Abstract
Two unchlorinated drinking water supplies were investigated to assess the potential of water treatment and distribution systems to support the growth of Legionella spp. The treatment plant for supply A distributed treated groundwater with a low concentration (<0.5 ppm of C) of natural organic matter (NOM), and the treatment plant for supply B distributed treated groundwater with a high NOM concentration (8 ppm of C). In both supplies, the water temperature ranged from about 10°C after treatment to 18°C during distribution. The concentrations of Legionella spp. in distributed water, analyzed with quantitative PCR (Q-PCR), averaged 2.9 (± 1.9) × 102 cells liter−1 in supply A and 2.5 (± 1.6) × 103 cells liter−1 in supply B. No Legionella was observed with the culture method. A total of 346 clones (96 operational taxonomical units [OTUs] with ≥97% sequence similarity) were retrieved from water and biofilms of supply A and 251 (43 OTUs) from supply B. The estimation of the average value of total species richness (Chao1) in supply A (153) was clearly higher than that for supply B (58). In each supply, about 77% of the sequences showed <97% similarity to described species. Sequences related to L. pneumophila were only incidentally observed. The Legionella populations of the two supplies are divided into two distinct clusters based on distances in the phylogenetic tree as fractions of the branch length. Thus, a large variety of mostly yet-undescribed Legionella spp. proliferates in unchlorinated water supplies at temperatures below 18°C. The lowest concentration and greatest diversity were observed in the supply with the low NOM concentration.
About 6,000 cases of Legionnaires' disease (LD) have been reported annually in Europe in recent years (19). The bacterium Legionella pneumophila is the causative agent in more than 90% of the LD cases (13, 19, 20). This organism is capable of thriving in a wide variety of natural freshwater habitats and in man-made water systems, including cooling towers, whirlpools, and installations for warm tap water (4, 5, 13, 14, 29, 34, 37). Currently, the genus Legionella comprises about 52 described species, and this number continues to increase (22, 23, 30). In recent years, cultivation-independent techniques revealed the presence of a large variety of Legionella (sequence) types in water (5, 8, 40). Most of these Legionella types have not been cultured and were identified by PCR and sequence analysis. Water temperature may be an important selective condition for the occurrence of these Legionella spp. in the environment (40).
In 1999, a large LD outbreak occurred among visitors to a flower show in the Netherlands (10). An unchlorinated whirlpool on display at the entrance of the show was identified as the source of amplification and exposure. This outbreak and observations of Legionella in water installations led to the initiation of investigations to determine the role of drinking water in disseminating these bacteria. A previous study using quantitative PCR (Q-PCR) revealed that treated groundwater and treated surface water contained a large variety of uncultured Legionella spp. in concentrations ranging from 1.1 × 103 to 7.8 × 105 cells liter−1 (40). The concentration of Legionella spp. in treated surface water was significantly higher than that in treated groundwater.
Sources for the drinking water supply in the Netherlands are groundwater (two-thirds) and surface water. Multiple barriers are applied in the treatment of surface water to remove contaminants, pathogenic microorganisms, and biodegradable organic compounds to achieve safe and biologically stable drinking water (39). Nevertheless, the concentrations of natural organic matter (NOM) in treated water vary extensively among different types of drinking water, with levels ranging from less than 0.5 ppm of C to about 8 ppm of C.
The objective of this study was to assess the potential of water supply systems to support the growth of Legionella by determining the concentrations and diversity of Legionella spp. in treated water and in the distribution systems of two groundwater treatment systems. Both systems distributed unchlorinated water, one with a low and one with a high concentration of NOM.
MATERIALS AND METHODS
Selected water supplies.
Two groundwater supplies were selected for the study, supply A, with an annual production of 5.6 × 106 m3 of drinking water, and supply B, with a production of 2.4 × 107 m3. For supply A, aerobic groundwater (oxygen concentration, 6.5 mg liter−1) with low concentrations of NOM (<0.5 ppm of C), methane (<0.01 mg liter−1), ammonia (<0.05 mg liter−1), iron (0.3 mg liter−1), and manganese (0.5 mg liter−1) is treated by aeration and limestone filtration for removal of CO2. For supply B, anaerobic groundwater (oxygen concentration, < 0.5 mg liter−1) with relatively high concentrations of NOM (8 ppm of C), methane (39 mg liter−1), ammonia (3.4 mg liter−1), iron (11.8 mg liter−1), and manganese (0.5 mg liter−1) is abstracted. Water treatment consists of intensive aeration, rapid sand filtration, and dosing with NaOH, followed by pellet softening, aeration, and a second stage of rapid sand filtration. The treated water of both treatment facilities is distributed without a disinfectant.
Sampling sites.
Raw-water samples of supply A were collected from four main wells feeding the treatment plant. The water was collected directly after abstraction and before it was transported to the treatment plant. Anaerobic groundwater of supply B was collected from the raw-water pipe before entering the treatment plant. Water samples were also collected after each treatment step. Treated water from both supplies was collected six times in the period from February until October to be analyzed for seasonal effects. Segments (about 30 cm) of unplasticized polyvinyl chloride (PVC) pipes (diameter, 110 mm) were taken from the distribution systems at various distances from the treatment facilities for biofilm analysis. A total of seven segments were removed from distribution system A and eight from distribution system B. Before being removed, the external surface of each segment was thoroughly cleaned, and the segment was subsequently placed in a plastic container filled with drinking water and transported to the laboratory. At the selected locations, water samples (2 liters) were collected in sterile glass containers. These samples were stored on ice and transported to the laboratory.
