Abstract
In the present work the effect of a mutation on tyrosine 33 residue (Y33G) of human cytidine deaminase (CDA) was investigated with regard to protein solubility and specific activity. Osmolytes and CDA ligands were used to increase the yield and the specific activity of the protein. The mutant enzyme was purified and subjected to a kinetic characterization and to stability studies. These investigations reinforced the hypothesis that in human CDA the side chain of Y33 is involved in intersubunit interactions with four glutamate residues (E108) forming a double latch that connects each of the two pairs of monomers of the tetrameric CDA.
Keywords: cytidine deaminase, site-directed mutagenesis, molecular modeling, circular dichroism
1.1 Introduction
Cytidine deaminase (CDA, cytidine/2'-deoxycytidine aminohydrolase; EC 3.5.4.5), plays an important role in the pyrimidine salvage pathways since it is involved in the hydrolytic deamination of cytidine or deoxycytidine to uridine or deoxyuridine, respectively. This enzyme also catalyzes the deamination of cytosine nucleoside analogues used in chemotherapy leading to the loss of their pharmacological activity.
Human cytidine deaminase consists of four identical subunits (4 × 14,900 Da) with one active site per subunit. The four active sites, which lie at the subunit interface, operate independently and each subunit is equipped with a zinc atom that is involved in the covalent addition of water to cytidine, followed by elimination of ammonia [1, 2]. The structure of the CDA active site was first elucidated by Betts et al. [3] crystallizing the dimeric E. coli enzyme. In this protein the active site of the monomer is formed with the contribution of residues coming from the so called “broken active site”. Successively the crystal structure of the tetrameric B. subtilis CDA evidenced that a complicate set of intersubunit interactions contributes to the building of the active site in each monomer [4]. This structure, as well as earlier molecular models of the human CDA [2] based on the E. coli enzyme [3], indicated that an essential zinc ion is tetrahedrally coordinated by the uridine O-4 and the thiolate side chains of Cys65, Cys99, and Cys102. Another important residue involved in the catalytic mechanism is Glu 67 that would be located in proximity of the zinc ion and the positions 4 and 3 of the nucleoside pyrimidine ring.
Concerning the quaternary arrangement of the tetrameric CDAs, it is evident that two of the monomers are assembled in a way that mimics the structure of a single monomer in the E. coli enzyme, which is larger and endowed with a pseudo two-fold symmetry [2]. In particular, the arrangement of two pairs of monomers yields a tetramer with an overall C222 symmetry that overlaps well with the E. coli dimer [2]. We dubbed the four subunits of the CDA tetramer A1, A2, B1, and B2, where the pair A1 and A2 corresponds to a single monomer of the E. coli dimer, and the pair B1 and B2 corresponds to the other monomer (see figure 4 of [2]).
The binding pocket for the substrate contained in each subunit is also lined by a few residues supplied by two additional subunits. For example, residues from subunits B1 and B2 contribute to the formation of the A1 binding pocket. In particular, our studies identified Phe137 of subunit B1, located at the interface between the subunits A1 and B1, as a notable residue that concurs to the formation of the binding pocket for the ligand pyrimidine moiety [2, 5]. In a further work [1], we investigated the role exerted by additional residues located at the subunit interface in human CDA, demonstrating that the enzyme is catalytically functioning only in the tetrameric form and that the monomer alone is not able to catalyze the substrate deamination. Nevertheless, despite the complex structure of the active site, cytidine deaminase does not show allosteric behaviour [5].
A residue of particular interest in human CDA is Y33, which is part of a motif (32PYSHF36) conserved in most tetrameric CDAs; the published structures of tetrameric CDAs, reveal that this region is located between the α-helix (α1) and the β-sheet (β1) in proximity to the active site and at the interface between the monomers that correspond to the catalytic domain and the “broken active site” of the dimeric E. coli enzyme. Interestingly, in dimeric E. coli CDA, a tyrosine is not found in the homologous 67PLSNF71 motif of the catalytic domain, but is present in the 205PYSKS209 homologous motif of the non-zinc-containing domain.
In a previous study we addressed the residue F36 belonging to the motif 32PYSHF36 finding out that, together with the residue F137, is involved in the hydrophobic interactions with the pyrimidine ring of the substrate [5].
To investigate on the functional role of Y33, by a series of site-directed mutagenesis on human CDA cDNA, we obtained the mutants Y33G, Y33F, and Y33S [1], but, in the prokaryotic expression system used, only Y33G produced a partially soluble protein without enzymatic activity. The other two mutant CDAs, Y33F and Y33S, showed no protein expression. The analysis of the very small amount of Y33G mutant enzyme found in the soluble fraction indicated that the Y33 residue may be involved in the stabilization of the quaternary structure. However, the difficulties encountered during the expression of Y33G mutant CDA did not allow to study in depth the role exerted by this residue.
One possibility to correct the protein folding and therefore to increase the solubility and the activity of an enzyme expressed in a heterologous system is the use of low molecular weight compounds added to the culture medium that stabilize proteins in their native conformations. In fact it has been shown by several research groups that a class of small molecules (osmolytes), called also chemical chaperones because of their influence on protein conformation [6], can sometimes correct protein folding defects that are present in mutant proteins [7–10]. More recently it was realized that substrates, inhibitors or other specific ligands for the wild-type proteins, called pharmacological (ligand-mediated) chaperones, are able to rescue the expression and function of mutant proteins at much lower concentration with respect to the chemical chaperones [8, 11, 12, 13]. Notably, chemical and pharmacological chaperones have been reported to restore the activity not only in cases of misfolding but also with mutants that impair the assembly of the quaternary structure of multimeric proteins [14, 15, 16]. In this work the effect of some osmolytes such as glycerol, DMSO, cyclodextrins, sorbitol and some specific CDA ligands, added to the culture medium during induction, was investigated with regard to protein solubility and specific activity of a CDA protein mutated in the tyrosine 33 residue (Y33G).
The mutant Y33G, that showed enzymatic activity after the addition of chaperones, was purified and a kinetic characterization was performed with some substrates and inhibitors. The effect of temperature and of small amounts of sodium dodecyl sulphate (SDS) on the protein stability was investigated and compared to the wild-type CDA. Furthermore circular dichroism (CD) experiments at different temperatures and in presence of small amounts of a denaturing agent were performed to get a deeper insight into the role exerted by the Y33 residue in the structure/function relationship of the Y33G mutated protein.
1.2 Materials and Methods
1.2.1 Chemicals
HiLoad 16/10 Phenyl Sepharose High Performance, Superdex 75 and the HPLC system Äkta Purifier was from GE Healthcare (Uppsala, Sweden). Mini Protean III electrophoresis apparatus, Western blot apparatus, nitrocellulose membrane, HRP conjugate substrate kit and low range protein markers were purchased from Bio-Rad (Hercules, CA). The polyclonal antibodies against cytidine deaminase (anti-CDA) were produced in rabbit by INBIOS (Seattle, WA) using as antigen human recombinant cytidine deaminase highly purified [17]. 6-[3, 5(Cytidyl)acryloylamino] hexanoic acid (CV6) has been synthesized by Prof. G. Cristalli (University of Camerino). 5-F-zebularine (5-FZEB) was the kind gift of Professor Victor E. Marquez (National Institute of Health, Bethesda). Other chemicals were reagent grade from J.T. Baker Chemicals B.V. (Deventer, The Netherlands). Escherichia coli SØ5201 (MC1061cdd∷Tn10 pyrD∷Kan) strain, lacking cytidine deaminase was used throughout. L-broth supplemented by ampicillin 100 μg/ml was used as rich medium.
