Abstract
Guanine nucleotide exchange is essential for Rab GTPase activities in regulating intracellular vesicle trafficking. This exchange process is facilitated by guanine nucleotide exchange factor (GEF). Previously, we identified Caenorhabditis elegans AEX-3 as a GEF for Rab3 GTPase. Here we demonstrate that AEX-3 regulates neural activities through a second, previously unrecognized pathway via interactions with the novel protein CAB-1. CAB-1 is 425 amino acids long and has an 80 amino acid motif in common with the mouse neural protein NPDC-1. cab-1 and rab-3 mutants have different behavioral defects, and RAB-3 localization and function are apparently normal in cab-1 mutants, indicating that the CAB-1 pathway is distinct from the RAB-3 pathway. The aex-3 mutant phenotype resembles the sum of the rab-3 and cab-1 mutant phenotypes, indicating that AEX-3 regulates two different pathways for neural activities. We propose that connection of multiple pathways may be an important feature of Rab GEFs to coordinate various cellular events.
Keywords: Caenorhabditis elegans/guanine nucleotide exchange/neurotransmission/Rab GTPases/secretion
Introduction
Rab GTPases regulate intracellular vesicle traffic from yeast to mammals (Novick and Zerial, 1997). Rab proteins are involved in membrane tethering and are thought to function upstream of the SNARE [soluble N-ethylmaleimide sensitive factor (NSF) attachment protein receptor] components (Lian et al., 1994; Sogaard et al., 1994; Lupashin and Walters, 1997). The SNARE proteins are essential in various membrane trafficking processes and are also conserved from yeast to mammals (Sudhof, 1995). These proteins include members of the syntaxin, synaptobrevin/VAMP and SNAP-25 families, and form stable ternary complexes. The formation of these complexes is proposed to be a prerequisite for membrane fusion (Hanson et al., 1997; Hohl et al., 1998; Poirier et al., 1998; Sutton et al., 1998; Weber et al., 1998). Rab GTPases regulate the formation of complexes between v-SNAREs and their cognate t-SNAREs. For example, recent studies have shown that Rab5 and its effector EEA1 mediate the docking/fusion of early endosomes by interacting with syntaxin 13 and NSF (Christoforidis et al., 1999; McBride et al., 1999). In addition, Rab3 is implicated in the fine-tune control of synaptic vesicle release. In Caenorhabditis elegans rab-3 mutants, synaptic vesicle clustering is partially disrupted at pre-synaptic terminals (Nonet et al., 1997). In Rab3a-deficient mice, pre-synaptic activities were quickly attenuated following a train of repetitive stimuli (Geppert et al., 1994). Further more, Rab3a is also implicated in synaptic plasticity, since Mossy fiber long-term potentiation was reduced in Rab3a-deficient mice (Castillo et al., 1997; Lonart et al., 1998). These observations support the idea that Rab GTPases are necessary for regulating docking/fusion in membrane trafficking pathways, in conjunction with the SNARE components.
GDP/GTP exchange is essential for Rab GTPase activities. The GTP-bound form of the Rab proteins recruits their effector proteins, and this exchange process is regulated by guanine nucleotide exchange factors (GEFs). GEFs for various Rab proteins have been identified including those for Rab3, Rab5 and Sec4 (Horiuchi et al., 1997; Iwasaki et al., 1997; Wada et al., 1997; Walch-Solimena et al., 1997). Surprisingly, these proteins show little similarity in amino acid sequence, implying that they have evolved from different ancestral genes. This raises the interesting question of why Rab GEFs are very diverse in spite of their common GEF function for conserved Rab GTPases.
Rab3 GEF was biochemically purified from rat and demonstrated to stimulate specifically the guanine exchange activity of Rab3, but not of other Rab proteins (Wada et al., 1997). The C.elegans protein AEX-3 is highly homologous to rat Rab3 GEF and is required for Rab3 function in vivo (Iwasaki et al., 1997). In aex-3 mutants, RAB-3 aberrantly accumulated in neural cell bodies instead of synapse-rich axons, and a variety of behavioral defects was observed (Iwasaki et al., 1997). These observations indicate that Rab3 GEF functions as a regulator of Rab3 both in vitro and in vivo.
Genetic analysis using C.elegans implied that AEX-3 has other functions in addition to that of a Rab3 GEF. aex-3 mutants showed defects in the defecation motor program that were not observed in rab-3 mutants (Iwasaki et al., 1997; Nonet et al., 1997). Furthermore, identification of the human protein MADD (MAP kinase activating protein containing death domain), an ortholog of rat Rab3 GEF and C.elegans AEX-3, which interacts with tumor necrosis factor receptor I (TNFR I) (Schievella et al., 1997), implies that AEX-3 may interact with other proteins besides RAB-3.
Here we show that the novel protein CAB-1 (C-terminus of AEX binding) is necessary for synaptic regulation by AEX-3. CAB-1 and AEX-3 bind to each other in the yeast two-hybrid system and in vitro. Both genes are expressed in a variety of neurons and their mutants show the same defecation defects, which are not seen with rab-3 mutants. aex-3 mutants show all the phenotypes of cab-1 and rab-3 mutants. Therefore, we conclude that the Rab3 GEF homolog AEX-3 is a regulator of multiple pathways for neural activities. We also propose that connection of multiple pathways may be an important feature of Rab GEFs, and that their sequence diversity may have arisen from a necessity to interact with different partners in addition to Rab GTPases.
Results
rab-3 and aex-3 mutants are different in the defecation behavior
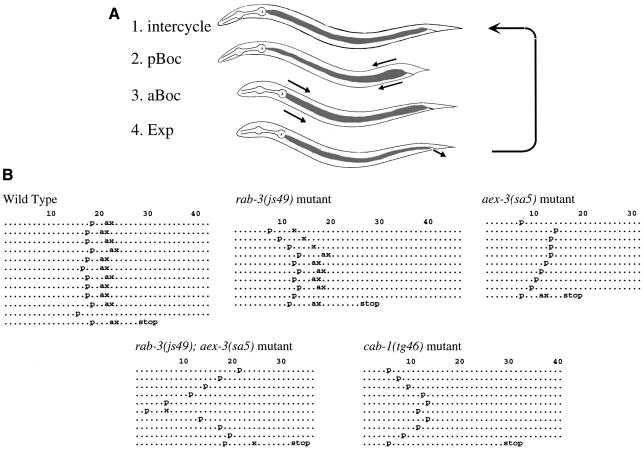
The defecation motor program consists of a stereotyped series of three muscle contractions (Figure 1A): the posterior body-wall muscle contraction (pBoc); the anterior body-wall muscle contraction (aBoc); and the enteric muscle contraction (Exp) (Thomas, 1990; Liu and Thomas, 1994). aex-3 mutants failed to express the aBoc and Exp steps (Aex standing for aBoc and Exp defective) while the wild type and rab-3 mutants showed all three steps regularly (Figure 1B, also see Figure 7A). We considered two possible explanations for the aex-3 phenotype being more severe than the rab-3 phenotype. First, the GDP-bound form of RAB-3 may inappropriately accumulate in the aex-3 mutant and interfere with other protein interactions. Alternatively, AEX-3 may have functions in addition to that of a Rab3 GEF.