Sample processing.
The water samples were processed within 24 h as previously described (40). A volume of about 500 ml of each water sample was filtered. An area of approximately 10 cm2 of the inner surface of each pipe segment was thoroughly swept with three sterile cotton swabs, which were subsequently placed in 10 ml of phosphate-buffered saline. Biomass was suspended by four low-energy sonification cycles of 2 min each in separate 10-ml volumes, yielding 40 ml of a suspension for further analysis. For the extraction of DNA, 20 ml of the suspension was concentrated by filtration (0.2 μm) and processed. The efficiencies of the filtration and DNA extraction and the inhibition of the PCR were analyzed by spiking an additional sample of each water and biofilm sample with 1.98 × 104 L. pneumophila cells. The cell suspension used for spiking was enumerated using acridine orange staining and fluorescence microscopy. Total concentrations of active biomass (ATP) in water and biofilm samples, representing the active microbial biomass, were determined by ATP analysis as described previously (26).
Detection of Legionella.
BCYE medium was used to detect culturable Legionella bacteria (11, 17). Sample concentrates without and with heat treatment (30 min at 50°C) were incubated on BCYE medium. DNA of Legionella spp. was detected by applying the primers LEG-225 and LEG-858, which amplify approximately 654 bp of the 16S rRNA gene (27). L. pneumophila was detected using a newly developed real-time TaqMan PCR assay with primers LpneuF (5′-CCGATGCCACATCATTAGC-3′) and LpneuR (5′-CCAATTGAGCGCCACTCATAG-3′) and a TaqMan probe, LpneuP (5′- 6-carboxyfluorescein [FAM]-TGCCTTTAGCCATTGCTTCCG-BHQ1-3′), which target the mip gene. The amplification mixture for the detection of Legionella spp. consisted of 25 μl of iQ SYBR green supermix (Bio-Rad Laboratories B.V., Veenendaal, Netherlands), 0.4 mg ml−1 of bovine serum albumin (BSA; PCR grade; Roche Diagnostics, Almere, Netherlands), 0.2 μM each primer, and 10 μl of DNA template in a total reaction volume of 50 μl. The amplification mixture for the detection of L. pneumophila using primers LpneuF and LpneuR and the probe LpneuP consisted of 25 μl of iQ supermix (Bio-Rad Laboratories B.V., Veenendaal, Netherlands), 0.4 mg ml−1 BSA, and 0.2 μM each primer and probe. The undiluted sample and a 10-fold dilution of each sample were analyzed in duplicate by Q-PCR. The concentrations of Legionella spp. or L. pneumophila DNA in water and biofilm were determined using a calibration curve obtained with known concentrations of L. pneumophila cells. The results of L. pneumophila detection in the spiked sample were used to calculate the percentages of spiked L. pneumophila cells recovered. The spiked samples were extracted in the same experiment, and the DNA of the reference suspension was used to calculate the calibration curve. Detection by Q-PCR and data analysis were performed with an iCycler iQ real-time PCR detection system (Bio-Rad Laboratories B.V., Veenendaal, Netherlands).
Cloning and sequence analysis.
The product of PCR using primers LEG 225 and LEG 858 on the undiluted DNA sample of the Legionella spp. was cloned into a pGEM-T Easy vector system (Promega, Leiden, Netherlands) according to the instructions of the manufacturer. Clone libraries were constructed for samples of aerobic groundwater and treated water of supply A, for samples of treated water of supply B, and for selected biofilm samples from both distribution systems. The clone libraries of groundwater of supply A were constructed using the Legionella PCR product amplified with DNA extracted from a 2-liter water sample. Clone inserts were sequenced in both forward and reverse directions with the primers T7 and SP6 (BaseClear, Leiden, Netherlands). The forward and reverse sequences of each analyzed cloned 16S rRNA gene were assembled and edited by using the software package Lasergene SeqMan II (DNAStar Inc., Madison, WI). The vector sequences were removed at both sides of the clone sequences, resulting in a final sequence of approximately 614 bp.
Sequence data analysis.
A BLASTN search (1) was performed to analyze the similarity of the clone sequences to all deposited 16S rRNA gene sequences. All sequences were included in the database of the ARB software package (25) and optimally aligned using the fast-aligner module. The number of operational taxonomical units (OTUs) and the total species richness (Chao1) (6) were determined using the software program DOTUR (33). OTUs were defined as sequences with at least 97% similarity (36).
The UniFrac program (24) was applied to measure the phylogenetic distances between the Legionella populations in the phylogenetic tree as fractions of the branch length of the tree. The Legionella populations were clustered by hierarchical clustering analysis (unweighted pair group method with arithmetic mean [UPGMA]) of the populations based on a distance matrix. For this purpose, a de novo parsimony tree was calculated in ARB with 82 full-length 16S rRNA gene sequences of 46 different Legionella species (>1,400 bp) and three out-group sequences (Piscirickettsia salmonis, Wolbachia persica, and Coxiella burnetii). The parsimony tree was calculated with the gamma_1_rr5_dec04 filter included in the ARB package, using positions 1106 to 42627. With the “quick add” option, all retrieved and analyzed Legionella clone sequences were added to the tree without a filter. Subsequently, all Legionella reference sequences were removed from the tree, leaving the clone sequences and the out-group sequences. This tree was imported into the UniFrac program. An environmental file in which each clone sequence was linked to the original sample from which it was isolated was created. These two files were used in the UniFrac program to cluster the clones of each environment. Jackknife values were calculated to determine how the numbers and evenness of sequences in the different samples affected the hierarchical clustering analysis. The values were calculated using 100 permutations and a minimum set of 46 sequences.