1.2.2 Expression of Y33G mutant CDA in presence of chemical chaperones and specific CDA ligands
Site directed mutagenesis experiments on cytidine deaminase tyrosine 33 aminoacidic residue were described in a previous work [1]. The chaperones stabilization studies on Y33G, mutant CDA were performed as follows: cultures (20 ml) of E. coli SØ5201 harboring pTrcHUMY33G were grown at 37°C in L-broth, supplemented with 100 μg/ml ampicillin, in presence of increasing concentrations of chemical chaperones (glycerol, ranging from 2 to 10 %; sorbitol, ranging from 0.2 to1.0 M; dimethyl sulphoxide, DMSO, ranging from 0.05 to 1.0%; cyclodextrins, ranging from 0.1 to 5.0 mM) or CDA substrates and inhibitors (cytidine and uridine, ranging from 1 to 50 mM; 5-fluoro-zebularine, 5FZEB, ranging from 0.001 to 0.1 mM). The expression was induced by the addition of 1mM IPTG. The negative control consisted of the same culture described above but in absence of chaperones. The positive control consisted of wild-type cytidine deaminase. After 16 hours of vigorous shaking at 37°C, cells were harvested by centrifugation at 8,000×g and washed with 0.9% NaCl, resuspended in buffer A (50 mM Tris-HCl, pH 7.5, 1mM DTT, 1 mM EDTA, 10% glycerol), lysed by sonic disruption and centrifugated at 27,000×g for 20 min. On each supernatant (crude extract) cytidine deaminase activity was determined as previously described [17], the total protein concentration by the Bradford protein assay [18] and the amount of expressed mutant protein in presence of a specific chaperone was evaluated by western blot analysis with anti-CDA polyclonal antibodies.
In order to discriminate if the presence of chaperones in the culture medium affects the stability or the expression of the Y33G mutant CDA, E. coli cells harboring the plasmid pTrcHUMY33G were incubated in LB broth (containing 1mM IPTG and 100 μg/ml ampicillin) in presence or in absence of 10% glycerol. After 16 h of vigorous shaking at 37°C, cells were harvested by centrifugation at 8,000×g, resuspended in 50 mM Tris/HCl pH 7.5, 1mM EDTA, 1mM DTT in presence or in absence of 10% glycerol and lysed by sonic disruption. After centrifugation at 27,000×g for 30 min, on each supernatant (with and without glycerol) the protein concentration and the CDA activity were determined. All pellets were resuspended in buffer A containing 8M urea. Finally the supernatants and the solubilized pellets were analyzed by western blotting with anti-CDA polyclonal antibodies.
1.2.3 Polyacrylamide gel electrophoresis experiments and western blot analysis
Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli, [19] using 15% acrylamide and the Mini Protean III apparatus (gel size 7 cm × 8 cm × 0.75 mm). The markers used were Bio-Rad low range.
For western blot analysis under denaturing conditions, the crude extract containing 20 μg of total proteins obtained by the titration of Y33G with each chaperone concentration, was subjected to 15 % SDS-PAGE. The proteins were transferred to nitrocellulose membrane using the Mini Trans-Blot cell apparatus (7.5 × 10 cm blotting area) at 50 V for 2h. The membrane was incubated with TBST buffer (10mM Tris/HCl pH 8.0, 150mM NaCl, 0.05% v/v Tween-20) containing 1% bovine serum albumine (BSA) for 30 min, washed with TBST and probed overnight with the polyclonal primary antibody anti-CDA diluted 1:200 in TBST. After a series of washing in TBST the membrane was incubated for 30 min with the horseradish peroxidase-conjugated anti-rabbit secondary antibody diluted 1:2500 in TBST. The complex antibody-protein was visualized by the chromogenic-substrate 3'-1' diamminobenzidine (HRP conjugate substrate kit).
Dissociation of Y33G mutant CDA into subunits was achieved as described previously for the wild-type enzyme [20], by a series of 15% PAGE in the presence of increasing concentrations of SDS, both in the gels and in the running buffer. An amount of 3.6 μg of each protein sample was incubated in presence of SDS to reach different SDS/enzyme molar ratios (50, 100 and 300). After 2 h incubation at 25°C, bromophenol blue was added and the samples were loaded on the gel. Electrophoresis was performed at 4°C and with a constant voltage of 200 V. The band corresponding to Y33G was visualized after the western blot analysis.
1.2.4 Purification of Y33G mutant CDA
Cultures (500 ml) of E. coli SØ5201 harboring pTrcHUMY33G were grown at 37°C in L-broth, supplemented with 100 μg/ml ampicillin, 10% glycerol, the expression was induced by the addition of 1mM IPTG. After 16 h of vigorous shaking at 37°C, cells were harvested by centrifugation at 8,000×g, washed with 0.9% NaCl, resuspended in buffer A and were lysed by sonic disruption. After centrifugation at 27,000×g for 30 min, the supernatant was loaded onto a DEAE-52 (column volume 25 ml) anionic exchange chromatography column equilibrated with buffer A and eluted by a linear gradient between buffer A and buffer B (50mM Tris/HCl pH 7.5, 1mM EDTA, 1mM DTT, 10% glycerol, 0.5M KCl). Fractions containing the enzyme were pooled, precipitated with 60% ammonium sulphate and, after a centrifugation at 27,000×g for 30 min, the pellet was resuspended in buffer A with 2M ammonium sulphate and loaded onto a Phenyl Sepharose High Performance Hiload 16/10 (20 ml, GE Healthcare) equilibrated with the same buffer. The column was eluted by a linear gradient between buffer A containing 2M ammonium sulphate and buffer A. The active fractions obtained from the hydrophobic interaction chromatography were pooled and concentrated by ultrafiltration. This partially purified preparation of Y33G mutant CDA was used in all experiments.
1.2.5 Kinetic studies on Y33G mutant CDA
CDA activity assays and kinetic studies on Y33G mutant were performed as previously described for the wild-type CDA in presence of several substrates and inhibitors of cytidine deaminase [1, 17].
The experimental error in the measure of the enzymatic activity has been determined by performing repeated assays at each 30 minutes in the 8 hours time span. From these measurements the mean value and standard deviation were calculated.
One enzyme unit is defined as the amount of enzyme which catalyses the deamination of 1 μ-mol of cytidine per minute at 37°C. Furthermore, Y33G stability was investigated in presence of small amounts of SDS ranging from 0.35 to 1.73 mM as previously described for the wild-type CDA [20].
The effect of temperature on the enzymatic activity of Y33G mutant and wild-type CDA was investigated at different temperatures ranging from 5 to 75°C and compared to the analogous studies performed on the wild-type CDA [1]. The enzymatic reaction was started by adding the sample to the assay medium adjusted to the specific temperature. The effect of temperature on Y33G stability was also checked by incubating the enzyme at different temperatures (from 10 to 75°C) for 6 min and then assaying it at 37 °C.
The temperature dependence of Vmax and Km values of Y33G and wild-type CDAs was also analyzed by using the empirical Arrhenius equation (ln Vmax= ln A - Ea/RT) and the van't Hoff equation (ln Km= -ΔH/RT + ΔS/R).
1.2.6 Spectroscopic measurements on Y33G mutant CDA
CD measurements on Y33G mutant enzyme were performed as already described for wild-type CDA [1] with a Jasco J710 spectropolarimeter in the far UV range (200 – 250 nm) at increasing temperatures or in presence of small amounts of SDS as denaturing agent. For the thermal unfolding experiments protein concentration was about 5 μM in 20mM phosphate buffer pH 7.5 in presence of 1mM DTT and 10% glycerol, cell length was 1 cm and temperatures were 5, 10, 20, 30, 40, 50, 60, 70, 80 and 90° C. Regarding the SDS unfolding experiments protein was pre-incubated for 1 h at 25°C with different SDS concentrations (0.035, 0.069, 0.17, 0.35, 0.52, 0.69, 1.04, 1.40, 1.70, 3.5, 5.2, 6.9 mM); protein concentration was about 2 μM in 20mM phosphate buffer pH 7.5 in presence of 1mM DTT and 10% glycerol, cell length was 1 cm and temperature was 20° C. All spectra were the average of four measurements and were corrected by baseline subtraction. The analysis and estimation of protein secondary structure content were carried out according to Yang et al [21].
Temperature reversibility experiments were performed by cooling the 90°C solution containing the mutant Y33G down to 5°C, whereas SDS reversibility experiments were done by removing the denaturing agent by extensive dialysis against buffer A; in both cases, the CD spectra of the protein samples obtained after the reversibility experiments were measured as described before. Furthermore the protein samples after reversibility experiments were submitted to a gel filtration analysis (Superdex 75 column equilibrated with buffer A) in order to assess the amount of monomeric and/or tetrameric form.
Temperature induced protein unfolding of the Y33G mutant was followed by plotting the CD signal at 222 nm between 5 and 90 °C. To calculate the thermodynamic parameters of the unfolding process a two-state van't Hoff analysis has been applied. At a given temperature the fraction α of unfolding was calculated from the ellipticity [CD]222:
where and are the CD signals obtained from the baseline extrapolation before and after the unfolding transition. At each temperature . Also in wild-type CDA temperature induced protein unfolding data were obtained by plotting the CD signal at 222 nm between 5 and 90°C obtained from a previous work (1).