Fig. 1. Defecation motor program and mutant phenotypes. (A) Schematic representation of the defecation motor program. (1) Intercycle; (2) pBoc, posterior body wall muscle contraction; (3) aBoc, anterior body wall muscle contraction; (4) Exp, enteric muscle contraction with expulsion of gut contents, then return to the intercycle. (B) Ethograms of defecation behavior in wild-type and mutant animals. Each dot or character represents 1 s. Seconds elapsed are indicated above each ethogram. p, a and x represent pBoc, aBoc and Exp, respectively. Ten animals were observed for each strain. The expulsion frequency (% ± SD) and defecation cycles (s ± SD) are as follows: 90.0 ± 6.3% and 48.8 ± 3.9 s for wild type; 91.6 ± 3.5% and 51.1 ± 3.7 s for rab-3; 23.3 ± 4.2% and 39.8 ± 2.9 s for aex-3; 19.8 ± 5.7% and 42.4 ± 7.6 s for rab-3; aex-3; and 8.9 ± 3.1% and 42.5 ± 4.5 s for cab-1.
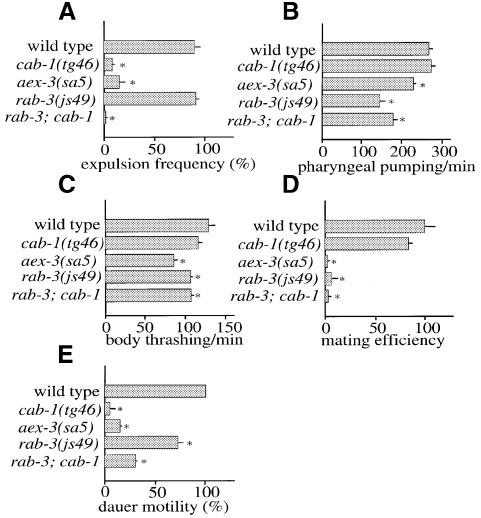
Fig. 7. Phenotypic analysis of cab-1 mutants. (A) Expulsion frequency. The x axis shows the percentage of Exp relative to pBoc. The error bars show the standard error. Ten animals were observed for each strain. An asterisk indicates statistical significance at P <0.05. (B) Pharyngeal pumping rate. Pharyngeal pumping in well-fed animals was counted for 1 min; n = 10 for each strain. (C) Body thrashing frequency. Body thrashing was counted in M9 solution for 1 min; n = 10 for each strain. (D) Male mating efficiency. Six males of each strain were mated with six dpy-11(e224) hermaphrodites for 24 h. All cross progeny (non-Dpy) were counted. Three trials were performed for each strain. The efficiency is expressed as the percentage of the wild-type mean. In all strains containing rab-3 mutations, him-8(e1489) was included in trans to the wild-type locus, therefore, him-8(e1489)/+. The mating efficiency of him-8/+ males was indistinguishable from that of the wild-type males (data not shown). (E) Dauer motility. Dauer motility was defined as the distance the dauer moved in 15 s, which was measured using a motion recorder system (Flovel Co., Japan); n >16 for each strain. These values are also expressed as a percentage normalized to wild type.
To test the first hypothesis, we examined the phenotype of rab-3; aex-3 double mutants. If GDP-bound RAB-3 interferes with other protein interactions, the loss of RAB-3 in the aex-3 mutant should convert the Aex phenotype to that of the rab-3 mutant. However, rab-3; aex-3 double mutants still showed the Aex phenotype (Figure 1B), indicating that this hypothesis is incorrect. Therefore, we pursued the second hypothesis that AEX-3 has functions in addition to that of a Rab3 GEF.
Identification of a novel AEX-3-binding protein
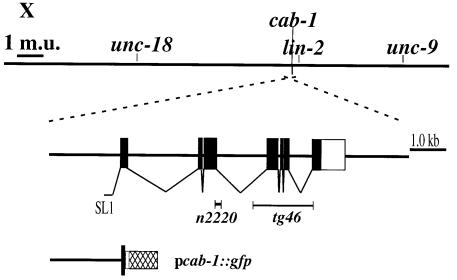
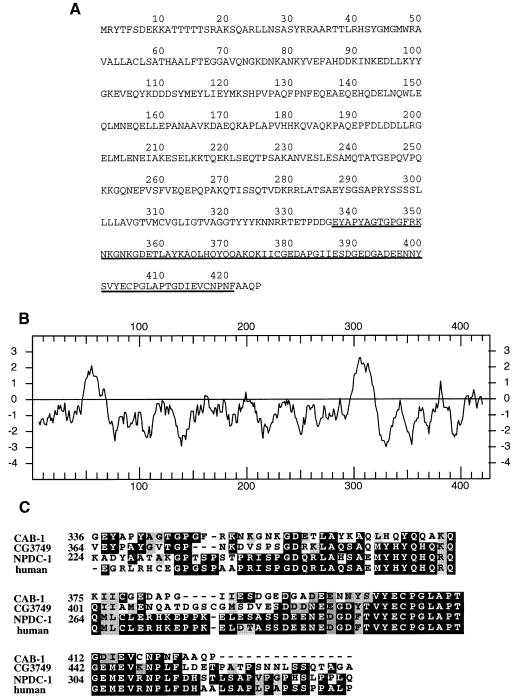
The human AEX-3 homolog MADD interacts with TNFR I through its C-terminus (Schievella et al., 1997), which is 52% identical in amino acid sequence to the AEX-3 C-terminus. We hypothesized that AEX-3 might also bind another protein (or proteins) via this domain, and that this interaction might be important for defecation regulation. To identify potential AEX-3-binding proteins, we used the yeast two-hybrid system with the 320 amino acid AEX-3 C-terminus as a bait (Fields and Song, 1989). After screening 1.6 × 106 clones, we recovered and sequenced 62 clones, of which 41 were derived from a single gene. This gene, which lies near lin-2 on chromosome X (Figure 2), is named cab-1 (C-terminus of AEX binding). cab-1 comprises 7 exons and is trans-spliced to the SL1 leader at the 5′ end (Figure 2). The predicted CAB-1 protein has 425 amino acids and contains two hydrophobic domains (Figure 3). An 80 amino acid domain at the C-terminus is weakly homologous to a region of mouse NPDC-1, whose expression correlates with differentiation of neurons (Galiana et al., 1995), the Drosophila melanogaster CG3749 translate and the human DKFZp586J0523 translate (Figure 3). We therefore named this region the NCH domain (NPDC-1 and CAB-1 homologous domain). To date, a biochemical function for this domain is unknown.