Nucleotide sequence accession numbers.
All partial 16S rRNA gene sequences determined in this study have been deposited in GenBank under accession numbers HQ111557 to HQ1112153.
RESULTS
Water temperature and active biomass.
Throughout the year, the temperature of treated groundwater ranged from 9.5 to 11.5°C in supply A and from 10.0 to 13.5°C in supply B (Table 1). The water temperatures at the sampling locations in the distribution systems were slightly higher in July, viz., 14.8 ± 2.3°C in supply A and 13.6 ± 1.5°C in supply B. Total ATP concentrations in treated water and in distributed water of supply A were all below the detection limit of 1 ATP ng liter−1. In supply B, the ATP concentrations in treated water were 10.6 ± 4.9 ATP ng liter−1, and in distributed water, 4.7 ± 1.2 ng ATP liter−1.
TABLE 1.
Quality characteristics of treated water at the treatment plants of supply A and supply Ba
| Parameter | Mean values (range) for treated water supply A | Mean values (range) for treated water supply B |
|---|---|---|
| Temp (°C) | 10.0 (9.5-11.5) | 11.5 (10.0-13.5) |
| pH | 7.8 (7.2-8.2) | 7.6 (7.4-8.1) |
| O2 concn (mg liter−1) | 6.4 (5.6-7.8) | 5.9 (3.9-8.3) |
| HCO3 concn (mg liter−1) | 98 (92-124) | 282 (273-308) |
| Cl concn (mg liter−1) | 13 (11-14) | 28 (27-31) |
| Ca concn (mg liter−1) | 35.4 (32.9-39.6) | 32.7 (25.7-52.8) |
| Mg concn (mg liter−1) | 2.4 (2.1-2.7) | 9.7 (8.4-10.9) |
| Total hardness (mmol liter−1) | 1.0 (0.9-1.1) | 1.2 (1.0-1.7) |
| Fe concn (μg liter−1) | <20 (<20-<20) | 25 (<20-73) |
| Mn concn (μg liter−1) | <10 (<10-<10) | <10 (<10-<10) |
| SO4 concn (mg liter−1) | 16 (13-19) | <10 (<10-<10) |
| NH4 concn (mg liter−1) | <0.05 (<0.05-<0.05) | <0.05 (<0.05-<0.05) |
| NPOC concn (mg C liter−1) | 0.33 (<0.3-0.49) | 7.9 (7.6-8.3) |
Values are based on routine monitoring over the period of a year.
Concentrations of Legionella spp. in water and biofilm.
Cultivable Legionella spp. were not observed in any of the water samples (<50 CFU liter−1) nor in the biofilm (<6 CFU cm−2). With Q-PCR, Legionella spp. were detected at concentrations ranging from 7.6 × 101 ± 1.2 × 101 to 3.9 × 102 ± 2.5 × 102 cells liter−1 in three of four samples of aerobic groundwater collected from different wells for supply A (Fig. 1 A). Limestone filtration caused a clear increase in the concentration of Legionella spp. in the treated water. No seasonal effect on the Legionella concentration was observed. L. pneumophila was not detected (detection limit, 1.7 × 101 cells liter−1) in any of the samples of groundwater and treated water of supply A. Legionella spp. were detected in six of seven biofilm samples from pipe segments removed from the distribution system of supply A at concentrations ranging from 1.8 ± 1.7 to 9.4 ± 4.2 cells cm−2 (average concentration, 5.2 ± 2.6 cells cm−2). Legionella concentrations in the water sampled at these locations ranged from 1.3 × 102 to 4.9 × 102 cells liter−1 (average, 2.9 [± 1.9] × 102 cells liter−1). The average concentration in distributed water was significantly lower than the average concentration in treated water (P < 0.05). L. pneumophila was detected in one biofilm sample, at a concentration of 5.9 cells cm−2 (detection limit, 1 cells cm−2), but was below the detection limit in all water samples collected from the distribution system (<2.0 × 101 cells liter−1).
FIG. 1.
Concentrations of Legionella spp. in water and in biofilms in two groundwater supplies. (A) Supply A (treated aerobic groundwater). (B) Supply B (treated anaerobic groundwater). Abbreviations: W1, W2, W3, and W4, raw-water wells; RW, raw water (combined); A, aeration; RSF, rapid sand filtration; PS, pellet softening; TW, treated water; D, water and biofilm sampled from distribution system. Bars show concentrations with standard deviations in duplicate samples, and for TW, with standard deviations in 8 (A) and 6 (B) samples collected over a period of 9 months. Open bars, water samples; solid bars, biofilm samples; stars, Legionella concentrations below the detection limit in water and in biofilm (D2).
Legionella spp. were not detected in the anaerobic groundwater entering treatment plant B nor in the water sampled directly after aeration (Fig. 1B). The concentration of 2.2 × 104 ± 8.2 × 103 cells liter−1 of Legionella spp. observed in water directly after rapid sand filtration indicates that Legionella spp. multiplied in the filter bed. Further treatment steps (softening and a second rapid sand filtration) had no clear effect on the concentration of Legionella spp. in the water. Seasonal fluctuations of the Legionella concentration were not observed in the treated water of supply B. Furthermore, the concentration of L. pneumophila was below the detection limit (<2.0 × 101 cells liter−1) in water during and after treatment.