1.2.7 Structural analyses and molecular modeling
All the molecular modeling and structural analysis procedures were carried out with the Molecular Operating Environment (MOE) [22]. Hydrogens were added to the crystal structure of the mouse tetrameric CDA [23] by means of the Protonate3D function of MOE [24]. Mutations were introduced by means of the mutate function. Cytidine arabinoside (Ara-C) was docked by means of the dock function, using the “triangle matcher” for the placement of the ligand and the London ΔG for the scoring.
1.3 Results
1.3.1 Expression of Y33G mutant CDA mediated by chemical and pharmacological chaperones
We previously reported that cytidine deaminase mutated in tyrosine 33 gave rise to a partially soluble and inactive Y33G protein and to a completely insoluble Y33F and Y33S mutant CDAs. Furthermore complementation test performed by inoculating E. coli SØ5201 strain requiring uracil as pyrimidine source and harboring Y33 mutant enzymes showed that Y33G, Y33F, Y33S mutant CDA did not allow the growth of bacterial cells because of their lack of enzymatic activity [1].
In this work we used low molecular weight compounds (osmolytes and ligands of cytidine deaminase), added to the culture medium as described in the Materials and Methods session, to overcome the misfolding or the impaired oligomerization of the Y33G mutant CDA. Once grown, the cells were resuspended in the proper lysis buffer and disrupted by sonication. After centrifugation each crude extract was assayed for cytidine deaminase activity and the expression of the mutant protein was evaluated by western blot analysis with polyclonal anti-CDA antibodies and by enzymatic assay.
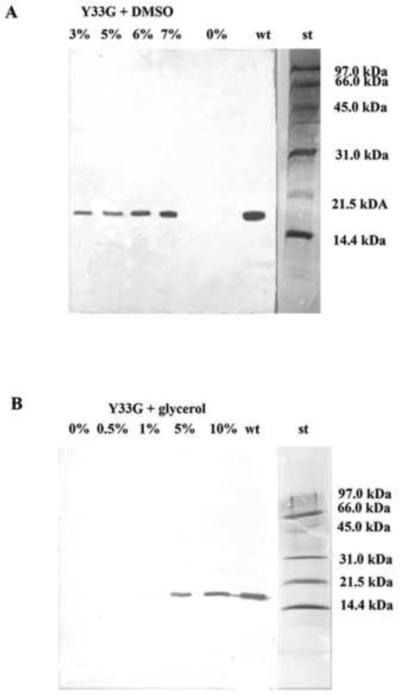
The results indicated that Y33G, in presence of low molecular weight compounds in the culture medium, increased its expression and its specific activity as shown in figures 1 and in table 1, respectively. In particular the western blot analysis of Y33G crude extracts evidenced that the addition of DMSO (3–7% final concentrations) or glycerol (5–10% final concentrations) to the culture medium increased the expression of the mutant CDA as shown in figure 1A and 1B respectively. The presence in the culture medium of sorbitol or cyclodextrins did not induce expression of Y33G (data not shown). Finally a mixture of glycerol (10% final concentration) and DMSO (7% final concentration) was added to the culture medium to test if the contemporary presence of two chaperons could have a synergetic effect on the expression of the Y33G mutant CDA, but the results indicated that the Y33G content, after the addition of the mixture to the culture medium, was similar to that obtained with glycerol and DMSO alone (data not shown). The specific activity assays performed on Y33G crude extracts (table 1) confirmed the data obtained with the western blot analyses indicating that the most effective osmolytes for the expression of Y33G mutant CDA were glycerol and dimethyl sulphoxide.
Figure 1.
Western blot analysis to evaluate the expression of the mutant CDA Y33G in presence of low molecular weight compounds in the culture medium.
A) DMSO 0%, 3%, 5%, 6%, 7%; B) Glycerol 0%, 0.5%, 1%, 5%, 10%, wt: wild-type CDA (positive control); st: Bio-Rad low molecular weight standard (97.4 kDa, phosphorylase b; 66.2 kDa, bovine serum albumin; 45.0 kDa, ovalbumin; 31.0 kDa, carbonic anhydrase; 21.5 kDa, soybean trypsin inhibitor; 14.4 kDa, lysozyme).
Table 1.
Specific activities of Y33G mutant CDA expressed in E.coli strain after addition of chemical chaperones or specific CDA ligands (substrates and inhibitors) in the growth medium. In bold are evidenced the molecules that increase Y33G specific activity.
| Chaperons or CDA ligands | CDA Specific activity (mU/mg) | |
|---|---|---|
| No Chaperons | 0.00 | |
| DMSO | 0.50% | 0.00 |
| DMSO | 1.00 % | 3.90 |
| DMSO | 5.00% | 4.60 |
| DMSO | 6.00% | 7.00 |
| DMSO | 7.00% | 5.20 |
| Glycerol | 2.00% | 0.00 |
| Glycerol | 4.00% | 1.40 |
| Glycerol | 6.00% | 1.60 |
| Glycerol | 8.00% | 3.10 |
| Glycerol | 10.0% | 4.30 |
| Cytidine | 1.00 mM | 0.00 |
| Cytidine | 5.00 mM | 0.00 |
| Cytidine | 10.0 mM | 12.0 |
| Cytidine | 20.0 mM | 19.6 |
| Cytidine | 50.0 mM | 36.2 |
| Uridine | 0.10 mM | 9.00 |
| Uridine | 0.50 mM | 9.80 |
| Uridine | 1.00 mM | 20.0 |
| Uridine | 10.0 mM | 7.90 |
| Uridine | 50.0 mM | 9.80 |
| 5-FZEB | 0.001 mM | 0.00 |
| 5-FZEB | 0.01 mM | 15.0 |
| 5-FZEB | 0.10 mM | 11.60 |
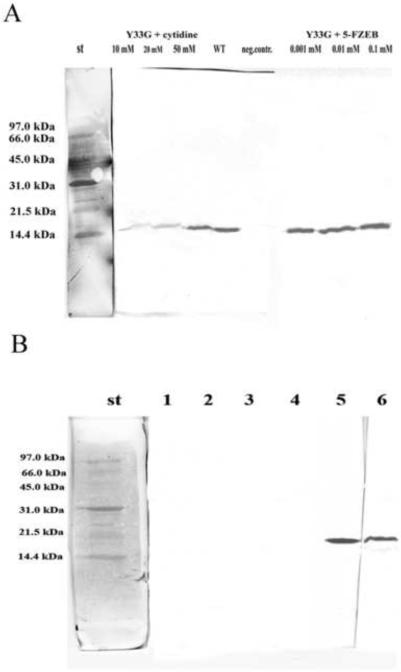
The presence of cytidine deaminase ligands (substrates and inhibitors) in the culture medium had a considerable effect on Y33G expression as evinced from figure 2A and in table 1: in particular the presence of cytidine, uridine and 5-fluorozebularine (5-FZEB) in the growth medium enhanced of 2–3 folds the specific activity of the Y33G mutant CDA, with respect to the chemical chaperones. Although the specific CDA ligands were present in the culture medium at lower concentration with respect to the chemical compounds, they resulted to be more efficient in increasing the yield of expression of the mutant CDA. The most effective pharmacological chaperone resulted to be the natural substrate cytidine at 50 mM concentration.
Figure 2.
A) Western blot analysis to evaluate the expression of the mutant CDA Y33G in presence of specific ligands of cytidine deaminase in the culture medium.
wt: wild-type CDA (positive control); neg. contr: no CDA ligands; st: Bio-Rad low molecular weight standard.
