Abstract
Nisin A is a pentacyclic peptide antibiotic produced by Lactococcus lactis. The leader peptide of prenisin keeps nisin inactive and has a role in inducing NisB- and NisC-catalyzed modifications of the propeptide and NisT-mediated export. The highly specific NisP cleaves off the leader peptide from fully modified and exported prenisin. We present here a detailed mutagenesis analysis of the nisin leader peptide. For alternative cleavage, we successfully introduced a putative NisP autocleavage site and sites for thrombin, enterokinase, Glu-C, and factor Xa in the C-terminal part of the leader peptide. Replacing residue F-18 with Trp or Thr strongly reduced production. On the other hand, D-19A, F-18H, F-18M, L-16D, L-16K, and L-16A enhanced production. Substitutions within and outside the FNLD box enhanced or reduced the transport efficiency. None of the above substitutions nor even an internal 6His tag from positions −13 to −8 had any effect on the capacity of the leader peptide to induce NisB and NisC modifications. Therefore, these data demonstrate a large mutational freedom. However, simultaneous replacement of the FNLD amino acids by four alanines strongly reduced export and even led to a complete loss of the capacity to induce modifications. Reducing the leader peptide to MSTKDFNLDLR led to 3- or 4-fold dehydration. Taken together, the FNLD box is crucial for inducing posttranslational modifications.
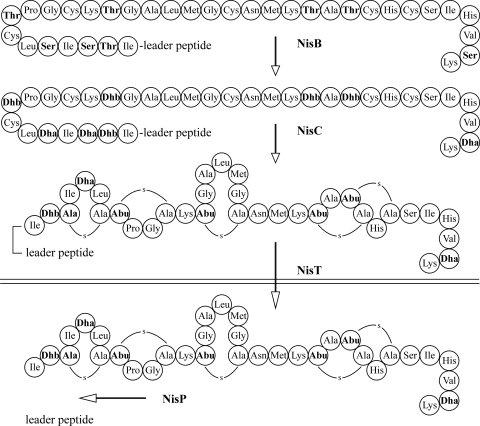
Nisin A is a pentacyclic peptide antibiotic produced by Lactococcus lactis (2, 9). It is applied worldwide as a food additive to prevent the growth of spoilage and pathogenic bacteria. Although it has already been widely used for a long time, virtually no resistance has been reported. Nisin is ribosomally synthesized as a 57-amino-acid prepeptide, comprising an N-terminal leader peptide of 23 amino acids (27). The leader peptide serves as a translocation signal, and it keeps the prepeptide inactive. In addition, it plays a dominant role in inducing the posttranslational modification of the C-terminal propeptide by interacting with the nisin modification enzymes (Fig. 1). The enzyme NisB dehydrates the serines and threonines present in the propeptide part but not those present in the leader peptide. Subsequently, the cyclase NisC catalyzes the coupling of the double bond in dehydro amino acids to the thiol groups of cysteines. The resulting (methyl)lanthionines make nisin a member of a group of antibiotics called lantibiotics (43). The fully modified prenisin is exported via the transporter NisT, and the leader peptidase NisP cleaves off the leader peptide.
FIG. 1.
Schedule of nisin biosynthesis involving NisB-mediated dehydration of Ser and Thr in the propeptide, NisC-catalyzed cyclization, NisT-mediated export, and NisP-mediated removal of the leader peptide.
Despite the crucial and intriguing roles in inducing the activities of both modifying and transporter enzymes and despite the diversity among lantibiotic leader peptides, only the leader peptides of nisin (49), Pep5 (31), mutacin II (3), lacticin 481 (32), and nukacin A (30) have been subjected to mutagenesis studies. Several distinct groups of lantibiotic leader peptides can be discerned (see Fig. S1 in the supplemental material). Figure 2 shows aligned leader peptides related to the nisin leader peptide. Many of these leader peptides end with a C-terminal PQ or PR and share a conserved F(N/D)LD box more or less in the central part. The F(N/D)LD box also seems to be present in the leader peptides of bacteriocin J46 (14) and lacticin 481 (34), though otherwise, these leader peptides bear a closer resemblance to an entirely different group of lantibiotic leader peptides (see Fig. S1 in the supplemental material). Leader peptides from this group often have alanines or glycines in position −1 or −2 or a double glycine motif, preceded by one or two small, hydrophobic residues. Strikingly, an L(D/E)E(L/V) box occurs frequently. Some leader peptides from two-component lantibiotics contain two processing sites, as is shown for cytolysin LL, cytolysin LS, plantaricin Wβ, and haloduracin A2 (8, 13, 28, 52). The length of most leader peptides varies from 19 to 48 amino acids. For a long time the only known exception has been cinnamycin, which contains 59 amino acids and which has a different sequence (17, 51). However, recently, a large group of lantipeptides, named prochlorosins, which contain leader peptides of up to 71 amino acids has been discovered (25). Even more remarkable, all 29 prochlorosins are produced by a single organism and are processed by using one promiscuous LanM homolog. The very diverse compositions of the leader peptides found so far point to the occurrence of several distinct leader peptide-enzyme interactions.
FIG. 2.
Leader peptides containing the F(N/D)LD box were aligned by use of the ClustalW2 program (European Bioinformatics Institute, Cambridge, United Kingdom). Streptin was aligned arbitrarily. Colons, conserved substitutions; periods, semiconserved substitutions; asterisk, identical residues. The numbers in parentheses on the right indicate the reference.