Legionella spp. were detected in all eight biofilm samples from the distribution system of supply B at concentrations ranging from 4.6 to 3.9 × 102 cells cm−2. In water samples collected from the distribution system, concentrations of Legionella spp. ranged from 5.7 × 102 to 5.7 × 103 cells liter−1, and the average concentration (2.5 [± 1.6] × 103 cells liter−1) was significantly lower (P < 0.05) than the average concentration in treated water. L. pneumophila was detected in seven of eight biofilm samples at concentrations ranging from 9.2 cells cm−2 (in one of the duplicate samples) to 4.9 × 101 ± 2.0 cells cm−2 but in none of the water samples (detection limit, 1.3 × 102 cells liter−1). Also in supply B, no cultivable Legionella spp. were observed in any of the samples of water (<50 CFU liter−1) and biofilm (<6 CFU cm−2).
Recovery of the spiked L. pneumophila cells ranged from 32 to 61% for groundwater to 55% ± 15% for the treated water of supply A. The efficiency of the analysis for raw groundwater of supply B was 14% and for aerated water only 2%, suggesting a substantial inhibition of the PCR. Recovery ranged from 12 to 28% for the samples of supply B taken after each treatment step to 32% ± 11% for the samples of treated water. Recovery was relatively high in the biofilm samples from the distribution systems of supplies A and B and slightly above 100% in three samples, possibly due to experimental variation.
Identification of Legionella spp. in water and in biofilm.
Analysis of clones retrieved from supply A resulted in 346 16S rRNA gene sequences (Table 2), and 251 sequences were obtained from supply B. All these sequences exhibited the highest levels of similarity to 16S rRNA sequences of Legionella spp. present in the GenBank database (Table 3). The clone sequences obtained from supply A were identified either to the species level (22%) or to previously defined OTUs (38%) (40) (Table 2; see the supplemental material). Sequences related to L. bozemanae predominated in clone libraries retrieved from the biofilm. Sequences related to L. pneumophila were found in both samples of untreated aerobic groundwater (eight clones) and also in the biofilm (three clones).
TABLE 2.
Identification of OTUs obtained from water supply Aa
| Species or type (GenBank accession no.)b | No. of sequences obtained from: |
Total no. (%) of sequences | |||||
|---|---|---|---|---|---|---|---|
| Raw water from well I | Raw water from well II | Treated water | Biofilm I | Biofilm II | Biofilm III | ||
| L. bozemanae | 1 | 10 | 15 | 26 (7.5) | |||
| Tag 1-4 (AY924177) | 5 | 8 | 6 | 19 (5.5) | |||
| Tang 7-4 (AY924082) | 3 | 11 | 1 | 15 (4.3) | |||
| Tag 3-4 (AY924175) | 9 | 3 | 1 | 1 | 14 (4.0) | ||
| L. pneumophila | 2 | 6c | 1 | 2 | 11 (3.2) | ||
| Tang 2-1 (AY924013) | 6 | 1 | 2 | 9 (2.6) | |||
| L. donaldsonii | 2 | 3 | 2 | 7 (2.0) | |||
| L. yabuuchiae | 6 | 1 | 7 (2.0) | ||||
| Sb 1-5 (AY924101) | 2 | 4 | 1 | 7 (2.0) | |||
| Tang 9-3 (AY924090) | 2 | 3 | 2 | 7 (2.0) | |||
| Tag 5-3 (AY924184) | 1 | 5 | 6 (1.7) | ||||
| Tsw 1-2 (AY923986) | 5 | 1 | 6 (1.7) | ||||
| S 7-9 (AY924171) | 5 | 5 (1.4) | |||||
| Tag 5-4 (AY924185) | 1 | 3 | 1 | 5 (1.4) | |||
| Tag 6-1 (AY924186) | 1 | 1 | 3 | 5 (1.4) | |||
| Tang 10-4 (AY924095) | 3 | 2 | 5 (1.4) | ||||
| L. anisa | 3 | 1 | 4 (1.2) | ||||
| L. lytica | 4 | 4 (1.2) | |||||
| Tsw 7-4 (AY924046) | 1 | 1 | 1 | 1 | 4 (1.2) | ||
| Sum of OTUs with <1% of total sequencesd | 6 | 8 | 4 | 7 | 10 | 9 | 44 (12.7) |
| Total no. of isolated clones | 48 | 46 | 94 | 54 | 49 | 55 | 346 |
| Total no. (%) of clones identified | 27 (56.3) | 31 (67.4) | 41 (43.6) | 32 (59.3) | 42 (85.7) | 37 (67.3) | 210 (60.7) |
| Total no. of OTUs | 23 | 16 | 25 | 25 | 23 | 20e | |
OTUs were defined as sequences with at least 97% similarity. The table is arranged with OTUs in decreasing order of occurrence.
Sequence types identified as Tag (treated aerobic/oxic groundwater), Tang (treated anaerobic/anoxic groundwater), Tsw (treated surface water), S (surface water), and Sb (surface water basin) were isolated in a previous study (40).
One clone sequence showed a sequence similarity of 99% to sequence Tag 4-3 (AY924179).
See the supplemental material for details.
Sequence similarity of 98%.
TABLE 3.