B) Western blot analysis to evaluate the presence of inclusion bodies in the crude extracts of Y33G. 1: no glycerol in the culture medium. E. coli cells were lysed and after centrifugation the supernatant was resuspended in buffer A without glycerol and analyzed; 2 and 4: no glycerol in the culture medium. E. coli cells were lysed and after centrifugation the pellet was resuspended in 8M urea and analyzed; 3: no glycerol in the culture medium. E. coli cells were lysed and after centrifugation the supernatant was resuspended in buffer A with10 % glycerol and analyzed; 5: 10% glycerol in the culture medium. E. coli cells were lysed and after centrifugation the supernatant was resuspended in buffer A with 10 % glycerol and analyzed; 6: 10% glycerol in the culture medium. E. coli cells were lysed and after centrifugation the pellet was resuspended in 8M urea and analyzed. St: Bio-Rad low molecular weight standard
Further experiments were done to distinguish if the presence of glycerol in the culture medium influences either the stability or the expression of the mutant enzyme. At this purpose E. coli cells harboring the plasmid pTrcHUMY33G were incubated in LB broth in the presence or absence of 10% glycerol, as described in the Materials and Methods session. After cell lysis and centrifugation each supernatant and each pellet (with and without glycerol) was analyzed by western blot with anti-CDA polyclonal antibodies and the specific activity was determined. In absence of glycerol in the growth medium the presence of a band corresponding to Y33G was not observed, whether in the supernatants (resuspended in buffer A with or without glycerol) or in the pellet resuspended in 8M urea (figure 2B, lanes 1–4). On the contrary, the presence of glycerol in the culture medium increased the expression of the mutant Y33G CDA as observed in figure 2B, lane 5. The presence of the protein also in the pellet resuspended in 8M urea (figure 2B, lane 6) may be due to the fact that the mutant protein may be not completely soluble.
1.3.2 Purification of Y33G mutant CDA
Y33G mutant CDA was expressed in the presence of glycerol 10% added to the culture medium and was purified according to the method described in the Materials and Methods session. The purified enzymes were judged >90% pure after 15% SDS-PAGE (data not shown). Partially purified Y33G mutant resulted to be unstable at +4°C for 24 hours (26% of residual activity) and maintained its enzymatic activity only at −20°C and in presence of 10% glycerol in the buffer A for at least one month.
1.3.3 Polyacrylamide gel electrophoresis experiments
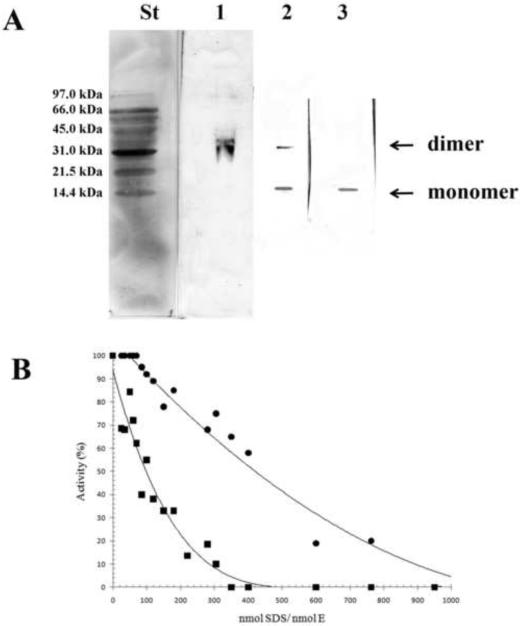
Dissociation of human CDA into subunits was studied by 15% PAGE performed at different SDS concentrations in order to reach the following SDS/enzyme molar ratios: 70, 120, 300. In a previous work [20] it has been shown that the wild-type CDA was present as a tetramer up to an SDS/enzyme ratio of 61 and completed the dissociation into monomers at an SDS/enzyme ratio of 616 without the presence of intermediate forms such as dimers and/or trimers. With the Y33G mutant CDA the dissociation process was faster and the tetrameric form of the enzyme was never observed. The dissociation process of Y33G started already at an SDS/enzyme ratio of 70, at which only the dimer was detected, continued at SDS/enzyme ratio of 120 with the presence of the dimer and the monomer and was completed at an SDS/enzyme ratio of 300, at which only the monomer was detected (figure 3A).
Figure 3.
A) 15% PAGE followed by western blot analysis of Y33G at different SDS concentrations. Each lane represents separate gels with different concentrations of the denaturing agent. The SDS/enzyme molar ratios in the individual lanes were as follows: lane 1, 70; lane 2, 120; lane 3, 300. St, Bio-Rad low molecular weight standard.
B) Percentage of enzymatic activity of wild-type CDA (●) andY33G mutant CDA (■) after incubations with SDS concentrations ranging from 0.35 to 1.73 mM in order to give different SDS/enzyme molar ratios. The data of wild type CDA are from [20].
1.3.4 Kinetic experiments
The experimental error in the measurement of the initial velocity has been estimated over a range of ten replicates and resulted to be 6.0 % of the observed value.
Kinetic experiments were performed by assaying the enzymatic activity of Y33G mutant CDA in the presence of several substrates or inhibitors of human CDA. In table 2 are given the Km and Vmax values for the wild-type CDA and for the mutant Y33G regarding the naturally occurring substrates, cytidine (CR) and deoxycytidine (CdR), and some cytosine nucleoside analogues used in chemotherapy as antitumor or antiviral agents. The kinetic parameters calculated in presence of CDA substrates revealed that deoxycytidine was the best substrate for Y33G and that the Km value for cytidine and 5-azadeoxycytidine was higher with respect to the wild-type CDA. The Vmax values calculated for all the assayed substrates resulted to be very low with respect to the wild-type CDA. Interestingly, cytosine arabinoside (Ara-C), an antileukemic agent, was not recognized as substrate by Y33G. The ability of a number of nucleosides and nucleotides to inhibit the mutant enzyme Y33G was tested (table 3): interestingly it was obtained a higher Ki value for the transition state analogue 3,4,5,6-tetrahydrouridine (THU) and a lower Ki value for uridine with respect to the wild-type cytidine deaminase. The Ki value for 5-fluorozebularine remained unchanged.
Table 2.
Kinetic parameters for substrates of wild-type and mutant Y33G CDA
| Substrates | wild-type CDAa | Y33G |
|---|---|---|
| Cytidine | ||
| Km (μM) | 39.0 | 74.5 |
| Vmax (U/mg) | 68.3 | 0.152 |
| Vmax/Km | 1.7 | 2.04×10−3 |
| Deoxycytidine | ||
| Km (μM) | 39.0 | 47.6 |
| Vmax (U/mg) | 45.0 | 0.127 |
| Vmax/Km | 1.2 | 2.67×10−3 |
| Cytosine arabinoside | ||
| Km (μM) | 110.0 | It is not substrate |
| Vmax (U/mg) | 42.0 | |
| Vmax/Km | 0.4 | |
| 5-Azadeoxycytidine b | ||
| Km (μM) | 74.8 | 349.0 |
| Vmax (U/mg) | 31.0 | 0.174 |
| Vmax/Km | 0.4 | 4.50×10−4 |
[17].
The wavelength used for 5-azadeoxycytidine assay was 270 nm.
Table 3.
Inhibition constants of wild type CDA and of Y33G mutant enzyme
| Ki (10−5 M) | ||
|---|---|---|
| Inhibitors | wild-type CDAa | Y33G |
| Uridine | 40.0 | 2.12 |
| 6-[3,5(cytidyl)acryloylamino] hexanoic acid (CV6)a | 4.50 | 0.34 |
| 3,4,5,6-Tetrahydrouridine (THU) | 0.67 | 2.56 |
| 5-Fluorozebularine | 0.03 | 0.02 |
Inhibition constants were obtained from the double-reciprocal plots of the deamination of deoxycytidine in the presence and absence of the inhibitors. All the inhibitors were competitive.
[5].
The behavior of Y33G in presence of small amounts of the denaturing agent SDS (ranging from 0.35 to 1.73 mM) was investigated in order to study the inactivation pattern of this mutant enzyme and compare it to the wild-type CDA [20]. At different SDS/enzyme molar ratios the profile of inactivation of the Y33G mutant CDA was an exponential curve (figure 3B), different to the linear one showed for the wild-type CDA (data from [20]) suggesting for the mutant enzyme a faster dissociation process, in fact it reached the complete inactivation at an SDS/enzyme molar ratio of 300 whereas the wild-type enzyme was completely inactivated at SDS/enzyme ratio of ~800 [20].