The nisin leader peptide can induce modifications and export of a variety of peptides fused to its C terminus (18, 39). For activity in most cases, the leader peptide has to be removed from the thioether-stabilized therapeutic peptide. However, the leader peptidase NisP does not have a broad substrate specificity since it cleaves neither dehydrated prenisin nor unmodified prenisin (20). Trypsin cleaves after R-1, thus removing the leader peptide. However, it also cleaves unprotected Lys and Arg, which may occur in the propeptide. Hence, we investigated the possibility of introducing alternative cleavage sites in the C terminus of the nisin leader peptide. We successfully processed prenisin with an engineered cleavage site and found that it is possible to replace multiple amino acids in the leader peptide without hampering the modification of nisin. We subsequently introduced a His tag in the leader peptide which could facilitate the purification of therapeutic peptides behind the nisin leader peptide. Furthermore, we aimed to modulate the interaction of the leader peptide with the modification and transporter enzymes by mutagenesis of the highly conserved FNLD box (Fig. 2).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Lactococcus lactis NZ9000 was used for expression of the modification enzymes and leader constructs. L. lactis was grown in M17 broth (47) supplemented with 0.5% glucose (GM17) or in minimal medium (15, 36) with or without chloramphenicol (5 μg/ml) and/or erythromycin (5 μg/ml). Cultures were grown on minimal medium after induction with nisin (1 ng/ml) prior to sample preparation for mass spectrometry. The strains and plasmids are listed in Table 1.
TABLE 1.
Lactococcus lactis strains and plasmids
| Strain or plasmid | Characteristic(s) | Reference(s) |
|---|---|---|
| Strains | ||
| Lactococcus lactis NZ9000 | nisRK+ | 23 |
| L. lactis LL108(pORI 280) | Emr Cmr | 23, 24 |
| Plasmids | ||
| pIL253-derived plasmid | 45 | |
| pIL3BTC | nisBTC, derived from pILangBTC by deletion of Emr gene and introduction of stop codon at start of ang gene | 36 |
| pNZ8048-derived plasmid | 23 | |
| pNZnisA-E3 | Nisin A, Emr | 20 |
Molecular cloning.
Standard genetic manipulations were performed using established procedures (40). Constructs coding for mutated prenisin were made by amplifying plasmid pNZnisA-E3 (20) via round PCR using a downstream sense primer and an upstream antisense primer. Each primer pair contained one 5′ phosphorylation and a (nonannealing) peptide-encoding tail (see Table S1 in the supplemental material for a list of the primers used in this study). The DNA amplification was performed with Phusion DNA polymerase (Finnzymes, Finland). Restriction enzymes used for cloning strategies were purchased from New England Biolabs Inc. Self-ligation of the resulting plasmids was carried out with T4 DNA ligase (Roche, Mannheim, Germany). Electrotransformation of L. lactis was carried out as previously described (12) using a Bio-Rad gene pulser (Richmond, CA). Nucleotide sequence analysis was performed by BaseClear (Leiden, Netherlands).
His tag-based peptide purification.
His-tagged prenisin was purified with nickel-nitrilotriacetic acid (Ni-NTA). Glycerol was added to the peptide-containing medium to reduce hydrophobic interactions, and β-mercaptoethanol was added to prevent multimerization.
Gel electrophoresis.
Peptides were isolated from the supernatant of minimal medium cell cultures by trichloroacetic acid (TCA) precipitation. Peptides were separated on a Tricine gel (41). Analysis was performed using a silver stain kit (Invitrogen) or by Coomassie staining (PageBlue). All experiments were repeated in at least three independent experiments, with identical results being obtained in each one.
Western blot analysis.
Polyclonal anti-leader peptide antibodies against the peptide H2N-STKDFNLDLVSVSKKDC-CONH2 coupled via the cysteine to keyhole limpet hemocyanin were raised in rabbits (18). Cell extracts were obtained by incubation of cells with 9% (vol/vol) acetone and 1% (vol/vol) toluene in 100 mM Tris, pH 7.0, for 30 min at 37°C and for 20 min at 100°C. Peptides were precipitated in 10% TCA, dissolved in sample buffer, and applied on a Tricine gel (41). Peptides were transferred to a polyvinylidene difluoride (PVDF) Western blotting membrane (Roche) using a Trans-Blot SD semidry transfer cell (Bio-Rad).
Mass spectrometry.
Samples were purified from the medium fraction by ziptip purification (C18 ziptip; Millipore), or samples were directly applied by spotting 1 μl of the supernatant on the target. After the spots were dried, they were washed once with 4 μl Millipore water to remove the salts. Subsequently, 1 μl of matrix (5 mg/ml α-cyano-4-hydroxycinnamic acid in 50% acetonitrile containing 0.1% [vol/vol] trifluoroacetic acid) was added to the target and allowed to dry. 1-Cyano-4-dimethylaminopyridinium tetrafluoroborate (CDAP) was used to react with free cysteine residues. Vacuum-dried sample was resuspended in 9 μl 25 mM citrate buffer, pH 3.0, and reduced with 1 μl Tris(2-carboxyethyl)phosphine (TCEP). After 10 min of incubation at room temperature, 2 μl of CDAP was added, followed by 15 min of incubation at room temperature. Mass spectra were recorded with a Voyager DE PRO matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Applied Biosystems). In order to maintain high sensitivity, an external calibration was applied.
Antimicrobial activity assay.
Indicator strain L. lactis LL108(pORI 280) (23, 24) was grown in GM17 containing chloramphenicol (5 μg/ml) and erythromycin (5 μg/ml) to an optical density of 0.1 at 600 nm. After 3 h of incubation in microwell plates with a series of 2-fold dilutions of the nisin mutant, growth inhibition was measured at 600 nm.
RESULTS
NisP cleaves an engineered proNisP cleavage site.