Fig. 2. Map position and gene structure of cab-1. The top bar shows a genetic map of a region of chromosome X. The scale bar represents one map unit (m.u.). cab-1 lies on the cosmid C23H4, which is three cosmids left from lin-2. The cab-1 gene is transcribed from right to left on the genetic map; the physical map below is reversed. Exons are represented on the physical map by boxes; the black boxes are coding regions and the white box is a non-coding region. The splice leader sequence of cab-1 is SL1. The n2220 and tg46 deletion mutations are represented under the physical map by bars and are 72 and 1269 nt, respectively. pcab-1::gfp is a fusion of a cab-1 promoter fragment, a nuclear localization signal (small white box) and the GFP coding region (hatched box).
Fig. 3. Sequence of the predicted CAB-1 protein. (A) CAB-1 amino acid sequence. The underlined sequence is the NCH domain (see C). The AEX-3-interacting domain (AID) is from amino acids 205–424. (B) Hydropathy of CAB-1. The y axis indicates the hydropathic scale; top to bottom is hydrophobic to hydrophilic. (C) Alignment between a region of CAB-1, Drosophila CG3749 translate, mouse NPDC-1 and human DKFZp586J0523 translate. Amino acid numbers are shown with the sequences. The human clone is a partial cDNA, the entire sequence is unknown. We named this region the NCH domain (NPDC-1 and CAB-1 homologous domain). The BLAST score between mouse NPDC-1 and CAB-1 was 70 and the probability was 6e–11.
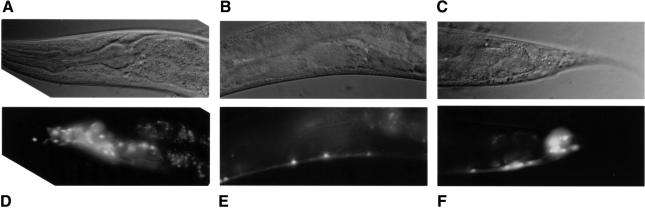
We previously reported that aex-3 is expressed in a variety of neurons (Iwasaki et al., 1997); if CAB-1 interacts with AEX-3 in vivo, then cab-1 should also be expressed in neurons. We constructed a reporter gene containing the green fluorescence protein (GFP) coding region under the control of the cab-1 promoter (Figure 2), and established transgenic lines carrying this gene. GFP fluorescence was detected in various neurons of transgenic animals, including the major ganglia near the nerve ring, ventral cord neurons and tail ganglion neurons (Figure 4). We also observed variation in fluorescence intensity between different neurons; for example, in the ventral cord only 1/3 of neurons were strongly fluorescent (Figure 4E). We are uncertain as to whether this variation reflects endogenous cab-1 expression levels. However, the expression of cab-1 in various neurons is consistent with the notion that CAB-1 and AEX-3 interact in vivo.
Fig. 4. Expression of a cab-1::gfp fusion construct. (A–C) Nomarski images of animals carrying pcab-1::gfp (see Figure 2). (D–F) Epifluorescence images of the corresponding Nomarski images. (A) and (D) show the head ganglia surrounding the nerve ring. (B) and (E) show a middle body segment with the ventral nerve cord on the ventral aspect. (C) and (F) show a posterior tail ganglion. In the head ganglia and the tail ganglia, the observed fluorescence haze arises from many fluorescent neurons positioned out of the focal plane of the photograph.
AEX-3- and CAB-1-interacting domains
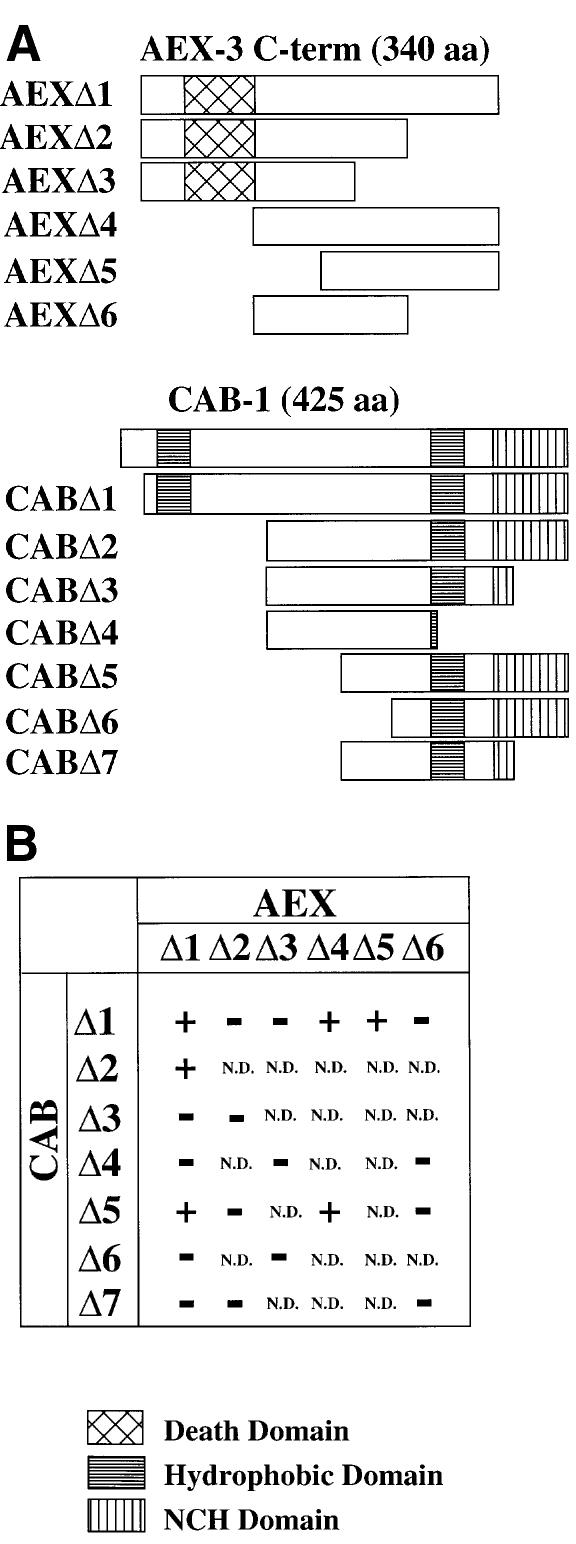
MADD and TNFR I interact via their death domains, a class of protein–protein interaction domain (Schievella et al., 1997). To determine whether the death domain-like motif in AEX-3 is responsible for binding to CAB-1, we constructed bait vectors containing different AEX-3 C-terminal domains, and prey vectors containing various CAB-1 domains for a yeast two-hybrid binding assay (Figure 5). The AEXΔ1 construct (the original bait) and the CABΔ1 construct (the longest clone isolated from the screen) were used as positive controls. The AEXΔ2 and AEXΔ3 constructs both contained the death domain-like motif but had no detectable interaction with CABΔ1 (Figure 5B). In contrast, AEXΔ4 (the C-terminal 233 amino acids) and AEXΔ5 (the C-terminal 178 amino acids), which did not contain the death domain-like motif, did interact with CABΔ1. AEXΔ6 (the central 142 amino acids) did not interact with CABΔ1 (Figure 5). These results indicate that AEX-3 does not bind to CAB-1 through its death domain-like motif. We defined the CAB-1-interacting domain (CID) as amino acids 1217–1405 in AEX-3.