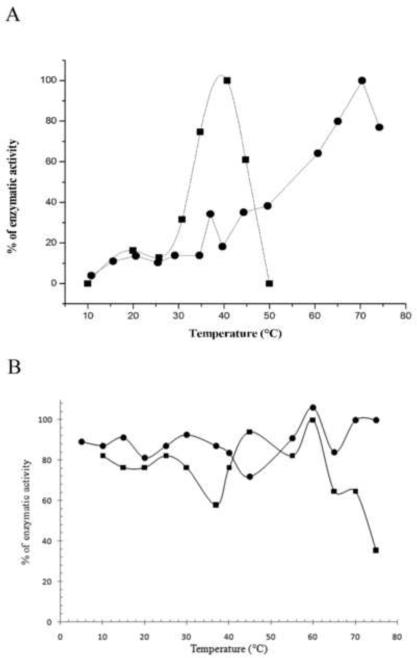
The effect of temperature on activity and stability of the wild-type CDA and the mutant Y33G was investigated as described under Materials and Methods. The optimal temperature was obtained by plotting the percentage of enzymatic activity as a function of increasing temperatures. The results, shown in figure 4A, indicated that Y33G is a thermolabile enzyme with an optimal temperature around 40°C. The rapid decrease of enzymatic activity at temperature upper to 60°C may be due to protein denaturation as showed by temperature stability experiments (figure 4B).
Figure 4.
A) Temperature effect on the activity of wild-type CDA (●) and Y33G mutant enzyme (■) obtained by plotting the percentage of enzymatic activity at temperatures ranging from 10 to 80°C.
B) Effect of temperature on enzyme stability of wild-type CDA (●) and Y33G mutant CDA(■).
On the contrary, wild-type CDA showed an activity optimum at about 70°C and the activity decreased above 75°C, as also shown in previous experiments [1].
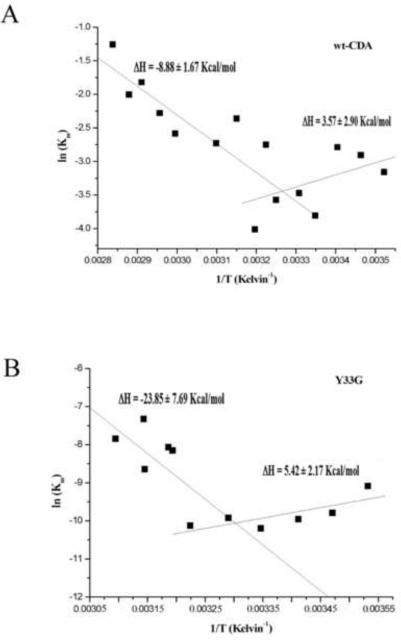
The temperature dependence of Vmax and Km values of Y33G and wild-type CDAs was also analyzed by Arrhenius equation (data not shown): the energy of activation (Ea) value calculated for the mutant enzyme was 13.812 ± 4.12 kcal/mol whereas for the wild-type CDA the Ea was 9.038 ± 0.61 kcal/mol. By the van't Hoff plot (figure 5) we have observed a discontinuity in the temperature dependence of Km for both the wild-type CDA and the Y33G mutant: the enthalpic values for the interaction between substrate and the enzyme, calculated for the wild-type CDA were ΔH = 3.57 ± 2.90 kcal/mol in the temperature range 10–35°C and ΔH = −8.88 ± 1.67 kcal/mol in the temperature range 35–80°C. The van't Hoff enthalpic value calculated for the interaction between the Y33G mutant and the substrate cytidine was ΔH = 5.42 ± 2.17 kcal/mol in the temperature range of 10–30°C and ΔH = −23.85 ± 7.69 kcal/mol in the temperature range of 30–50°C.
Figure 5.
Van't Hoff plot for wild-type CDA (A) and for Y33G mutant CDA (B).
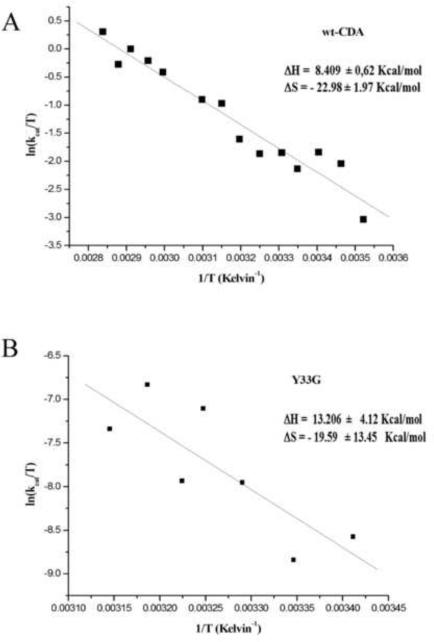
The Eyring plot (figure 6) allowed to calculate the values of entropy and enthalpy of activation. The enthalpy of activation calculated for wild-type and Y33G CDAs were 8.409 ± 0.62 kcal/mol and 13.206 ± 4.12 kcal/mol respectively whereas the values for the entropy of activation for wild-type CDA and Y33G resulted to be almost similar (−22.98 ± 1.97 cal/mol and −19.599 ± 13.45 cal/mol respectively).
Figure 6.
Eyring plot for wild-type CDA (A) and Y33G mutant CDA (B).
1.3.5 Circular Dichroism experiments on Y33G mutant enzyme
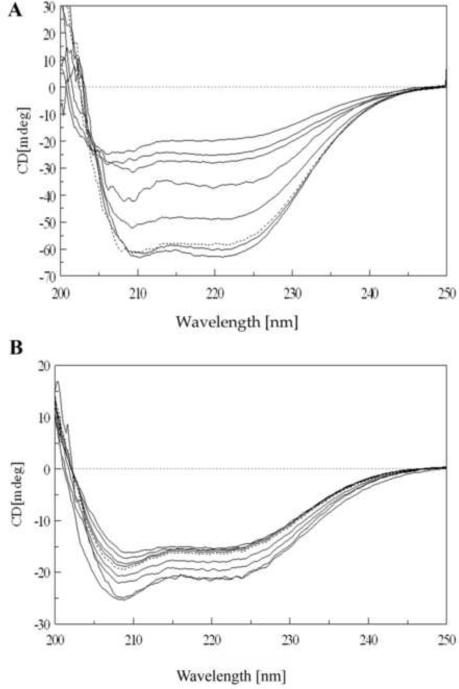
Besides functional experiments, circular dichroism (CD) measurements performed under different denaturing conditions allowed us to investigate the secondary structure and, indirectly, the whole quaternary structure of the mutated protein. At 5°C the CD spectrum of Y33G (figure 7A) was quite similar in shape to that of wild-type CDA (see figure 6 of [1]) showing that the rescued mutant folds properly. By increasing temperature, the mutant protein structure seemed sensibly influenced, although the whole secondary structure was not greatly affected: in the range from 5 to 40°C an evident decrease of the ratio 222/208 was observed, along with a slight decrease of the 222 nm ellipticity value (figure 7A). These data could indicate a change in the aromatic residues pattern [1, 25, 26] which are greatly involved in the quaternary structure formation so confirming a monomerization process as seen in the temperature dependent activity data. From 50°C to 70°C the whole ellipticity rapidly decreased but, surprisingly the shape of the spectrum remained enough unchanged with respect to that recorded at low temperature. No transition from well ordered secondary structures to random coil was evident, as it was observed for wild-type CDA from the increased negative value of the 200 nm peak, whereas the relative percentages of α-helix, β-sheet and random coil [21] resulted to be somewhat similar to those measured from the 5°C spectrum (table 4). Only at 80 and 90°C the shift to the 200 nm peak became evident. Furthermore no protein precipitation or aggregation was detected.
Figure 7.
A) CD spectra of Y33G temperature unfolding experiment: protein concentration was 5 μM in 20 mM phosphate buffer pH 7.5 in the presence of 1 mM DTT and 10% glycerol. Temperatures are from bottom to top (at 222 nm) 5, 40, 50, 60, 70, 80 and 90° C. Dotted line is the spectrum of the protein sample refolded by cooling the solution from 90° C to 5° C. Spectra obtained at 10, 20 and 30°C are omitted for the sake of clarity (see text).
B) CD spectra of Y33G SDS unfolding experiment: protein concentration was 2 μM in 20 mM phosphate buffer pH 7.5 in the presence of 1 mM DTT and 10% glycerol. Temperature was 20°C. SDS concentrations are from top to bottom (at 210 nm) 0.035, 0.69, 1.04, 1.40, 1.70, 3.5, 5.2, 6.9 mM. Dotted line is the spectrum of protein sample refolded after extensive dialysis against buffer A. Spectra obtained at SDS concentrations of 0.069, 0.17, 0.35, 0.52 mM are omitted for the sake of clarity (see text).
Table 4.