The high specificity of NisP (20) precludes its processing of therapeutic peptides fused to the native nisin leader peptide. Interestingly, NisP is likely autoactivated by autocleaving proNisP, which does not contain thioether rings. Therefore, we investigated the capacity of NisP to cleave its putative autocleavage site, VSLR↓QP (where the down arrow indicates the location of peptide cleavage) (44), engineered in prenisin (Table 2). The cleavage site ASPR↓IT from the nisin precursor was exchanged for VSLR↓IT and for VSLR↓QP by mutagenesis. The two prenisin mutants were produced by L. lactis containing pIL3BTC, which encodes the ring-forming enzymes NisB and NisC and the transporter NisT. Incubation of the supernatants with NisP-expressing L. lactis cells released active nisin only from the VSLR↓IT mutant. Apparently, the VSLR↓IT site can be recognized or the presence of lanthionine rings is determining NisP cleavage. The mutant containing VSLR↓QP was also cleaved but did not release active nisin, due to incomplete modification. We subsequently produced both mutants in an unmodified form by L. lactis containing NisP and NisT. MALDI-TOF mass spectrometric analysis demonstrated that only prenisin containing VSLR↓QP was cleaved. These data demonstrate that, in the absence of lanthionine rings, NisP can cleave off the leader peptide from unmodified nisin when the cleavage site is identical to the putative proNisP autocleavage site, VSLR↓QP.
TABLE 2.
Cleavage of prenisin with engineered peptidase sites exported out of L. lactis containing pIL3BTCa
| Peptidase | Prenisin with engineered cleavage siteb | Dehydrations | Cleavage |
|---|---|---|---|
| NisP | MSTKDFNLDLVSVSKKDSGASPR↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | + |
| NisP | MSTKDFNLDLVSVSKKDSGVSLR↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | + |
| NisP | MSTKDFNLDLVSVSKKDSGVSLR↓QPSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | Incomplete | + |
| Thrombin | MSTKDFNLDLVSVSKKDSGAVPR↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | + |
| Thrombin | MSTKDFNLDLVSVSKKDSGLVPR↓GSSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 7 | + |
| Enterokinase | MSTKDFNLDLVSVSKKDSGDDDK↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | + |
| Enterokinase | MSTKDFNLDLVSVSKKDSDDDDK↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | + |
| Glu-C | MSTKDFNLDLVSVSKKDSGASPD↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | − |
| Glu-C | MSTKDFNLDLVSVSKKDSGASPE↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | + |
| Factor Xa | MSTKDFNLDLVSVSKKDSGIDGR↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | ± |
| Factor Xa | MSTKDFNLDLVSVSKKDSGIEGR↓ITSISLCTPGCKTGALMGCNMKTATCHCSIHVSK | 8 | ± |
Peptides in the supernatant were identified by MALDI-TOF mass spectrometry.
Substituted amino acids are indicated in boldface type.
Introducing proteolytic cleavage sites.
The successful introduction of the VSLR↓IT site suggests a mutational freedom in the C terminus of the leader peptide. We further investigated the engineering of other cleavage sites in the leader peptide by changing the ASPR↓IT site in prenisin via mutagenesis. The cleavage sites for thrombin (preferably LVPR↓GS), enterokinase (DDDDK↓), Glu-C (D↓ or E↓), and factor Xa (IDGR↓ or IEGR↓) were introduced (Table 2).
When the original ASPR↓IT site was changed to AVPR↓IT, cleavage of the precursor by thrombin resulted in antimicrobially active nisin. Similar activity was observed when the IT at positions +1 and +2 was replaced by GS. In the latter case, one propeptide residue escaped dehydration. This could be Ser2, since glycine as a flanking residue of serine does not favor dehydration (1).
Prenisin containing (D)DDDK↓IT was efficiently cleaved by enterokinase. However, the production in the medium of this prenisin mutant was strongly reduced, presumably as a result of the presence of negatively charged amino acids. Clearly, an enterokinase site is therefore not preferred.
Glu-C cleavage was efficient for ASPE↓IT but not for ASPD↓IT. Cleavage of the latter primarily resulted in nisin with truncated leader peptides: LVSVSKKDSGASPD-nisin and SGASPD-nisin. Production of prenisin with the engineered Glu-C cleavage site was slightly reduced compared to that of wild-type prenisin.
Unaltered production was observed when a factor Xa cleavage site, IDGR↓ or IEGR↓, was engineered. Cleavage efficiency, however, was low, especially for nisin containing IDGR↓IT (Table 2).
Together these data demonstrate that alternative cleavage sites can be effectively introduced in the nisin leader peptide without interfering with the leader peptide's capacity to induce posttranslational modifications. However, engineered cleavage sites may affect the production and/or export efficiency of the prepeptide (Table 2).
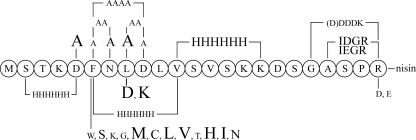
Introduction of His tags within the leader peptide.
The successful engineering of several cleavage sites demonstrated a large mutational freedom in the C terminus of the nisin leader peptide. We subsequently investigated whether other parts of the leader peptide could also be replaced by introducing His tags into the leader peptide. His tags were genetically introduced in prenisin N terminally in place of the STKD, leading to prenisin containing MH6FNLDLVSVSKKDSGASPR, termed pNZE3HisA, directly behind the highly conserved FNLD box, replacing VSVSKK, leading to prenisin containing MSTKDFNLDLH6DSGASPR, termed pNZE3HisB, and replacing the FNLDLV sequence, resulting in MSTKDH6SVSKKDSGASPR, termed pNZE3HisC. Plasmids encoding these constructs were coexpressed with pIL3BTC. Production and modification were analyzed by MALDI-TOF mass spectrometry and measurement of antimicrobial activity.