Identification of OTUs obtained from water supply Ba
| Species or type (GenBank accession no.)b | No. of sequences obtained from: |
Total no. (%) of sequences | |||
|---|---|---|---|---|---|
| Treated water | Biofilm I | Biofilm II | Biofilm III | ||
| Tang 5-5 (AY924055) | 47 | 1 | 48 (19.1) | ||
| Tsw 3-3 (AY924006) | 27 | 5 | 5 | 37 (14.7) | |
| L. worsleiensis | 14 | 2 | 4 | 20 (8.0) | |
| L. pneumophila | 5 | 5 | 3 | 2 | 15 (6.0) |
| L. anisa | 5 | 7 | 2 | 14 (5.6) | |
| Tang 3-1 (AY924018) | 12 | 12 (4.8) | |||
| Sb 2-1 (AY924107) | 11 | 1 | 12 (4.8) | ||
| L. bozemanae | 4 | 2 | 1 | 1 | 8 (3.2) |
| Tsw 8-5 (AY924059) | 3 | 1 | 4 (1.6) | ||
| LLAP 11 | 1 | 1 | 1 | 3 (1.2) | |
| L. adelaidensis | 1 | 2 | 3 (1.2) | ||
| Sum of OTUs with <1% of total no. of sequencesc | 1 | 1 | 1 | 9 | 12 (4.8) |
| Total no. of isolated clones | 96 | 50 | 53 | 52 | 251 |
| Total no. (%) of clones identified | 89 (92.7) | 48 (96.0) | 24 (45.3) | 27 (51.9) | 188 (74.9) |
| Total no. of OTUs | 10 | 7d | 16 | 21 | |
OTUs were defined as sequences with at least 97% similarity.
Sequence types identified as Tang (treated anaerobic/anoxic groundwater), Tsw (treated surface water), and Sb (surface water basin) were isolated in a previous study (40).
See the supplemental material for details.
Sequence similarity of 98%.
The clones obtained from supply B could be identified either as a described species (25%) or as a previously defined OTU (50%) (Table 3; see details in the supplemental material). A total of 47 clones (49%) isolated from treated water and 1 clone from the biofilm showed ≥97% sequence similarity to clone type Tang 5-5, previously isolated from treated water of a similar groundwater supply (40). Sequence type Tsw 3-3, isolated from treated surface water, predominated in the clone library of biofilm I with 27 (54%) clones. Sequences related to L. pneumophila were found in all libraries in numbers ranging from two to five clones.
All L. pneumophila-related sequences (>97%) were added to a de novo phylogenetic tree calculated with 82 complete 16S rRNA gene sequences of 46 described Legionella species (Fig. 2). Two clone sequences isolated from untreated groundwater of supply A had similarities of 100% (La-RwB50) and 99% (La-RwB51) to L. pneumophila sequences in the ARB database. Clones La-BfA47 and La-BfA48 were positioned just outside the L. pneumophila cluster, with a sequence similarity of 98%, whereas all other L. pneumophila-related clone sequences were positioned outside the L. pneumophila cluster.
FIG. 2.
Phylogenetic tree showing positions of the partial 16S rRNA gene sequences with ≥97% sequence similarities to described L. pneumophila sequences. A de novo parsimony tree was calculated in ARB with 82 16S rRNA gene sequences from described Legionella spp. (>1,400 bp) and three out-group sequences (Piscirickettsia salmonis, Wolbachia persica, and Coxiella burnetii). The clone sequences were added to the tree using the “Quick add” tool. A number of reference sequences were removed from the final tree to reduce its size.
Legionella diversity and clustering.
The libraries of clones retrieved from supply A yielded a higher number of OTUs than the libraries of clones retrieved from supply B (Table 4). Also, the total richness of Legionella-related OTUs, estimated with Chao1, is clearly higher in supply A than in supply B. The clone sequences retrieved from supplies A and B included a total of 130 different OTUs, with an estimated average total richness of 198 OTUs.
TABLE 4.
Numbers of retrieved clones and OTUs and richness of Legionella-related sequences with similarities of >97% in water supplies A and B
| Origin of sample | No. of clones | No. of OTUs | Total species richnessb |
||
|---|---|---|---|---|---|
| Avg | Lower limit | Upper limit | |||
| Raw water supply A | 94 | 33 | 42 | 35 | 71 |
| Treated water supply A | 94 | 25 | 30 | 26 | 52 |
| Biofilm supply A | 158 | 53 | 108 | 68 | 256 |
| Total supply A | 346 | 96 | 153 | 120 | 229 |
| Treated water supply B | 96 | 10 | 14 | 10 | 56 |
| Biofilm supply B | 155 | 36 | 42 | 38 | 74 |
| Total supply B | 251 | 43 | 58 | 47 | 98 |
| Supply A + supply B | 597 | 130 | 198 | 162 | 274 |
| Supply A + supply B + treated water from previous researcha | 721 | 161 | 218 | 190 | 274 |
See reference 40.
Chao1 estimation.
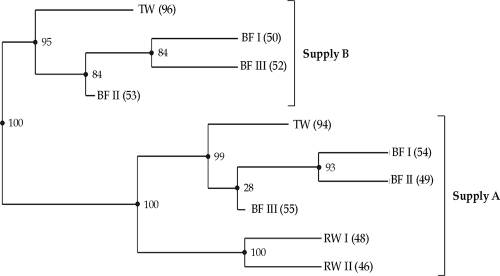
The library of clones retrieved from treated water of supply A included 25 OTUs; the highest contribution of a single OTU (Tag 3-4) was 9.5% (Table 2). However, one specific OTU (Tang 5-5) represented about 50% of the sequences retrieved from treated water of supply B (Table 3). L. bozemanae comprised 20 to 27% of the sequences in the clone libraries of two biofilm samples of supply A but represented only a few percent of the biofilms in supply B. Only a few sequences of clones retrieved from biofilm samples of supply B showed >97% sequence similarities to the clone sequences from supply A. Calculation of the phylogenetic distances between the Legionella populations in the phylogenetic tree as fractions of the branch length resulted in two distinct clusters (Fig. 3). The high jackknife value of the major nodes shows the robustness of the presented topography of the cluster. The Legionella populations in the raw groundwater of supply A subclustered with the populations in treated water and the biofilms of supply A. Clone sequences retrieved from the biofilms did not cluster with those from treated water of supplies A and B, indicating that the Legionella communities in the biofilms differ genetically from those in treated water. Hence, water treatment and water composition affected both the concentration and the genetic diversity of Legionella bacteria in treated water and in distribution system biofilms. The lowest concentrations and the greatest diversity were observed in the supply with the low NOM concentration.