Percentage of secondary structure measured in the Y33G mutant CDA at different temperatures and SDS concentrations
| Sample | α-helix % | β-sheet % | random % | others % |
|---|---|---|---|---|
| Y33G (5°C) | 37 | 29 | 28 | 6 |
| Y33G (70°C) | 34 | 28 | 30 | 8 |
| Y33G (90°C) | 27 | 22 | 36 | 5 |
| Y33G (0.035 mM SDS) | 37 | 29 | 28 | 6 |
| Y33G (6.90 mM SDS) | 36 | 32 | 25 | 7 |
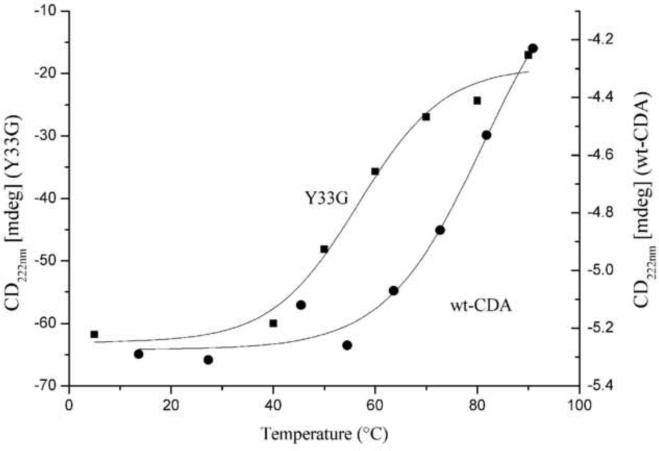
The thermal stability of Y33G mutant CDA was measured by monitoring the CD spectrum as a function of temperature. From the thermal unfolding analysis of the CD signal at 222 nm plotted against the temperature (figure 8), we have measured the thermodynamic parameters of the transition according to a two state model, obtaining a Tm of 52.16 ± 8.71 °C and a enthalpy of transition ΔH = 24.08 ± 14.89 kcal/mol. By comparison we have plotted in the same figure the unfolding curve obtained by CD spectra of the wild-type CDA previously published (see figure 6 of [1]): the highly stable wild-type protein exhibits a non-complete unfolding curve from which we couldn't obtain a good fit, (the best fit apparent transition temperature Tm is 79.25 ± 8.36 °C). Indeed, as shown in the CD spectra of the wild type CDA, reported in our previous paper [1], the protein maintains a large amount of secondary structure up to 70 °C, then the decrease of signal observed at 80–90 °C seems to reflect a loss of quaternary structure (tetramer-monomer transition) rather than the population of a fully unfolded state. A comparison of the wild-type CDA and Y33G unfolding curves indicated that the percentage of CD signal at 90°C, relatively to the pre-transition value, for the wild-type CDA is substantially higher than the one observed for the mutated protein. CD spectra of the Y33G protein undergone SDS induced unfolding revealed that the secondary structure of the protein was almost unchanged by increasing denaturing agent concentration (figure 7B). As previously shown for thermal denaturation, a change in the 222/208 ratio from 0.035 mM SDS to 6.90 mM SDS, indicated a tetramer to monomer transition quite complete at SDS/enzyme ratio of 250 according to electrophoresis and enzymatic activity experiments performed in presence of the denaturing agent (figure 3A and B). Unexpectedly, at the highest SDS concentrations used the ellipticity value became greater than that measured in the absence of denaturing agent. Also in this case, as in the aforementioned thermal denaturation data, the relative percentages of α-helix, β-sheet and random coil were similar to those measured from the initial spectrum. Reversibility experiments, followed by CD measurements, gave us more insights into the amazing structural behavior of the Y33G protein. Looking at the thermal refolding process, cooling the 90°C solution containing the Y33G mutant enzyme down to 5°C, a partial reversibility was observed: the CD spectrum showed a structure similar to that observed at 40–50°C when the monomerization process probably is complete as indicated by activity data. Similarly, removing SDS from the solution led to a CD spectrum quite similar to that of a SDS/enzyme ratio of 350, when most of the mutated protein is in the monomeric form. By means of gel filtration chromatography performed on the two unfolded/refolded proteins we have observed that both samples are monomeric (data not shown). As a control we have performed the same reversibility experiments on wild-type CDA obtaining a 90% recovery of native tetrameric protein. CD spectra of both SDS and temperature unfolded/refolded proteins are similar but not superimposable (data not shown).
Figure 8.
Thermal unfolding analysis of the CD signal of the wild type (●) and Y33G (■) CDAs (performed as described in Materials and Methods) plotted against the temperature (from 5 to 90°C). The difference of the Y axis scale is due to a different protein concentration and cell length used on the two series of experiments.
1.4 Discussion
In this work we succeeded in overcoming the above mentioned problem of the partial solubility of the mutant Y33G, by the use of molecules such as glycerol and DMSO (chemical chaperones), as well as specific ligands of cytidine deaminase (pharmacological chaperones) added to the culture medium during induction of the mutant CDA enzyme.
As shown in figure 1 and in table 1, glycerol and DMSO, when added to the culture medium as chemical chaperons, caused an increase of the solubility and the specific activity of the Y33G mutant. Substrates and inhibitors of CDA, added to the culture medium during the induction of Y33G, had a greater effect assisting the correct folding of the Y33G mutant CDA at very low concentrations, when compared to the above mentioned chemical chaperones (figure 2A and table 1): their activity is probably due to their preferential binding to the enzyme in the native and active state with its consequent stabilization [13]. CD experiments confirmed that the rescued Y33G mutant CDA is properly folded, in fact as shown in figure 7A, the CD spectrum of Y33G recorded at 5°C is quite similar in shape to that of the wild-type CDA [1].
Thanks to the help of the chaperones, the Y33G mutant was obtained as functional protein, allowing its purification and characterization, as well as a deeper study of the actual role exerted by the Y33 residue in the stabilization of the quaternary structure of the enzyme.
Kinetic studies performed on the purified protein showed that, differently from the wild-type CDA, Y33G mutant protein is a thermolable enzyme: above 60°C it is completely inactivated in an irreversible manner (see figure 4A and B).
From an analysis of the data obtained from the Km temperature dependence experiments, it may be supposed that the substitution of Y33 residue with glycine in human CDA introduces strong interactions with the ligands as confirmed by the strong negative enthalpic value calculated by the van't Hoff plot in the temperature range 30–50°C (see figure 5B). This may in part explain the greater effect exerted by the CDA ligands in the rescuing of Y33G mutant. On the other hand, the Y33G mutant enzyme has to overcome a higher energy barrier in converting the substrate into the product because of the increasing Ea value that leads to a decrease of the Vmax value with respect to the wild-type CDA.
The increase of the activation energy for the reaction catalyzed by the Y33G mutant CDA can be due either to a decrease in free energy of the enzyme-substrate complex or to an increase of the free energy of the enzyme-transition state complex. Energetically this would imply that the Y33G mutant enzyme makes stronger interactions with the substrate than with the transition state, as shown also by the higher Ki value for THU and the lower Ki value for uridine of Y33G with respect to the wild-type enzyme. Structurally THU is a transition state analogue while uridine is the product of the reaction, geometrically resembling the substrate.
The van't Hoff plot for the wild-type CDA (figure 5A) shows two different temperature ranges corresponding to different binding energies for the substrate: in the temperature range of 10–35°C the enthalpy of binding is endothermic (ΔH=3.57 ± 2.9 kcal/mol) whereas in the temperature range of 35–80°C is exothermic (ΔH=−8.88 ± 1.67kcal/mol). The fact that in the temperature range of 10–35°C the enthalpic value is slightly positive seems to indicate that the binding process is leaded entropically; whereas in the temperature range of 35–80°C the binding process with the substrate may be due to the formation of favorable non-covalent bonds. Therefore a conformational modification may occur in the temperature range around 35°C that leads to a strong interaction with the substrate deoxycytidine.
A similar behavior was observed for the Y33G mutant CDA (see figure 5B) indicating that at 30°C the binding of cytidine switched from an endothermic to a exothermic process, as a conformational change had occurred.
Considering the data obtained from the Eyring plot (figure 6), the increase of the enthalpy of activation (13.206 ± 4.12 kcal/mol) of about 5 kcal/mol for the mutant Y33G with respect to the wild-type CDA (8.409 ± 0.62 kcal/mol) is indicative that a greater number of chemical bonds needs to be broken to reach the transition state of the reaction because of the substitution of tyrosine 33 with a glycine residue. This substitution seems not to have effect on the geometry of the enzymatic reaction since the value of the entropy of activation for the Y33G mutant CDA was not changed with respect to the wild-type enzyme.