Expression of pNZE3HisA and pNZE3HisB resulted in the production of prenisin 8-fold dehydrated (see Fig. S2 in the supplemental material). pNZE3HisC encoding the His tag to replace the FNLD(LV) box gave no production at all. In case of expression of pNZE3HisA, the Met at position 1 stayed attached, which is hardly ever the case with produced fusion peptides (18, 20). Apparently, the methionine aminopeptidase cleaves less efficiently when Met is followed by a His tag. Importantly, both mutants that were produced displayed antimicrobial activity after trypsin-mediated removal of the leader peptide. pNZE3HisB was produced at higher levels than pNZE3HisA and could be efficiently purified by Ni-NTA. This demonstrates that the leader peptide with a His tag in positions VSVSKK can be effectively used for purification of leader fusion peptides. This replacement might, furthermore, constitute an advantage, since we observed that the VS sites are sensitive to cleavage (see Fig. S2 in the supplemental material). Elimination of the two V↓S cleavage sites would facilitate mass spectrometry data analyses. Taken together, the introduction of the His tag replacing VSVSKK in the leader peptide surprisingly shows that simultaneously altering 6 positions of the leader peptide does not significantly affect modification and production of the propeptide.
Mutagenesis of FNLD box in nisin leader peptide.
Engineering of the leader peptide may modulate its interaction with the modification enzymes and transporter. This may lead to enhanced or reduced modification and/or transport. Alignment of nisin-related leader peptides shows a highly conserved box consisting of F(N/D)LD(L/V) (Fig. 2). In a previous study using a different expression system, no secretion or intracellular accumulation of nisin or its precursors could be detected after mutagenesis of F-18, L-16, or D-15 (49). Here we applied a more robust two-plasmid expression system and aimed at elucidating the effects of mutagenesis of the FNLD box on modification and production.
In a first step, all 4 conserved amino acids were exchanged separately for alanines, thus yielding four prenisin mutants: F-18A, N-17A, L-16A, and D-15A. Mutant peptides were produced by L. lactis containing pIL3BTC. Full NisB-mediated dehydration was observed by analysis of the supernatant with MALDI-TOF mass spectrometry. Visualization of the prenisin mutants on a Tricine gel showed that nisin mutants F-18A and D-15A were produced to a much lower level than wild-type prenisin, whereas mutant L-16A showed a level of production equal to or increased over that for the wild type (Fig. 3 A). After trypsin-mediated cleavage of the leader peptide, the N-17A, L-16A, and D-15A mutants showed antimicrobial activity comparable to that of wild-type nisin. It therefore appears that none of the amino acid substitutions interferes with maturation, whereas they do interfere with production or export efficiency. This contrasts with the above mentioned study of Van der Meer et al. (49), in which mutagenesis of each of the F18, L16, and D15 residues completely abolished production. This difference may result from the difference in expression system or from the difference in the substituting amino acids.
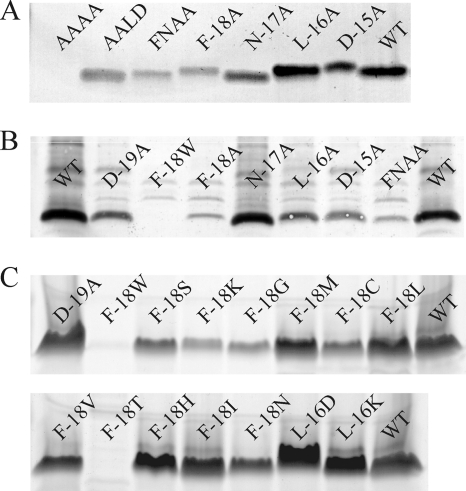
FIG. 3.
Production analysis of FNLD mutants. (A) Supernatants of L. lactis containing pIL3BTC producing wild-type prenisin and FNLD box mutants. Peptide was analyzed by PageBlue-stained gel electrophoresis. Lanes: AAAA, quadruple mutation F-18A/N-17A/L-16A/D-15A; AALD, double mutation F-18A/N-17A; FNAA, double mutation L-16A/D-15A; WT, wild type prenisin A. (B) Prenisin mutants detected by anti-leader peptide antibody in cell extracts from L. lactis containing pIL3BTC. WT, wild type prenisin A; FNAA, double mutation L-16A/D-15A. (C) Supernatants of L. lactis containing pIL3BTC producing wild-type prenisin (WT) and prenisin containing a point mutation. Peptide was analyzed by PageBlue-stained gel electrophoresis.
We subsequently introduced double Ala mutations in the FNLD box and even replaced the whole sequence by alanines (Fig. 3A). Consistent with the low level of production of F-18A and D-15A prenisin, a low level of production was observed for double mutant F-18A/N-17A and an even lower level of production was observed for double mutant L-16A/D-15A. Fully modified prenisin and prenisin lacking one or two dehydrations were observed. The quadruple mutant F-18A/N-17A/L-16A/D-15A was extracellularly present at barely detectable levels and was not dehydrated (Fig. 4).
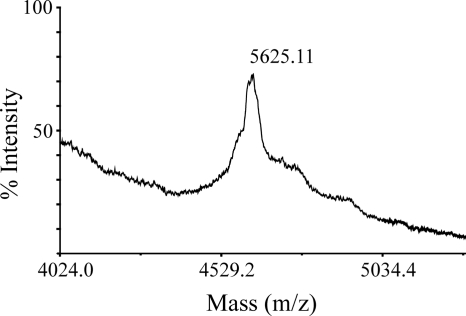
FIG. 4.
Export of prenisin with quadruple mutations (F-18A/N-17A/L-16A/D-15A) without dehydration by L. lactis containing pIL3BTC. Peptide export was detected by MALDI-TOF mass spectrometric analysis of the culture supernatant, after incubation with 1 mg/ml TCEP to prevent cysteinylation. The [M + H+] peak of 5,625.11 corresponds to unmodified prenisin without an initial methionine (5,627.58 Da). The difference of −2 Da could be a result of disulfide bond formation.
To investigate whether the low level of production might be due to reduced transport leading to intracellular accumulation, extracts of cells producing prenisin mutants were subjected to blotting with antileader antibodies. Figure 3B shows that intracellular levels of mutant prenisin are comparable to or lower than the level of wild-type prenisin. A low level of production measured extracellularly corresponds to a low level of production measured intracellularly. Replacement of the phenylalanine by another aromatic residue, F-18W, resulted in even lower levels of production than for F-18A. Taken together, the data indicate that the FNLD box mutants mostly do not accumulate intracellularly.