Fig. 5. Binding domains of AEX-3 and CAB-1. (A) Schematic diagrams of AEX-3 and CAB-1 domains used for the yeast two-hybrid binding assays. Construct AEXΔ1 contained the 340 C-terminal amino acids of AEX-3 (amino acids 1066–1405); AEXΔ2, 1066–1309; AEXΔ3, 1066–1259; AEXΔ4, 1163–1405; AEXΔ5, 1217–1405; AEXΔ6, 1163–1309. Construct CABΔ1 contained amino acids 23–425 of CAB-1; CABΔ2, 136–425; CABΔ3, 138–353; CABΔ4, 138–299; CABΔ5, 205–424; CABΔ6, 266–424; CABΔ7, 205–353. Hatched boxes, death domains; horizontally striped boxes, hydrophobic domains; vertically striped boxes, NCH domain. (B) Results of binding assays. + and – indicate positive and negative results, respectively. N.D., not determined. The β-galactosidase units (± standard errors), which were detected by liquid assays using chlorophenolred-d-galacto-pyranoside (CPRG), were 2.6 ± 0.2 for AEXΔ1/CABΔ1; 7.5 ± 0.7 for AEXΔ1/CABΔ2; 0.4 ± 0.0 for AEXΔ1/CABΔ5; 4.1 ± 0.2 for AEXΔ4/CABΔ1; and 1.5 ± 0.1 for AEXΔ5/CABΔ1.
All 41 CAB-1 clones isolated from the yeast two-hybrid screen contained the C-terminus in full while their N-terminal ends varied between amino acids 23 and 136 (the shortest isolate, CABΔ2 in Figure 5). To define further the AEX-3-binding domain, we constructed prey vectors containing CAB-1 subdomains (Figure 5). Removal of the C-terminus (CABΔ3 and CABΔ4) eliminated binding to AEXΔ1. The C-terminal 220 amino acids (CABΔ5) bound to AEXΔ1, although CABΔ5 binding was ∼1/6 of CABΔ1 binding (Figure 5, legend). The smaller constructs, CABΔ6 (159 amino acids) and CABΔ7 (146 amino acids), did not bind AEXΔ1. Therefore, the minimum AEX-3-binding domain of CAB-1 resides in the C-terminal 220 amino acids. We named this binding domain AID (AEX-3-interacting domain).
CAB-1 and AEX-3 interact in vitro
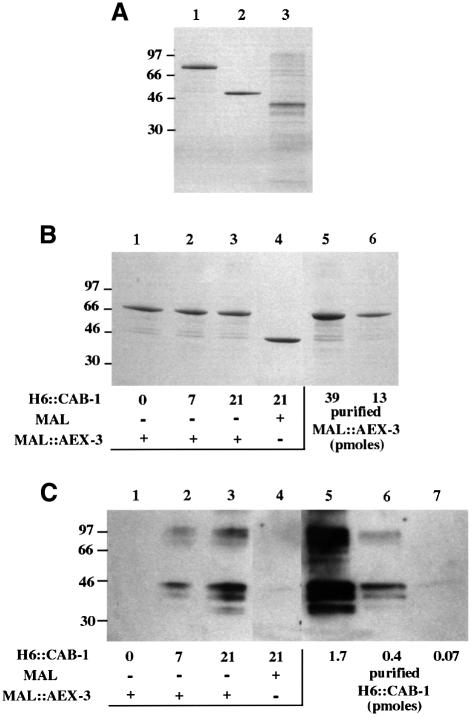
To determine whether CAB-1 binds directly to the AEX-3 C-terminus, we performed an in vitro binding assay using bacterially expressed proteins. The AEXΔ1 domain was cloned into the pMAL expression vector to generate an in-frame fusion (MAL::AEX-3) with maltose-binding protein (MAL). We also cloned the CABΔ2 domain into the pQE vector, which encodes a histidine hexamer peptide (H6::CAB-1). MAL::AEX-3, MAL and H6::CAB-1 were purified using affinity columns and analyzed by SDS–PAGE (Figure 6A). To test the interaction between AEX-3 and CAB-1, purified H6::CAB-1 was incubated for 18 h with amylose resin bound to either MAL::AEX-3 or MAL. After extensive washing, proteins bound to these resins were eluted and separated by SDS–PAGE. Both MAL::AEX-3 and MAL were visible on a Coomassie Blue-stained gel (Figure 6B). However, H6::CAB-1 bands were faint; therefore, immunoblot analysis was performed using an anti-H5 antibody (Figure 6C). Purified H6::CAB-1 protein was included in this gel as a reference. H6::CAB-1 was detected when MAL::AEX-3, but not MAL, was present in the binding mixture. The band intensity of H6::CAB-1 depended on the concentration of H6::CAB-1 added (lanes 1–3 in Figure 6C). The molar ratio between MAL::AEX-3 and H6::CAB-1 was <10:1 under these conditions (Figure 6B and C). These results demonstrate that AEX-3 directly interacts with CAB-1 without additional factors in C.elegans.