FIG. 3.
UPGMA clusters of Legionella populations in aerobic groundwater, treated aerobic and anaerobic groundwater, and biofilm samples from pipe segments of the distribution network. The number of sequences that represents each environment is indicated next to the name. Calculated jackknife values (percentages) are indicated in each node. RW, raw water, TW, treated water; BF, biofilm from distribution system.
DISCUSSION
Genetic diversity of Legionella.
L. pneumophila, the causative organism of the large majority of LD cases, was not cultured from the samples collected in our study, but sequences with ≥97% similarities to this organism were observed in a few samples from supplies A and B (cf. Tables 2 and 3). Moreover, L. pneumophila was detected with Q-PCR in a number of biofilm samples of supply B at concentrations below the detection level of the culture method. The use of different targets for these methods, viz., 16S rRNA gene sequencing and the mip gene, confirms that sequences highly related to L. pneumophila were present at low concentrations in water at a temperature below 15°C. The retrieved sequences showed a high level of diversity and differed from commonly observed L. pneumophila sequences, which make laboratory contamination highly unlikely. Clone sequences that are positioned outside the L. pneumophila cluster exhibited sequence similarities of 97 to 99% to published L. pneumophila sequences (Fig. 2). Sequence-based typing (SBT) using seven genes shows that L. pneumophila is a highly diverse species (15, 31). Certain sequence types (ST) include either environmental isolates, isolates from patients, or both. Hence, differentiation based on the sequence of the 16S rRNA fragment does not give information about a possible health risk related to the presence of L. pneumophila.
The observations of uncultured Legionella spp. in the two groundwater supplies confirm that such organisms are a natural component of the microbial communities of treated water at temperatures below 15°C (40). Uncultured, yet-unidentified Legionella spp. have been observed in a variety of freshwater environments, e.g., Antarctic lakes, treated water, and geothermal habitats (4, 5, 8, 9, 40). In the present study, Legionella spp. were detected using the Legionella-specific primers LEG 225 and LEG 858, which target the 16S rRNA gene and are commonly used to study Legionella spp. in the environment (2, 4, 5, 32). We observed a greater diversity of Legionella spp. in all water and biofilm samples (Table 4). Approximately 43% of the clone sequences showed the highest levels of similarity (>97%) to clone sequences obtained from samples of treated water in the previous study (40). Still, a total of 33% of the sequences retrieved from water and biofilm samples were not related to previously retrieved sequences (5, 40). The Chao1 estimation yielded an average species richness of 198 OTUs for supplies A and B. Combining the clone sequences of this study with sequences of clones previously retrieved from samples of treated water (40) resulted in an average species richness of 218 OTUs (Table 4). The small difference between the values for estimated species richness suggests that most OTUs related to Legionella spp. in drinking water at temperatures <18°C have been obtained. However, observation of different sequences in other freshwater studies indicates that the total diversity of Legionella spp. in fresh water environments is much greater (35). Hence, classification of species of the genus Legionella in the aquatic environment remains a challenge. Two further main questions are (i) which conditions in the aquatic environment determine the concentration of Legionella bacteria, and (ii) which conditions determine the nature of the predominant Legionella species.
Legionella concentrations in water and biofilm.
Multiplication of L. pneumophila and other defined Legionella spp. depends on protozoan hosts (21). The observation of a number of Legionella-like amoebal pathogen (LLAP) types (cf. Table 2 and 3) indicates that the uncultured Legionella spp. also need protozoan hosts for proliferation. In water supplies A and B, a large variety of free-living protozoa, including species serving as hosts for L. pneumophila, e.g., Hartmannella vermiformis, was observed in a study conducted simultaneously with this investigation (38). However, the identities of protozoa serving as hosts for the observed Legionella spp. remain unknown. The availability of host protozoa most likely depends on conditions affecting bacterial growth, e.g., the presence of oxygen, the concentration and nature of biodegradable compounds, hydraulic conditions, and temperature.
Uncultured Legionella spp. were not observed in anaerobic groundwater (supply B), confirming that these bacteria and their protozoan hosts require oxygen for growth. Aeration of the anaerobic groundwater at a temperature of about 10°C in the treatment facility of supply B did not lead to proliferation of Legionella spp., but a strong increase was observed in water after subsequent sand filtration (Figure 1). This increase may be attributed to biofilm formation in the filter beds induced by the presence of NOM, ammonia, and methane in the raw water. Additional treatment steps for supply B did not clearly affect the total Legionella concentration, which remained at a level of about 104 cells liter−1. The presence of Legionella spp. in the aerobic groundwater of supply A suggests that biological processes in the sand aquifer support the growth of these bacteria, despite the high level of oligotrophic conditions (NOM, <0.5 ppm of C). Legionella spp. have also been observed in biofilms on surfaces exposed to water from surface water embankment filtration (12). The elevated concentration of Legionella spp. observed after limestone filtration may be attributed to more intensive biofilm formation as a result of a high level of hydraulic loading of the filter beds. Still, the concentration of Legionella spp. in treated water of supply A was about 1 log unit below the concentration in supply B, which is consistent with the difference in the concentration of ATP in treated water of these supplies. The large difference in ATP concentrations between the two types of treated water corresponds with the large difference in NOM concentrations. Therefore, differences between the concentrations and levels of diversity of Legionella spp. in these supplies may be related to the difference in NOM concentrations.