Prenisin residue F-18 is highly conserved (Fig. 2). We randomized the codons for position −18 and screened for new mutants (Fig. 3C). We obtained several well-produced, fully or nearly completely modified mutants (F-18M, F-18L, F-18V, F-18H, F-18I) and other mutants which were less efficiently produced and/or which lacked more dehydrations (F-18W, F-18K, F-18N, F-18T, F-18S, F-18C, F-18G). Mutants F-18M and F-18H appeared to be produced better than wild-type prenisin. Also, replacing residues in other positions, D-19A, L-16D, and L-16K, appeared to lead to levels of production higher than the level for the wild type (Fig. 3C). An overview of the production of previously mentioned leader peptide mutants is given in Fig. 5.
FIG. 5.
Effect of mutations on export efficiency. Production levels are relative to the level of wild-type prenisin A production. Small characters indicate lower levels of production, medium-size characters indicate levels of production similar to the level of production of wild-type nisin, and large characters indicate higher levels of production.
Leader truncations.
As reported above, replacing the FNLD box with four alanines abolished modifications but still allowed export. To investigate whether the conserved box alone or the N-terminal domain of the leader might be sufficient for modification, we made a construct coding for MFNLDLR-nisin and a Δ(−13 to −2) prenisin variant, with the leader being reduced to MSTKDFNLDLR. The corresponding peptides were produced by L. lactis containing pIL3BTC and analyzed by MALDI-TOF mass spectrometry. Analysis of the supernatant revealed unmodified MFNLDLR-nisin with an initial methionine and up to 4-fold dehydrated STKDFNLDLR-nisin without an initial methionine (Fig. 6 A and B). Most of the MFNLDLR-nisin mutant contained an additional 28 Da. This corresponds to N-terminal formylation and indeed is seen only in combination with the initial methionine (35). The 4-fold dehydration of the STKDFNLDLR-nisin mutant raises the question of whether Ser and Thr residues need to be located at a minimal distance from the FNLD box in order to be dehydrated by NisB. Fragmentation of the STKDFNLDLR-nisin mutant by trypsin digestion or MALDI-TOF/TOF mass spectrometry to investigate the location of the dehydro amino acids was hampered by very low yields of the mutant peptide. We subsequently engineered a double-truncated nisin mutant, MSTKDFNLDLR-ITSISLCTPGCKTG, which is composed of the shortened leader peptide and only the N-terminal part of pronisin. The resulting peptide was poorly produced but contained only one dehydro amino acid (Fig. 6C). This implies that in the above case of the STKDFNLDLR-nisin peptide, at least three of the observed four dehydrations occur in the C-terminal part of the nisin propeptide. These data demonstrate that the MSTKDFNLDLR sequence alone is indeed sufficient for dehydration by NisB.
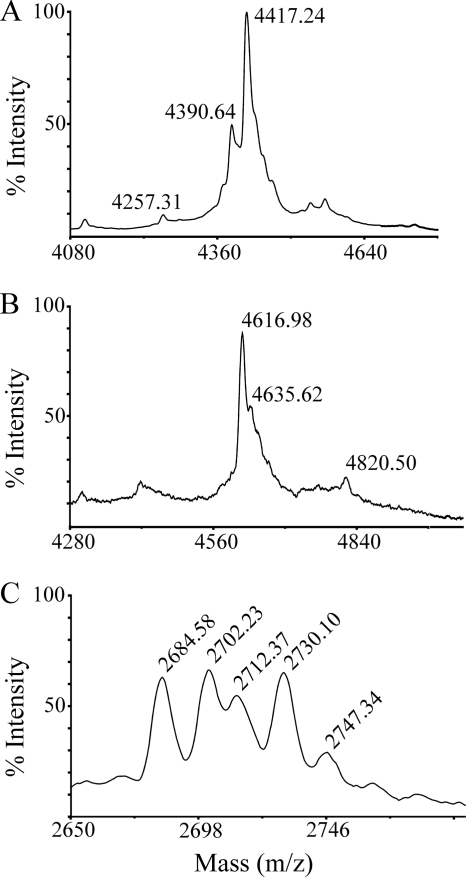
FIG. 6.
Export of prenisin with shortened leader peptides containing the FNLD box by L. lactis containing pIL3BTC. Peptide export was detected by MALDI-TOF mass spectrometric analysis of the culture supernatant, after incubation with 1 mg/ml TCEP to prevent cysteinylation. (A) MFNLDLR-nisin. The [M + H+] peaks of 4,390.64 and 4,257.31 correspond to unmodified prenisin with an initial methionine (4,389.28 Da) and without an initial methionine (4,258.08 Da), respectively. Peak 4,417.24 corresponds to formylated (—CHO) MFNLDLR-nisin (4,417.29 Da). (B) MSTKDFNLDLR-nisin. The [M + H+] peaks of 4,616.98 and 4,635.62 correspond to prenisin without an initial methionine 4-fold dehydrated (4,617.47 Da) and 3-fold dehydrated (4,635.49 Da), respectively. The peak of 4,820.50 corresponds to undehydrated prenisin with an initial methionine (4,820.72 Da). (C) MSTKDFNLDLR-ITSISLCTPGCKTG is dehydrated once. Peptide masses correspond to prenisin with an initial methionine dehydrated once (2,685.14 Da) and not dehydrated (2,703.16 Da). Subsequent peaks are again once and not dehydrated prenisin but contain a formylated initial methionine, resulting in an additional mass of 28 Da. The [M + H+] peak of 2,747.34 could represent either an oxidation (+16 Da) or another dehydration (+18 Da).