Fig. 6. In vitro binding between AEX-3 and CAB-1. (A) Purified MAL::AEX-3 (lane 1), MAL (lane 2) and H6::CAB-1 (lane 3) were separated by SDS–PAGE, and the gel was then stained with Coomassie Blue. The positions of molecular size markers (kDa) are shown to the left of the gel. The predicted molecular weights of MAL::AEX-3, MAL and H6::CAB-1 were 79.4, 50.7 and 33.6 kDa, respectively. (B) Amylose-resin bound to either MAL::AEX-3 or MAL was incubated with purified H6::CAB-1. After extensive washing, proteins were eluted from the resin, separated by SDS–PAGE, and subjected to Coomassie Blue staining. Lane 1, MAL::AEX-3 (resin-bound) incubated with a mock buffer; lane 2, MAL::AEX-3 with 7 µM H6::CAB-1; lane 3, MAL::AEX-3 with 21 µM H6::CAB-1; lane 4, MAL with 21 µM H6::CAB-1; lanes 5 and 6, 39 and 13 pmol of purified MAL::AEX-3, respectively. (C) Eluted proteins were subjected to immunoblot analysis using an anti-H5 antibody. Lanes 1–4, elutants as described for lanes 1–4 in (B), respectively; lanes 5–7, 1.7, 0.4 and 0.07 pmol of purified H6::CAB-1, respectively.
cab-1 functions in rab-3-independent pathways
To test whether CAB-1 functions in the same pathway as AEX-3, we analyzed the phenotype of cab-1 mutants. Wild-type animals were treated with EMS (ethylmethanesulfonate), and animals carrying cab-1 deletions were identified by PCR (Jansen et al., 1997). The cab-1(tg46) mutant carried a deletion of exons 4, 5 and 6, which was necessary for AEX-3 binding (Figure 2), and showed defects in defecation (an Aex phenotype), suggesting that CAB-1 functions in the AEX-3 pathway (Figures 1B and 7A). A new aex (n2220) mutant isolated by E.Jorgensen (personal communication) was mapped near lin-2, therefore we tested whether n2220 is an allele of cab-1. The tg46/n2220 heterozygote showed the Aex phenotype (data not shown) and, furthermore, n2220 carried a 72 nt deletion at the third cab-1 exon–intron boundary (Figure 2). Therefore, we conclude that n2220 is an allele of cab-1. Both tg46 and n2220 strains showed nearly identical phenotypes.
cab-1 mutants had no detectable defects in pharyngeal pumping, body thrashing or male mating in adult animals; however, they exhibited altered dauer motility in addition to defecation defects (Figure 7). In contrast, rab-3 mutations affected pharyngeal pumping, body thrashing and male mating in adult animals, slightly affected dauer motility, but did not affect the defecation motor program (Figure 7), suggesting that rab-3 is required for behaviors that differ from those regulated by cab-1. aex-3 mutants were defective in all these behaviors, suggesting that aex-3 regulates both cab-1 and rab-3 pathways. Furthermore, rab-3; cab-1 double mutants showed a combination of both rab-3 and cab-1 phenotypes, again consistent with the notion that these two pathways are distinct (Figure 7).
It has been shown that rab-3 and aex-3 mutations affect synaptic transmission and confer resistance to the acetylcholinesterase inhibitor aldicarb (Iwasaki et al., 1997; Nonet et al., 1997); such resistance is well correlated with defects in synaptic transmission (Nonet et al., 1993; Miller et al., 1996). We found that cab-1 mutants were also resistant to aldicarb, indicating reduced acetylcholine transmission (Figure 8A). Interestingly, rab-3; cab-1 double mutants were far more resistant to aldicarb than either single mutant, suggesting that rab-3 and cab-1 mutations affect different pathways in synaptic transmission, consistent with their different behavioral phenotypes.
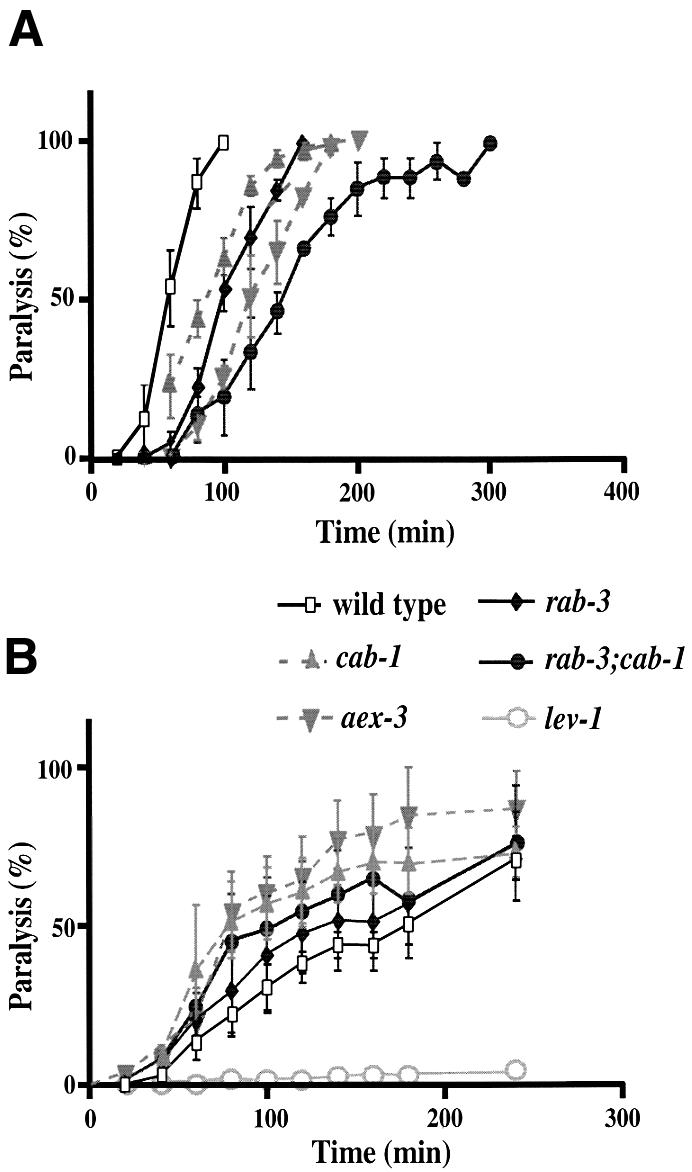
Fig. 8. cab-1 mutations confer pre-synaptic defects. (A) Aldicarb sensitivity of wild-type and mutant strains. Animals were exposed to 1.0 mM aldicarb, and paralyzed animals were counted periodically. Twenty-five animals/plate were tested (three trials/strain). The standard error is shown at each point. (B) Levamisole sensitivity of wild-type and mutant strains. Animals were exposed to 200 µM levamisole. Twenty-five animals/plate were tested (three trials/strain).