Electrophoresis experiments indicated that the Y33G mutant is more sensitive to the subunits dissociation induced by SDS than the wild-type enzyme. Already at very low SDS concentrations, the Y33G mutant was detected only as a dimer. From there, the enzyme proceeded very rapidly to a complete dissociation, as evinced by the dissociation curve obtained by plotting the percentage of enzymatic activity vs. the SDS/enzyme ratios (figure 3B) and by the electrophoretic pattern (figure 3A). On the contrary the wild-type CDA was detected as a tetramer at low SDS concentrations and its dissociation to the monomeric state proceeded without the intermediate formation of trimers or dimers, via a non-allosteric mechanism [20].
CD experiments confirmed that a monomerization process occurs for the Y33G mutant in the presence of increasing SDS concentrations, but also indicated that the secondary structure of the mutant enzyme does not undergo changes (figure 7B). The same behavior was observed recording the CD spectra for Y33G mutant at increasing temperatures. In particular, from 5°C to 40°C a rearrangement of the aromatic side chains of the amino acids involved in the formation of the active quaternary structure was observed, in agreement with a clear decrease of the activity of the enzyme (figure 7A). However, from 40°C to 90°C, no substantial changes in the secondary structure of the Y33G mutant were observed. The transition enthalpy calculated for the thermal unfolding of the Y33G CDA (ΔH = 24.08 ±14.89 kcal/mol) is in fact quite low for a complete unfolding event, especially when considering a tetrameric protein. Our temperature studies of the Y33G mutant suggested the population of an inactive partly folded intermediate after the transition centered at the Tm of 52.16 °C (figure 8).
These results emphasize the important role exerted by the Y33 residue in the intersubunit contacts: the substitution of the tyrosine with a glycine residue favors the monomerization process at low denaturant concentrations or temperatures above 50 °C, however, it also leads to a more compact and stable secondary structure as confirmed by the temperature reversibility experiments. It is possible to speculate that the smaller volume of Gly confers to the protein a more rigid packing, especially at the level of the β-sheet and the α-helix close to the mutated residues, leaving the protein with an increased secondary structure rigidity and stronger hydrophobic contacts. This is confirmed by the increase of the whole ellipticity in the SDS micelle-entrapped monomeric protein.
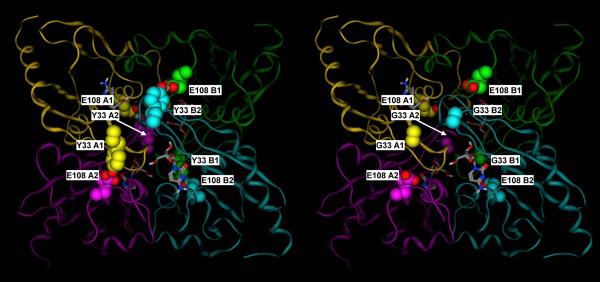
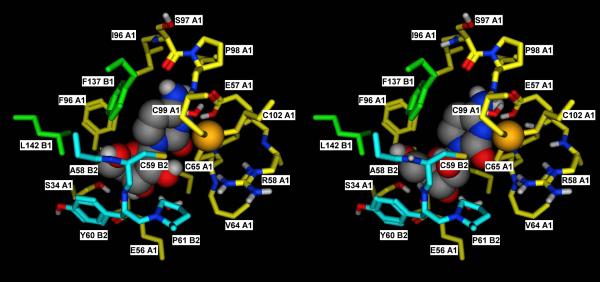
As discussed, the CDA homotetramer shows an overall C222 symmetry, with two subunits (A1 and B1) corresponding to the catalytic subunits of the E. coli dimer and the other two (A2 and B2) corresponding to the C-terminal “broken site” of the E. coli dimer [2]. Looking at the published crystal structures (PDB IDs: 1JTK, 1MQ0, 2FR6, and 2D30) [4, 23, 27], it appears evident that the Tyr and the Glu residues that correspond to Y33 and E108 in human CDA play a relevant role in the stabilization of the quaternary structure of the enzyme. In particular, as shown in figure 9, Y33 of subunit A1 is hydrogen bonded to E108 of subunit A2, while Y33 of subunit A2 is hydrogen bonded to E108 of subunit A1, thus forming a double latch that keeps together the two subunits corresponding to a single E. coli monomer. Exactly the same thing happens for the subunits B1 and B2. Notably, the same pair is found in each single monomer of the E. coli enzyme, where Y206 (homologous of the human Y33 in the E. coli C-terminal domain) is hydrogen bonded to E138 (homologous of the human E108 in the E. coli catalytic domain). The functional role of this residue pair is also suggested by its high conservation: it was detected in 69 out of the 75 sequences of tetrameric CDAs in our published sequence alignment [2]. This observation is in line with the discussed experimental data, which all suggest that the Y33G mutation makes the homotetramer significantly more labile, thus explaining the significantly lower Vmax values for all of the tested substrates with respect to the wild-type CDA. Most likely, the prevalent form of the Y33G mutant is the homodimer (A1–B1) rather than the catalytically active homotetramer. As discussed, the importance of the quaternary structure of the enzyme for its catalytic activity is well known, as residues from three different subunits contribute to the formation of each of the four active sites. Figure 10 shows how residues of subunits B1 and B2 contribute to the formation of the catalytic site of subunit A1. In the Y33G mutant, as mentioned, the interactions of the two subunits 1 with the two subunits 2 are likely to be weaker, thus compromising the stability of the intact tetramer. This view is also consistent with the fact that Ara-C is not a substrate for the Y33G mutant, as the 2'-hydroxyl group sterically interferes with the already weakly bound third subunit (B2 in the figure) and further contributes to the destabilization of the tetramer.
Figure 9.
A) Crystal structure of the mouse cytidine deaminase in complex with cytidine (2FR6, (Teh et al., 2006).
B) Corresponding molecular model of the Y33 mutant. Y33 of subunits A1, A2, B1 and B2 form hydrogen bonds with E108 of subunits A2, A1, B2 and B1, respectively, thus double latching each of the two monomer pairs corresponding to the catalytic site and the C-terminal broken site in the E. coli dimeric enzyme (A1–A2 and B1–B2). Subunit A1, A2, B1 and B2 are in yellow, purple, green and cyan, respectively. Y33 and E108 residues are shown in space filling representation, with the carbon atoms colored by subunit, and the heteroatoms colored by element. Cytidine is shown in sticks representation, with the atoms colored by element.
Figure 10.
A) Detailed view of the catalytic site of subunit A1 in the crystal structure of the mouse cytidine deaminase in complex with cytidine (2FR6, (Teh et al., 2006)).
B) Corresponding docking complex obtained in silico for Ara-C. The catalytic importance of the tetrameric structure derives from the fact that residues of subunits B1 and B2 contribute to the formation of the catalytic site. The docking complex obtained for Ara-C suggests that the orientation that the 2'-OH group assumes in this compound may exert some steric hindrance with the residues of subunit B2, thus adding a destabilizing element for the quaternary structure. In the Y33 mutant, this is probably sufficient to disrupt the already weak tetramer, thus explaining its inability to catalyze the deamination of Ara-C.
Subunit A1, B1 and B2 are in yellow, green and cyan, respectively. Residues lining the binding pocket are shown in sticks representation, with the carbon atoms colored by subunit, and the heteroatoms colored by element. Cytidine and Ara-C are shown in space filling representation, with the atoms colored by element.
1.5 Conclusions
Human CDA is a functional enzyme only when it is assembled into a tetramer of four identical subunits, each containing a zinc atom. The monomer alone is not able to catalyze the deamination of the substrate and no cooperativity is observed between monomers.
Mutations of Y33 cause a lack of expression of the enzyme. Through the addition of chemical or pharmacological chaperones (CDA ligands), we rescued the native form of the Y33G mutant enzyme. Furthermore, the high sensitivity of Y33G to SDS-induced dissociation and a wealth of additional experiments suggest that in human CDA the side chain of Y33 is involved in the intersubunit interactions and affects the stability of the quaternary structure of the enzyme. This is confirmed by the published crystal structures of tetrameric CDAs, which reveal that, the four tyrosine residues corresponding to Y33 in human are always hydrogen bonded to the four glutamate residues corresponding to E108 in human. In particular, Y33 residues of subunits A1, A2, B1 and B2 are paired with E108 residues of subunits A2, A1, B2, and B1, forming two double latches that connect the monomers, corresponding to the catalytic subunits in the dimeric CDAs, to the monomers related to the C-terminal “broken active sites”.