Higher concentrations of uncultured Legionella spp. have been reported for drinking water produced from treated surface water, for lake water in the United States, and for treated sewage (14, 28, 40). Concentrations of L. pneumophila in warm-water installations generally range from 103 to 107 CFU liter−1 (3, 16). The concentration of NOM is one of the water quality parameters affecting the concentration of Legionella. The concentrations of Legionella spp. observed at low temperatures in supplies A and B may be indicative of the growth potential of culturable L. pneumophila and L. anisa in these water types at elevated temperatures. However, quantitative data are not available to demonstrate such a relationship.
Diversity of the Legionella community in relation to environmental conditions.
The temperature of water directly after treatment of supplies A and B ranged from 9.5 to 13.5°C throughout the year (Table 1). The increase in the concentration of Legionella spp. in water during treatment therefore suggests that the identified OTUs represent organisms which most likely had multiplied at these temperatures. L. pneumophila or L. anisa was observed in these environments but have been cultured from samples from installations for warm tap water and natural warm-water habitats. Legionella spp., e.g., L. birminghamensis, L. maceachernii, and L. parisiensis, and clone sequences retrieved from slow sand filtrate at 20°C (5) differed from those found in the present investigation and in treated water at 10 to 15°C in an earlier study (40). Furthermore, Legionella spp. observed in a geothermic habitat at 30°C, with L. sainthelensi predominating, differed from the species observed at 35 and 38°C (35). These observations indicate that temperature differences of 5 to 10°C have a clear impact on the Legionella species predominating in specific aquatic habitats.
A specific clone type (Tang 5-5), previously retrieved from treated anaerobic groundwater (40), predominated in the treated water of supply B. The clone library of treated water of supply A is also dominated by certain sequence types previously obtained from treated aerobic groundwater (Tag sequences). Furthermore, a specific clone type that had been retrieved from treated surface water (TSW 3-3) predominated in two biofilm samples from the distribution system of supply B. These results suggest that certain yet-unknown environmental conditions in different ecosystems support growth of the same Legionella species, which have been described as ecotypes (7).
All water and biofilm samples contained a great diversity of Legionella spp. (Tables 2 to 4). UPGMA clustering of the Legionella populations shows that different populations were observed in different water types and biofilms, although populations observed within a water supply clustered together (Fig. 3). Environmental conditions in treated water differ from those in biofilms in distribution systems and may be responsible for the observed differences in the community structures of Legionella. Table 4 shows that the average total species richness in the biofilm of supply A is clearly higher than that in the biofilm of supply B. This observation is consistent with the difference in richness of OTUs of free-living protozoa in the biofilms in these supplies (38). Apparently, a low concentration of NOM corresponds with greater diversity of Legionella and protozoa species compared to those in water with a high NOM concentration. This inverse relationship between the availability of food or energy sources and genetic diversity is consistent with observations on the diversity of (micro)organisms in natural aquatic environments (18).
In conclusion, this study confirmed that a great diversity of Legionella spp. is able to multiply at a low temperature in biofilms on surfaces exposed to treated water, even at a low NOM concentration.
Supplementary Material
Acknowledgments
This research was conducted within the framework of the Joint Research Program of Water Supply Companies in the Netherlands.
We are indebted to Remco Voogt for excellent technical assistance and to Paul van der Wielen for critical reading of the manuscript.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bobeldijk, I., A. Brandt, B. Wullings, and T. Noij. 2001. High-performance liquid chromatography—ToxPrint: chromatographic analysis with a novel (geno)toxicity detection. J. Chromatogr. A 918:277-291. [DOI] [PubMed] [Google Scholar]
- 3.Borella, P., et al. 2005. Legionella contamination in hot water of Italian hotels. Appl. Environ. Microbiol. 71:5805-5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, T., et al. 2004. Detection and identification of Legionella species from groundwaters. J. Toxicol. Environ. Health A 67:1845-1859. [DOI] [PubMed] [Google Scholar]
- 5.Calvo-Bado, L. A., J. A. Morgan, M. Sergeant, T. R. Pettitt, and J. M. Whipps. 2003. Molecular characterization of Legionella populations present within slow sand filters used for fungal plant pathogen suppression in horticultural crops. Appl. Environ. Microbiol. 69:533-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao, A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 7.Cohan, F. M., A. Koeppel, and D. Krizanc. 2005. Sequence-based discovery of ecological diversity within Legionella, p. 367-376. In N. Cianciotto (ed.), Legionella: state of the art 30 years after its recognition. ASM Press, Washington, DC.