To determine whether the cab-1 defect is pre-synaptic or post-synaptic, we measured the sensitivity of cab-1 mutants to levamisole, an acetylcholine receptor agonist in nematodes (Lewis et al., 1980). If cab-1 mutations cause post-synaptic defects in synaptic transmission, cab-1 mutants would be expected to be resistant to levamisole. However, cab-1 mutants were slightly more sensitive to levamisole than wild-type animals (Figure 8B). This suggests that cab-1 mutations most likely cause pre-synaptic defects. As a control for post-synaptic defects, a lev-1 mutant showed strong resistance to levamisole (Figure 8B); lev-1 encodes a non-alpha subunit of nicotinic acetylcholine receptor (Fleming et al., 1997).
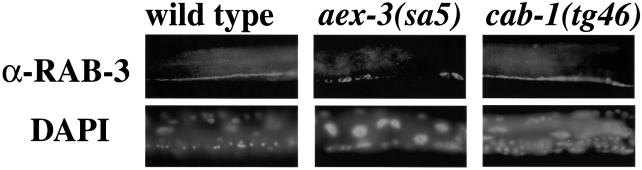
Previously, we observed that RAB-3 protein is aberrantly localized in neural cell bodies instead of synapse-rich axons in aex-3 mutants (Iwasaki et al., 1997), and the same RAB-3 mislocalization was observed when the GTP-binding domain of RAB-3 itself was mutated (Nonet et al., 1997). Therefore, RAB-3 localization correlates well with its GTP-bound state or the GEF function of AEX-3. Using immunostaining we observed that in cab-1 mutants, RAB-3 was distributed along synapse-rich axons in the ventral cord, which was indistinguishable from wild type (Figure 9). This suggests that CAB-1 is not required for RAB-3 function or AEX-3 GEF function.
Fig. 9. Localization of RAB-3 in the nervous system. Animals were stained with an anti-RAB-3 (α-RAB-3) antibody. DAPI staining shows the cell bodies. Animal genotypes are shown above each panel. In each photograph, the ventral nerve cord is at the bottom. In both wild-type and cab-1 mutants, RAB-3 immunoreactivity was restricted primarily to the ventral cord where many neuromuscular synapses are found. In contrast, in aex-3 animals much of the RAB-3 immunoreactivity was located in neural cell bodies.
Discussion
AEX-3 regulates two different pathways
Here we report the identification and characterization of a novel C.elegans gene, cab-1. cab-1 was isolated in a yeast two-hybrid screen aimed at the identification of AEX-3-binding proteins. A physical interaction between AEX-3 and CAB-1 was confirmed, both in a yeast two-hybrid binding assay and in an in vitro binding assay. We also demonstrated that the C-terminal 178 amino acids of AEX-3 are minimally required for CAB-1 binding in yeast; we named this domain CID (CAB-1-interacting domain). The similar expression patterns of aex-3 and cab-1 provide support for interaction between their gene products in vivo. Furthermore, this interaction is likely to be functionally relevant because their mutant phenotypes overlap.
Studies of rat Rab3 GEF demonstrated that almost the entire protein (1409 out of 1602 amino acids) is necessary for GEF activity (Oishi et al., 1998). The following evidence supports the theory that AEX-3 directly interacts with, and facilitates the GDP/GTP exchange of RAB-3. First, amino acids 355–371 of AEX-3 contain the G-protein binding consensus sequence. Secondly, AEX-3 and RAB-3 are expressed in the same cells, i.e. nearly all neurons (Iwasaki et al., 1997; Nonet et al., 1997). Thirdly, aex-3 and rab-3 mutants share similar phenotypes including pharyngeal pumping defects, reduced male mating and low body-thrashing frequency (Figure 7; Iwasaki et al., 1997; Nonet et al., 1997). Finally, RAB-3 aberrantly localized in neural cell bodies instead of synapse-rich axons in aex-3 mutants. The same RAB-3 mislocalization was observed when the GTP-binding domain of RAB-3 itself was mutated, indicating that RAB-3 in aex-3 mutants fails to exchange GDP with GTP (Nonet et al., 1997).
Thus, AEX-3 has at least two distinct domains for protein binding, one interacts with RAB-3 and the other with CAB-1. This notion is supported by the finding that Drosophila CRAG (calmodulin-binding protein related to a Rab3 GDP/GTP exchange protein) also has two distinct domains, one similar to Rab3 GEF and the other, absent in Rab3 GEF, which is the calmodulin-binding domain (Xu et al., 1998).
RAB-3 and CAB-1 seem to regulate different pathways for neural activities even though they interact with AEX-3; cab-1 and rab-3 mutants have different behavioral defects. Interestingly, the aex-3 mutant phenotype resembles the sum of the rab-3 and cab-1 mutant phenotypes, suggesting that AEX-3 regulates these two different pathways. However, either rab-3 or cab-1 mutants showed more severe defects in some phenotypes than the aex-3 mutant. Loss of AEX-3 may not completely abolish RAB-3 function, since RAB-3 without AEX-3 could still exchange GDP with GTP at a slower rate and, similarly, CAB-1 may still have residual function without AEX-3.
AEX-3, RAB-3 and CAB-1 are commonly expressed in many neurons, implying that these two pathways co-exist in a single cell; however, only one pathway seems to be active at a time because of the phenotypic difference between rab-3 and cab-1 mutants. AEX-3 might function as a switch to activate either pathway, depending on environmental or developmental conditions. For example, locomotion of dauer larva is mainly controlled by CAB-1, while that of non-dauers is controlled by RAB-3 (see Figure 7C and E).
To test whether the CAB-1-interacting domain of AEX-3 functions independently of RAB-3 binding, we expressed a truncated AEX-3 protein (amino acids 333 to the C-terminus) in the aex-3(sa5) mutant under the control of the aex-3 promoter (see Materials and methods). This region contains the CAB-1-binding domain but should have little GEF activity (Oishi et al., 1998). We observed that 2/10 transgenic animals carrying this truncated AEX-3 protein showed potenital rescue for defecation (expulsion frequency at 70 and 50%) but not for pharyngeal pumping (180 and 174 pumps/min). This observation may indicate that two AEX-3 domains function independently. However, the frequency of obtaining transgenic animals was very low: 10 transgenic animals were isolated out of 203 parental animals injected and no heritable lines were established. We previously encountered difficulties in obtaining rescued transgenic animals using aex-3 genomic DNAs (Iwasaki et al., 1997); this effect was more pronounced with this truncated gene than with the full-length gene.