From our data it is possible to hypothesize that the mutation of tyrosine 33 to glycine may impede the proper assembly of the quaternary structure of the enzyme, leading to protein aggregation and degradation as shown also for other mutant proteins [7]. This hypothesis is supported by the evidence that in absence of glycerol in the growth medium, the presence of Y33G whether in the supernatant or in the pellet resuspended in 8M urea was not detected, after western blot analysis (see figure 2B, lanes 1–4). On the other hand, Y33G, once rescued thanks to the help of the chaperones, is a functional protein endowed with catalytic activity, suggesting that it adopts a native quaternary structure.
Intersubunit residues are directly involved in the cooperative folding step and, according to our results, in human CDA, the Y33G mutation seems to affect the cooperativity of the folding process, as it can be assumed by the presence of dimers in the dissociation equilibrium of the Y33G mutant. Y33 can be considered essential for the cooperative folding of CDA, enabling the protein to reach the soluble native quaternary structure and thus avoiding the population of partly unfolded states prone to aggregation. Thanks to chemical or pharmacological chaperones the Y33G mutant CDA was able to overcome the energy barrier necessary to reach the quaternary structure either by destabilizing the partly folded intermediate (chemical chaperones), or by promoting the cooperative folding by stabilizing the native tetramer (pharmacological chaperones).
Acknowledgements
This research was supported, in part, by the Intramural Research Program of the NIH, NIDDK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Vincenzetti S, Quadrini B, Mariani P, De Sanctis G, Cammertoni N, Polzonetti V, Pucciarelli S, Natalini P, Vita A. Modulation of human cytidine deaminase by specific aminoacids involved in the intersubunit interactions. Proteins. 2008;70:144–156. doi: 10.1002/prot.21533. [DOI] [PubMed] [Google Scholar]
- [2].Costanzi S, Vincenzetti S, Cristalli G, Vita A. Human cytidine deaminase: a three-dimensional model of a tetrameric metallo-enzyme inferred from a crystal structure of a distantly related dimeric homologue. J. Mol. Graph. Model. 2006;25:10–16. doi: 10.1016/j.jmgm.2005.10.008. [DOI] [PubMed] [Google Scholar]
- [3].Betts L, Xiang S, Short SA, Wolfenden R, Carter CW. Cytidine deaminase. The 2.3 Ǻ crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- [4].Johansson E, Mejlhede N, Neuhard J, Larsen S. Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 Å resolution. Biochemistry. 2002;41:2563–2570. doi: 10.1021/bi011849a. [DOI] [PubMed] [Google Scholar]
- [5].Vincenzetti S, Cambi A, Maury G, Bertorelle F, Gaubert G, Neuhard J, Natalini P, Salvatori D, De Sanctis G, Vita A. Possible role of two phenylalanine residues in the active site of human cytidine deaminase. Protein Eng. 2000;13:791–799. doi: 10.1093/protein/13.11.791. [DOI] [PubMed] [Google Scholar]
- [6].Tatzel J, Prusiner SB, Welch WJ. Chemical chaperones interfere with the formation of scrapie prion protein. EMBO J. 1996;15:6363–6373. [PMC free article] [PubMed] [Google Scholar]
- [7].Singh LR, Chen X, Kožich V, Kruger WD. Chemical chaperone rescue of mutant cystationine b-synthase. Mol. Genet. Metab. 2007;91:335–342. doi: 10.1016/j.ymgme.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Morello JP, Petäjä-Repo UE, Bichet DG, Bouvier M. Pharmacological chaperones: a new twist on receptor folding. TIPS. 2000;21:466–469. doi: 10.1016/s0165-6147(00)01575-3. [DOI] [PubMed] [Google Scholar]
- [9].Leandro P, Lechner MC, Tavares de Almeida I, Konecki D. Glycerol increases the yield and the activity of human phenylalanine hydroxylase mutant enzymes produced in a prokaryotic expression system. Mol. Genet. Metab. 2001;73:173–178. doi: 10.1006/mgme.2001.3172. [DOI] [PubMed] [Google Scholar]
- [10].Sato S, Ward CL, Krouse ME, Wine JJ, Kopito RR. Glycerol reverses the misfoling phenotype of the most common cystic fibrosis mutation. J. Biol. Chem. 1996;271:635–638. doi: 10.1074/jbc.271.2.635. [DOI] [PubMed] [Google Scholar]
- [11].Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- [12].Ulloa-Aguirre A, Janovick JA, Brothers SP, Conn PM. Pharmacologic rescue of conformationally-defective proteins: implications for the treatment of human disease. Traffic. 2004;5:821–837. doi: 10.1111/j.1600-0854.2004.00232.x. [DOI] [PubMed] [Google Scholar]
- [13].Arakawa T, Ejima D, Kita Y, Tsumoto K. Small molecule pharmacological chaperones: from thermodynamic stabilization to pharmacological drugs. Biochim. Biophys. Acta. 2006;1764:1677–1687. doi: 10.1016/j.bbapap.2006.08.012. [DOI] [PubMed] [Google Scholar]
- [14].Song JL, Chuang DT. Natural osmolyte trimethylamine N-oxide corrects assembly defects of mutant branched-chain alpha-ketoacid decarboxylase in maple syrup urine disease. J. Biol. Chem. 2001;276:40241–4026. doi: 10.1074/jbc.M107242200. [DOI] [PubMed] [Google Scholar]
- [15].Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, Kotler M, Hizi A, Loyter A, Friedler A. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc. Natl. Acad. Sci. U S A. 2007;104:8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Canals M, Lopez-Gimenez JF, Milligan G. Cell surface delivery and structural re-organization by pharmacological chaperones of an oligomerization-defective alpha(1b)-adrenoceptor mutant demonstrates membrane targeting of GPCR oligomers. Biochem. J. 2009;417:161–172. doi: 10.1042/BJ20081227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Vincenzetti S, Cambi A, Neuhard J, Garattini E, Vita A. Recombinant human cytidine deaminase: expression, purification and characterization. Protein Expres. Purif. 1996;8:247–253. doi: 10.1006/prep.1996.0097. [DOI] [PubMed] [Google Scholar]
- [18].Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:249–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- [19].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [20].Vincenzetti S, De Sanctis G, Costanzi S, Cristalli G, Mariani P, Mei G, Neuhard J, Natalini P, Polzonetti V, Vita A. Functional properties of subunit interactions in human cytidine deaminase. Protein Eng. 2003;16:1055–1061. doi: 10.1093/protein/gzg117. [DOI] [PubMed] [Google Scholar]
- [21].Yang JT, Wu CSC, Martinez HM. Calculation of protein conformation from circular dichroism. Method Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- [22].MOE . version.10 Chemical Computing Group Inc.; 2008. [Google Scholar]
- [23].Teh AH, Kimura M, Yamamoto M, Tanaka N, Yamaguchi I, Kumasaka T. The 1.48 A resolution crystal structure of the homotetrameric cytidine deaminase from mouse. Biochemistry. 2006;45:7825–7833. doi: 10.1021/bi060345f. [DOI] [PubMed] [Google Scholar]
- [24].Labute P. Protonate3D: assignment of ionization states and hydrogen coordinates to macromolecular structures. Proteins. 2009;75:187–205. doi: 10.1002/prot.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Santucci R, Polizio F, Desideri A. Formation of a molten-globule like state of cytochrome c induced by high concentration of glycerol. Biochimie. 1999;81:745–751. doi: 10.1016/s0300-9084(99)80132-1. [DOI] [PubMed] [Google Scholar]
- [26].Manning MC, Woody RW. Theoretical study of the contribution of the aromatic side chains to the circular dichroism of basic bovine pancreatic trypsin inhibitor. Biochemistry. 1989;28:8609–8612. doi: 10.1021/bi00447a051. [DOI] [PubMed] [Google Scholar]
- [27].Chung SJ, Fromme JC, Verdine GL. Structure of human cytidine deaminase bound to a potent inhibitor. J. Med. Chem. 2005;48:658–660. doi: 10.1021/jm0496279. [DOI] [PubMed] [Google Scholar]