DISCUSSION
Two decades ago it was suggested in a paper in Nature (43) that lantibiotic enzymes could be used for the stabilization of therapeutic peptides. In line with this suggestion, we previously constructed genetically engineered peptides composed of an N-terminal nisin leader peptide and a C-terminal therapeutic peptide and subjected these fusion peptides in L. lactis to the nisin modification and export machinery. In 2004, we were the first to report lantibiotic enzyme-mediated dehydration of a therapeutic peptide analog (20). Subsequently, we have demonstrated that not only NisB but also the nisin cyclase NisC has broad substrate specificity (39). We recently demonstrated the enhanced stability, activity, and bioavailability (19) and oral and pulmonary delivery (5) of thioether-bridged angiotensin-(1-7).
To eventually obtain a thioether-stabilized therapeutic peptide, the leader peptide has to be cleaved off. The leader peptidase NisP has restricted substrate specificity (20), which is in contrast to the broad cleavage substrate specificity of the bifunctional leader peptidase/transport protein of the lacticin 481 system (6). NisP removes the leader peptide only from fully modified prenisin (20). A large variety of fully modified prenisins with a mutated ring A were also substrates for NisP (38). Van der Meer et al. (49) demonstrated that some residues in the C-terminal part of the nisin leader peptide are required for NisP-mediated cleavage; R-1Q and A-4D prevented cleavage. We demonstrated here that engineering the presumed proNisP cleavage site (44) in prenisin allows NisP-mediated cleavage in the absence of the lanthionine rings. We successfully engineered a functional recognition site for thrombin, enterokinase, Glu-C, and factor Xa in the C-terminal part of the leader peptide without compromising NisB- and NisC-mediated modifications. However, the introduction of multiple negatively charged amino acids in the C-terminal part of the leader peptide, as in the enterokinase cleavage site (D)DDDK↓, appeared to reduce production. The possibility demonstrated here to insert a cleavage site of choice in the nisin leader peptide is crucial for liberating thioether-stabilized therapeutic peptides.
Studies on the nisin leader (49), the Pep5 leader (31), the mutacin II leader (3), the nukacin A leader (30), and the lacticin 481 leader (32) have demonstrated that, apart from a few conserved residues, a large mutational freedom exists to replace single amino acids. Here we simultaneously altered 6 positions by engineering in three positions a His tag within the leader peptide, of which MSTKDFNLDLH6DSGASPR appeared to be fully functional. We observed no loss of this engineered leader's capacity to induce NisB- and NisC-mediated modifications of prenisin. In contrast, MSTKDH6SVSKKDSGASPR-pronisin, in which the conserved FNLD box was replaced, was not produced.
Simultaneous replacement of the FNLD residues in prenisin by alanines, yielding MSTKDAAAALVSVSKKDSGASPR-pronisin (F-18A/N-17A/L-16A/D-15A), allowed some production but abolished the capacity to induce modifications. Double and single amino acid replacements of the FNLD residues by alanines did not lead to loss of the capacity to induce modifications. Production, however, was reduced for most of the alanine mutants. Interestingly, the single substitution L-16A resulted in an increase in prenisin production. These data point to an important role of the FNLD box in NisB- and NisC-mediated modification. Although the level was low, the export of the quadruple mutant F-18A/N-17A/L-16A/D-15A suggests that NisT-mediated export is not completely dependent on the conserved FNLD box.
In a previous study by Van der Meer et al. (49), none of the three mutants F-18L, L-16K, and D-15A was observed either intra- or extracellularly. Here we observed production and modification of the three mutants F-18A, L-16A, and D-15A. This apparent inconsistency is likely due to the improved two-plasmid expression system in L. lactis NZ9000 that we used, with the nisBTC genes being on one plasmid and the mutant prenisin being on a second, compared to the one-plasmid system in the nisin producer L. lactis NZ9700, which contains the whole gene cluster on the chromosome. Alternatively, the differences might be caused by different detection methods. In the case of Pep-5, reduced production was observed for mutants F-19S and D-16K (31), which is more in concordance with our findings.
The reduced production of prenisin F-18A and the even more reduced production of F-18W points to a specific role of the phenylalanine. We therefore randomized the codons for this amino acid and obtained a large number of mutants. All mutants allowed prenisin production, but F-18W and F-18T gave very low levels of production. In contrast, mutants F-18H, F-18M, and also some substitutions in different positions (D-19A, L-16D, and L-16K) enhanced production.
Reducing the nisin leader to just MFNLDLR allowed some production but no NisB-mediated dehydration. Also, processing of the N terminus of this prenisin mutant seemed impeded, as the peptide still contained a (formylated) initial methionine. Interestingly, replacing the nisin leader by MSTKDFNLDLR resulted in 4-fold dehydrated prenisin, indicating that residues −13 to −2 are not essential for interaction with NisB. A maximum of four dehydrations further suggests that the 12 missing leader peptide residues might function as a spacer to bridge a distance between the NisB interaction site and its modification site. Deleting this spacer would preclude access to 4 of the 8 residues that can be dehydrated. It would also explain why the Ser or Thr residues inside the leader peptide are not dehydrated by NisB. This hypothesis was supported by one or no dehydration of the N-terminal fragment of pronisin preceded by the MSTKDFNLDLR leader. Further experiments are being conducted to investigate this hypothesis in more detail.
NisB (21), NisC (26), and NisT (20) can each function independently of the other two enzymes but not in the absence of the N-terminally located leader peptide in the substrate peptide. It therefore seems that the leader peptide is capable of functionally interacting with three different enzymes. The question arises whether one motif is responsible for three different interactions or whether different parts of the nisin leader determine a distinct interaction with a different enzyme. Here we demonstrated that many residues of the nisin leader peptide can be altered simultaneously without affecting modification or export. Moreover, reducing the leader peptide to MSTKDFNLDLR still allows partial NisB-mediated dehydration. The leader peptide has been demonstrated to function when it is present internally between a Sec signal sequence and a modifiable peptide (21). The possibility to use a short modification-inducing leader sequence demonstrated here opens the perspective to incorporate such a sequence in a protein to induce very local modifications. The present data indicate that complete removal of the FNLD box eliminates modification, whereas removal of more than this sequence is needed to preclude export.