Sequence diversity and bridging function of GEFs
Rab GEFs are very diverse in their amino acid sequences. Such diversity is surprising since they all interact with conserved Rab GTPases and stimulate GDP/GTP exchange. For example, while Rab3 and Rab5 are 53% homologous in amino acid sequence, their GEFs have almost no similarity (Horiuchi et al., 1997; Wada et al., 1997). One possible explanation is that a diverse sequence confers interaction with a specific Rab partner. Moreover, as seen in AEX-3, if Rab GEFs also interact with unique non-Rab partners, then additional sequence diversity becomes important for these interactions. Another example is found with the Rab5 GEF, Rabex-5. This protein also interacts with Rabaptin-5, a proposed Rab5 effector, and this interaction presumably contributes to effective signal transduction (Horiuchi et al., 1997). Although it is not known whether other Rab GEFs interact with non-Rab proteins, many GEFs of other ras superfamily G-proteins have been shown to do so. The Rac1–RhoA–GEF trio interacts with the membrane-bound form of phosphotyrosine phosphatase LAR and seems to participate in signal transduction between LAR and Rac1–RhoA (Debant et al., 1996). Ral GDS is a GEF for Ral G-protein as well as an effector of another G-protein, TC21 (Lopez-Barahona et al., 1996). Interaction of Ral GDS with TC21 only occurs when TC21 is in the active GTP-bound state. Epac, a GEF for Rap1 G-protein, also binds cAMP and is proposed to integrate the cAMP pathway with the Rap1 pathway for cell proliferation/differentiation (de Rooij et al., 1998). These findings support the hypothesis that GEFs interact with specific non-Rab partners, including small signaling molecules. Such interactions bridge, and presumably coordinate, multiple regulatory pathways. We propose that connection of multiple pathways is an important function of Rab GEFs, and that their sequence diversity may have arisen from a necessity to interact with different partners in addition to Rab GTPases. Interaction of Rabex-5 with Rabaptin-5 can be considered to be a derivative of the bridging function, but within the same pathway. Further investigation is necessary to test whether other Rab GEFs interact with non-Rab partners or bridge multiple pathways.
How similar are the AEX-3 and the MADD pathways?
The human protein MADD was identified as a TNFR I-binding protein, and its over-expression activated the ERK- and JNK-MAP kinases in COS cells (Schievella et al., 1997). These observations indicate that MADD is a component of the TNF signaling cascade. Intriguingly, MADD is an ortholog of AEX-3 and rat Rab3 GEF (Iwasaki et al., 1997; Schievella et al., 1997; Wada et al., 1997). Thus, it is possible that Rab3 GEF/AEX-3/MADD functions in both synaptic transmission and TNF pathways.
In C.elegans, however, several lines of evidence indicate that AEX-3 does not function in the TNF pathway. First, our BLAST search failed to find homologs for TNF, TNFR or the TNFR-interacting protein TRADD in the C.elegans genome. Secondly, no abnormal apoptosis was observed in aex-3 mutants (M.Hengartner, personal communication.). Thirdly, all phenotypes of aex-3 mutants are so far consistent with defects in synaptic transmission (Iwasaki et al., 1997).
Another interesting question is whether or not MAP kinases function in the AEX-3 pathway for defecation. In addition to MADD-activating MAP kinases (Schievella et al., 1997), JNK3 physically interacted with, and phosphorylated, an alternatively spliced form of MADD (Zhang et al., 1998). However, we observed that defecation of mek-2(n2678) and jkk-1(km2) mutants (Kornfeld et al., 1995; Kawasaki et al., 1999), in which the expulsion frequencies were 93.0% (±3.8, standard error) and 92.8% (±3.8), respectively (n = 10 for both), suggests that neither MAPK kinase is involved in the AEX-3/CAB-1 pathway for defecation. Kawasaki et al. (1999) also reported no defecation defects in jkk-1 mutants. We also observed that mpk-1(oz140) mutants were not defective in defecation (98.3 ± 1.7%, n = 5), although this mutation was unlikely to be null (Lackner et al., 1994). We failed to obtain solid evidence for the involvement of MAP kinases in the AEX-3/CAB-1 pathway for defecation.
Materials and methods
Genetics
Methods for C.elegans culture, genetic analysis and nomenclature were as previously described (Horvitz et al., 1979; Sulston and Hodgkin, 1988). The following mutations were used: LGI, mek-2(n2678), sup-11(n403) dpy-5(e61); LGII, rab-3(js49, y250, y251), clr-1(e1745), rol-1(e91); LGIII, mpk-1(oz140), dpy-17(e164), unc-79(e1068); LGIV, him-8 (e1489); LGV, dpy-11(e224); LGX, aex-3(ad418, sa5, ad696, n2166, y255), lin-2(e1453), lon-2(e678), lin-15(n765ts), cab-1(tg46, n2220), jkk-1(km2).
Isolation of cab-1 mutants
Deletion of the cab-1 locus (tg46) was generated following a standard gene knock-out protocol (Jansen et al., 1997). cab-1(n2220) was isolated based on the Aex phenotype by Erik Jorgensen (personal communication). We sequenced these cab-1 loci and found a 1269 nt deletion for tg46 (including exons 4, 5 and 6) and a 72 nt deletion for n2220 (at the third exon–third intron junction). We also confirmed that n2220 failed to complement tg46 for the Aex phenotype.
Construction of double mutant strains
For rab-3; aex-3 double mutants, clr-1 rol-1/+ males were mated with aex-3 hermaphrodites. From doubly heterozygous F1 hermaphrodites, F2 Aex progeny were picked singly and at the F3 generation Rol progeny were then cloned (the Clr phenotype was confirmed at the next generation), resulting in the clr-1 rol-1; aex-3 triple mutant. Then, rab-3; him-8 males were mated with the triple mutants, producing rab-3/clr-1 rol-1; him-8/+; aex-3/+ heterozygotes. rab-3; aex-3 animals were picked from their progeny, and their genotypes were confirmed based on progeny phenotypes.
The same method as that used for rab-3; aex-3 strain construction was used for generation of the first rab-3; cab-1 mutant. Once the phenotypes of rab-3; cab-1 were known, a second construction method was used. cab-1 males were mated with rab-3 mutants. From F1 cross progeny, pale and starved-looking F2 animals were singly picked. After confirming their Aex phenotype, F3 progeny with slow pharyngeal pumping were cloned. Their genotypes were confirmed by immunostaining for rab-3, and by PCR for cab-1.