- 8.Costa, J., I. Tiago, M. S. da Costa, and A. Verissimo. 2005. Presence and persistence of Legionella spp. in groundwater. Appl. Environ. Microbiol. 71:663-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Declerck, P., J. Behets, V. van Hoef, and F. Ollevier. 2007. Detection of Legionella spp. and some of their amoeba hosts in floating biofilms from anthropogenic and natural aquatic environments. Water Res. 41:3159-3167. [DOI] [PubMed] [Google Scholar]
- 10.Den Boer, J. W., et al. 2002. A large outbreak of Legionnaires' disease at a flower show, the Netherlands, 1999. Emerg. Infect. Dis. 8:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelstein, P. H. 1981. Improved selective medium for isolation of Legionella pneumophila from contaminated clinical and environmental samples. J. Clin. Microbiol. 14:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emtiazi, F., T. Schwartz, S. M. Marten, P. Krolla-Sidenstein, and U. Obst. 2004. Investigation of natural biofilms formed during the production of drinking water from surface water embankment filtration. Water Res. 38:1197-1206. [DOI] [PubMed] [Google Scholar]
- 13.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fliermans, C. B., et al. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaia, V., et al. 2005. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 43:2047-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habicht, W., and H. E. Muller. 1988. Occurrence and parameters of frequency of Legionella in warm water systems of hospitals and hotels in Lower Saxony. Zentralbl. Bakteriol. Mikrobiol. Hyg. B 186:79-88. [PubMed] [Google Scholar]
- 17.International Organization for Standardization. 2004. Detection and enumeration of Legionella. Part 2: direct membrane filtration for waters with low bacterial counts. ISO standard 11731-2:2004. International Organization for Standardization, Geneva, Switzerland.
- 18.Jiang, J. G., and Y. F. Shen. 2007. Development of the microbial communities in Lake Donghu in relation to water quality. Environ. Monit. Assess. 127:227-236. [DOI] [PubMed] [Google Scholar]
- 19.Joseph, C. A., and K. D. Ricketts. 2010. Legionnaires disease in Europe 2007-2008. Euro Surveill. 15:19493. [PubMed] [Google Scholar]
- 20.Joseph, C. A., K. D. Ricketts, and the European Working Group for Legionella Infections. 2005. Legionnaires' disease in Europe 2003-2004. Euro Surveill. 10:256-259. [DOI] [PubMed] [Google Scholar]
- 21.Kuiper, M. W., B. A. Wullings, A. D. Akkermans, R. R. Beumer, and D. van der Kooij. 2004. Intracellular proliferation of Legionella pneumophila in Hartmannella vermiformis in aquatic biofilms grown on plasticized polyvinyl chloride. Appl. Environ. Microbiol. 70:6826-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroki, H., et al. 2007. Legionella impletisoli sp. nov. and Legionella yabuuchiae sp. nov., isolated from soils contaminated with industrial wastes in Japan. Syst. Appl. Microbiol. 30:273-279. [DOI] [PubMed] [Google Scholar]
- 23.La Scola, B., et al. 2004. Legionella drancourtii sp. nov., a strictly intracellular amoebal pathogen. Int. J. Syst. Evol. Microbiol. 54:699-703. [DOI] [PubMed] [Google Scholar]
- 24.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig, W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magic-Knezev, A., and D. van der Kooij. 2004. Optimisation and significance of ATP analysis for measuring active biomass in granular activated carbon filters used in water treatment. Water Res. 38:3971-3979. [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto, H., et al. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer, C. J., et al. 1995. Detection of Legionella species in reclaimed water and air with the EnviroAmp legionella PCR kit and direct fluorescent antibody staining. Appl. Environ. Microbiol. 61:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer, C. J., Y. L. Tsai, C. Paszko-Kolva, C. Mayer, and L. R. Sangermano. 1993. Detection of Legionella species in sewage and ocean water by polymerase chain reaction, direct fluorescent-antibody, and plate culture methods. Appl. Environ. Microbiol. 59:3618-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, M. Y., K. S. Ko, H. K. Lee, M. S. Park, and Y. H. Kook. 2003. Legionella busanensis sp. nov., isolated from cooling tower water in Korea. Int. J. Syst. Evol. Microbiol. 53:77-80. [DOI] [PubMed] [Google Scholar]
- 31.Ratzow, S., V. Gaia, J. H. Helbig, N. K. Fry, and P. C. Luck. 2007. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 45:1965-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riffard, S., et al. 2001. Occurrence of Legionella in groundwater: an ecological study. Water Sci. Technol. 43:99-102. [PubMed] [Google Scholar]
- 33.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwartz, T., S. Hoffmann, and U. Obst. 1998. Formation and bacterial composition of young, natural biofilms obtained from public bank-filtered drinking water systems. Water Res. 32:2787-2797. [Google Scholar]
- 35.Sheehan, K. B., J. M. Henson, and M. J. Ferris. 2005. Legionella species diversity in an acidic biofilm community in Yellowstone National Park. Appl. Environ. Microbiol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 37.Tison, D. L., and R. J. Seidler. 1983. Legionella incidence and density in potable drinking water supplies. Appl. Environ. Microbiol. 45:337-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valster, R. M., B. A. Wullings, G. Bakker, H. Smidt, and D. van der Kooij. 2009. Free-living protozoa in two unchlorinated drinking water supplies, identified by phylogenic analysis of 18S rRNA gene sequences. Appl. Environ. Microbiol. 75:4736-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van der Kooij, D., J. H. M. Lieverloo, J. Schellart, and P. Hiemstra. 1999. Maintaining quality without a disinfectant residual. J. Am. Water Works Assoc. 91:55-64. [Google Scholar]
- 40.Wullings, B. A., and D. van der Kooij. 2006. Occurrence and genetic diversity of uncultured Legionella spp. in drinking water treated at temperatures below 15°C. Appl. Environ. Microbiol. 72:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.