Both NisB and NisC perform multiple reactions on a single substrate. Mechanistic models have been integrated for the modification of nisin (22) and put forward for, among others, LctM (32), a bifunctional enzyme that houses both the dehydration and the cyclization catalytic sites. In this model the binding of the leader peptide is shifting the equilibrium from an inactive enzyme to an active conformation of the enzyme. Alternatively, leader peptide binding might just bring the modifiable peptide close to the catalytic sites and modified peptides might be released by having a lower affinity for the catalytic sites. So far no maximum distance from the leader peptide with respect to modification has been demonstrated. NisB has been demonstrated to dehydrate farther than position 33, which is the most distal position in nisin, to even position 42 (37). If the leader peptide binds in a fixed position, one would expect that modification would not take place at a great distance from the leader peptide. Alternatively, the leader peptide might move along or through the modification enzyme. Since bifunctional lantibiotic modification enzymes as well as NisC are active in vitro, such movement cannot be induced by transporters. Cocrystallization of modification enzymes and leader peptides and/or substrate peptides will likely yield important mechanistic insights into the highly intriguing leader peptide-enzyme interactions.
Supplementary Material
Acknowledgments
Leon D. Kluskens was supported by STW project 06927.
Saffira Kiewiet is gratefully acknowledged for technical assistance.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Chatterjee, C., G. C. Patton, L. Cooper, M. Paul, and W. A. van der Donk. 2006. Engineering dehydro amino acids and thioethers into peptides using lacticin 481 synthetase. Chem. Biol. 13:1109-1117. [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee, C., M. Paul, L. Xie, and W. A. van der Donk. 2005. Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105:633-684. [DOI] [PubMed] [Google Scholar]
- 3.Chen, P., F. X. Qi, J. Novak, R. E. Krull, and P. W. Caufield. 2001. Effect of amino acid substitutions in conserved residues in the leader peptide on biosynthesis of the lantibiotic mutacin II. FEMS Microbiol. Lett. 195:139-144. [DOI] [PubMed] [Google Scholar]
- 4.De Kwaadsteniet, M., K. ten Doeschate, and L. M. T. Dicks. 2008. Characterization of the structural gene encoding nisin F, a new lantibiotic produced by Lactococcus lactis subsp. lactis isolated from freshwater catfish (Clarias gariepinus). Appl. Environ. Microbiol. 74:547-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Vries, L., et al. 2010. Oral and pulmonary delivery of thioether-bridged angiotensin-(1-7). Peptides 31:893-898. [DOI] [PubMed] [Google Scholar]
- 6.Furgerson Ihnken, L. A., C. Chatterjee, and W. A. van der Donk. 2008. In vitro reconstitution and substrate specificity of a lantibiotic protease. Biochemistry 47:7352-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furmanek, B., et al. 1999. Identification, characterization and purification of the lantibiotic staphylococcin T, a natural gallidermin variant. J. Appl. Microbiol. 87:856-866. [DOI] [PubMed] [Google Scholar]
- 8.Gilmore, M. S., et al. 1994. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relation to lantibiotic determinants. J. Bacteriol. 176:7335-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross, E., and J. L. Morrell. 1971. The structure of nisin. J. Am. Chem. Soc. 93:4634-4635. [DOI] [PubMed] [Google Scholar]
- 10.Gross, E., H. H. Klitz, and E. Nebelin. 1973. Die Struktur des Subtilins. Hoppe-Seylers Z. Physiol. Chem. 354:810-812. [PubMed] [Google Scholar]
- 11.Heidrich, C., et al. 1998. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 64:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 13.Holo, H., Z. Jeknic, M. Daeschel, S. Stevanovic, and I. F. Nes. 2001. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology 147:643-651. [DOI] [PubMed] [Google Scholar]
- 14.Huot, E., J. Meghrous, C. Barrena-Gonzalez, and H. Petitdemange. 1996. Bacteriocin J46, a new bacteriocin produced by Lactococcus lactis subsp. cremoris J46: isolation and characterization of the protein and its gene. Anaerobe 2:137-145. [Google Scholar]
- 15.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellner, R., G. Jung, C. Kaletta, K.-D. Entian, and H.-G. Sahl. 1989. Pep5: structure elucidation of a large lantibiotic. Angew. Chem. Int. Ed. Engl. 28:616-619. [Google Scholar]
- 17.Kessler, H., et al. 1988. The structure of the polycyclic nonadecapeptide Ro 09-0198. Helv. Chim. Acta 71:1924-1929. [Google Scholar]
- 18.Kluskens, L. D., et al. 2005. Post-translational modification of therapeutic peptides by NisB, the dehydratase of the lantibiotic nisin. Biochemistry 44:12827-12834. [DOI] [PubMed] [Google Scholar]
- 19.Kluskens, L. D., et al. 2009. Angiotensin-(1-7) with thioether bridge: an angiotensin-converting enzyme-resistant, potent angiotensin-(1-7) analog. J. Pharmacol. Exp. Ther. 328:849-854. [DOI] [PubMed] [Google Scholar]
- 20.Kuipers, A., et al. 2004. NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J. Biol. Chem. 279:22176-22182. [DOI] [PubMed] [Google Scholar]
- 21.Kuipers, A., et al. 2006. Sec-mediated transport of posttranslationally dehydrated peptides in Lactococcus lactis. Appl. Environ. Microbiol. 72:7626-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuipers, A., R. Rink, and G. N. Moll. Genetics, biosynthesis, structure and mode of action of lantibiotics. In D. Drider and S. Rebuffat (ed.), Prokaryotic antimicrobial peptides: from genes to applications, in press. Springer-Verlag, Berlin, Germany.
