, p. 104-126. In R. Calendar (ed.), The bacteriophages, 2nd ed. Oxford University Press, Oxford, United Kingdom.