- 23.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1997. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 15:135-140. [DOI] [PubMed] [Google Scholar]
- 24.Leenhouts, K., A. Bolhuis, G. Venema, and J. Kok. 1998. Construction of a food-grade multi-copy integration system for Lactococcus lactis. Appl. Microbiol. Biotechnol. 49:249-256. [DOI] [PubMed] [Google Scholar]
- 25.Li, B., et al. 2010. Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 107:10430-10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, B., J. P. Yu, J. S. Brunzelle, G. N. Moll, W. A. van der Donk, and S. K. Nair. 2006. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311:1464-1467. [DOI] [PubMed] [Google Scholar]
- 27.Lubelski, J., R. Rink, R. Khusainov, G. N. Moll, and O. P. Kuipers. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65:455-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClerren, A. L., et al. 2006. Discovery and in vitro biosynthesis of haloduracin, a new two-component lantibiotic. Proc. Natl. Acad. Sci. U. S. A. 103:17243-17248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulders, J. W. M., I. J. Boerrigter, H. S. Rollema, R. J. Siezen, and W. M. de Vos. 1991. Identification and characterization of the lantibiotic nisin Z, a natural nisin variant. Eur. J. Biochem. 301:581-584. [DOI] [PubMed] [Google Scholar]
- 30.Nagao, J., et al. 2009. Mapping and identification of the region and secondary structure required for the maturation of the nukacin ISK-1 prepeptide. Peptides 30:1412-1420. [DOI] [PubMed] [Google Scholar]
- 31.Neis, S., et al. 1997. Effect of leader peptide mutations on biosynthesis of the lantibiotic Pep5. FEMS Microbiol. Lett. 149:249-255. [DOI] [PubMed] [Google Scholar]
- 32.Patton, G. C., M. Paul, L. E. Cooper, C. Chatterjee, and W. A. van der Donk. 2008. The importance of the leader sequence for directing lanthionine formation in lacticin 481. Biochemistry 47:7342-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen, J., et al. 2009. Identification and characterization of a bioactive lantibiotic produced by Staphylococcus warneri. Biol. Chem. 390:437-444. [DOI] [PubMed] [Google Scholar]
- 34.Piard, J. C., O. P. Kuipers, H. S. Rollema, M. J. Desmaazeaud, and W. M. de Vos. 1993. Structure, organization, and expression of the lct gene for lacticin 481, a novel lantibiotic produced by Lactococcus lactis. J. Biol. Chem. 268:16361-16368. [PubMed] [Google Scholar]
- 35.Rajagopalan, P. T. R., A. Dattta, and D. Pei. 1997. Purification, characterization, and inhibition of peptide deformylase from Escherichia coli. Biochemistry 36:13910-13918. [DOI] [PubMed] [Google Scholar]
- 36.Rink, R., et al. 2005. Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry 44:8873-8882. [DOI] [PubMed] [Google Scholar]
- 37.Rink, R., et al. 2007. Production of dehydroamino acid-containing peptides by Lactococcus lactis. Appl. Environ. Microbiol. 73:1792-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rink, R., et al. 2007. Dissection and modulation of the four distinct activities of nisin by mutagenesis of rings A and B and by C-terminal truncation. Appl. Environ. Microbiol. 73:5809-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rink, R., et al. 2007. NisC, the cyclase of the lantibiotic nisin, can catalyze cyclization of designed nonlantibiotic peptides. Biochemistry 46:13179-13189. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and A. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Laboratory Press, Cold Spring Harbor, NY.
- 41.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 42.Schnell, N., et al. 1989. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine containing antibiotic. FEMS Microbiol. Lett. 58:263-268. [DOI] [PubMed] [Google Scholar]
- 43.Schnell, N., et al. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276-278. [DOI] [PubMed] [Google Scholar]
- 44.Siezen, R. J., H. S. Rollema, O. P. Kuipers, and W. M. de Vos. 1995. Homology modelling of the Lactococcus lactis leader peptidase NisP and its interaction with the precursor of the lantibiotic nisin. Protein Eng. 8:117-125. [DOI] [PubMed] [Google Scholar]
- 45.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 46.Stein, T., et al. 2002. Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J. Bacteriol. 184:1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van de Kamp, M., et al. 1995. Elucidation of the primary structure of the lantibiotic epilancin K7 from Staphylococcus epidermis K7—cloning of the epilancin-K7-encoding gene and NMR analysis of mature epilancin K7. Eur. J. Biochem. 230:587-600. [DOI] [PubMed] [Google Scholar]
- 49.Van der Meer, J. R., et al. 1994. Influence of amino acid substitutions in the nisin leader peptide on biosynthesis and secretion of nisin by Lactococcus lactis. J. Biol. Chem. 269:3555-3562. [PubMed] [Google Scholar]
- 50.Wescombe, P. A., N. C. K. Heng, J. P. Burton, and J. R. Tagg. 2010. Something old and something new: an update on the amazing repertoire of bacteriocins produced by Streptococcus salivarius. Probiotics Antimicrob. Proteins 2:37-45. [DOI] [PubMed] [Google Scholar]
- 51.Widdick, D. A., et al. 2003. Cloning and engineering of the cinnamycin biosynthetic gene cluster from Streptomyces cinnamoneus cinnamoneus DSM 40005. Proc. Natl. Acad. Sci. U. S. A. 100:4316-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willey, J. M., and W. A. van der Donk. 2007. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61:477-501. [DOI] [PubMed] [Google Scholar]
- 53.Wirawan, R. E., N. A. Klesse, R. W. Jack, and J. R. Tagg. 2006. Molecular and genetic characterization of a novel nisin variant produced by Streptococcus uberis. Appl. Environ. Microbiol. 72:1148-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zendo, T., et al. 2003. Identification of the lantibiotic nisin Q, a new natural nisin variant produced by Lactococcus lactis 61-14 isolated from a river in Japan. Biosci. Biotechnol. Biochem. 67:1616-1619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.