Behavioral assays
Defecation assays were performed as previously described (Liu and Thomas, 1994). Ten cycles from 10 animals were observed for each strain. For the mek-2(n2678) mutant, sterile animals from the mek-2/sup-11 dpy-5 heterozygotes were used for the defecation assay. The mek-2(n2678) mutant showed a longer defecation cycle of 57.1 ± 8.9 s (n = 10). Similarly, for the mpk-1(oz140) mutant, sterile animals from mpk-1(oz140)/dpy-17(e164) unc-79(e1068) heterozygotes grown at 25°C were used for the defecation assay. The mpk-1(oz140) mutant at 25°C showed stronger vulva-developmental defects (Lackner et al., 1994). The defecation cycle of the mpk-1(oz140) mutant was 54.6 ± 9.9 s (n = 5). The expulsion frequency (%) and defecation cycles (s) of the rab-3; cab-1 mutant were 1.7 ± 1.2 and 59.2 ± 12.3, respectively (n = 10). These numbers for other strains are described in the legend of Figure 1. Male mating assays were also performed as previously described (Hodgkin, 1983). Briefly, six males of the genotypes described in Figure 7 were mated with six dpy-11(e224) hermaphrodites for 24 h, and then the males were removed. The dpy-11 hermaphrodites were transferred to fresh plates every day. Cross progeny on these plates were counted 3 days later. The experiment was performed in triplicate for each strain. For strains containing rab-3, him-8(e1489) was included in trans to the wild-type locus, therefore, him-8/+. him-8/+ males were mated as efficiently as the wild-type males (data not shown).
For body-thrashing activity, young adults were placed in M9 solution and their body thrashing was observed for 1 min (Miller et al., 1996). Pharyngeal pumping was counted for 20–60 s using young adults that were well fed. Dauer motility was measured as follows. Starved animals were treated with 1% SDS to isolate dauers (Riddle, 1988). Dauers were washed three times with distilled water, and then placed on agar plates with no bacteria. The motility of these animals was observed for 15 s using the Motion Recording system (Flovel Co., Japan) and their tracks were traced using the Image Filing System (Flovel Co., Japan).
Pharmacological assays
Just before pouring 35 mm plates, aldicarb (100 mM in 70% ethanol) or levamisole (100 mM in water) was added to NGM agar at the appropriate concentration (Nonet et al., 1993). After seeding the plates with bacteria, 25 young adult worms were placed on each of triplicate plates. Acute drug effects were assessed in a blind paralysis assay (Iwasaki et al., 1997).
Microinjection
Microinjection was performed as previously described (Mello et al., 1991). To generate strains carrying the cab-1::gfp construct, lin-15 (n765ts) animals were injected with pcab-1::gfp and pbHL98 (lin-15 marker) at 200 and 20 ng/µl, respectively (Huang et al., 1994). The progeny were grown at 25°C to isolate non-LIN animals. To generate strains carrying the truncated aex-3: gene construct, aex-3(sa5) animals were injected with pKI150 and pRF4 (rol-6 marker) at 10 ng/µl and 80–150 ng/µl, respectively (Mello et al., 1991).
Immunocytochemistry
Immunocytochemistry was performed after methanol–acetone fixation as previously described (Duerr et al., 1999). Rabbit antiserum against RAB-3 was raised using the synthetic peptide l-6-1A DKDPQQ QPKGQKLEAC. The antibody was affinity-purified using this peptide, and its specificity was confirmed by analysis of the rab-3(js49) mutant; no staining in the nervous system was observed.
Yeast two-hybrid screening and assay
Yeast two-hybrid screening was after the method of Fields and Song (1989) and was carried out using the Matchmaker System (Clontech). The Y190 yeast strain was co-transformed with pJ1 1-1 (pAS2-1 plasmid containing an aex-3 fragment), and C.elegans cDNAs cloned into the pACT vector. This cDNA library was a generous gift from Robert Barstead.
To identify AEX-3 domains that bind CAB-1, and CAB-1 domains that bind AEX-3, the Y190 yeast strain was transformed (Frederick, 1987) in appropriate combinations with bait and prey constructs (see Figure 5). Transfected cells were grown on selection plates [SD –Leu, –Trp –His +3AT (3-amino-1,2,4-triazole)]. Ten to 14 days later, the resulting colonies were re-plated on fresh plates with the same selection medium. Colonies from the second plates were assayed for β-gal activity. The criteria for scoring an interaction between bait and prey constructs were that >80% of the first-plate colonies grew on the second plates and turned blue in the β-gal assay.
Liquid assays for β-gal were performed using chlorophenolred-d-galacto-pyranoside as previously described (Kokkola et al., 1998).
Nucleic acid analysis
Molecular biological methods were essentially as described (Frederick, 1987; Sambrook et al., 1989). Reverse transcription PCR (RT–PCR) was performed using a 5′ RACE system (Life Technologies) following the manufacturer’s protocol. First strand cDNAs were generated using 5 µg of total C.elegans RNA and the cab-1 specific primer OLG9 (ATG CACTGGAGCCAAAGCAC), and then amplified by PCR OLG10 primer (CGGGATCCAAAGGTGCCTTCTGTTCAG) with either the SL1, SL2, SL3, SL4 or SL5 primer (Ross et al., 1995). Only the primer combination OLG10 and SL1 generated a DNA fragment suitable for cloning; this fragment was cloned into the Bluescript SK+ plasmid and sequenced. Sequences of both PCR and sequencing primers will be provided upon request.
The cab-1::gfp fusion plasmid pcab-1::gfp was constructed as follows. A cab-1 promoter fragment was generated by PCR using the OLG25 (TGACATCGACACACGCTATC) and OLG26 (TCCCCCGGGCTA GCTGCTCGGCGGTAG) primers, which contained XhoI and SmaI enzyme sites. The cab-1 promoter fragment was cloned into pPD95.69 (a GFP expression vector, gift from A.Fire) using SmaI and SalI restriction enzyme sites. Sequencing confirmed that the 36th codon of cab-1 was fused in-frame with gfp.
Plasmids used for the yeast two-hybrid assay were constructed as follows. Using either aex-3 or cab-1 cDNA as a template, PCR was performed with primers containing either a NcoI, EcoRI or BamHI site, and the resulting PCR products were cloned into the appropriate site of the pAS2-1 or pACT vector. The amino acids included in AEXΔ and CABΔ constructs series are described in the legend of Figure 5. Primers used for plasmid construction will be provided upon request.
Plasmids used for the in vitro binding assays were constructed as follows. Using either aex-3 or cab-1 cDNA as a template, PCR was performed using primers containing either a BamHI or EcoRI site. These PCR products were cloned into either the pQE30 or pMAL-c2 expression vector using the appropriate enzyme sites. All inserts were sequenced to confirm no mutations. pMAL::AEX-3 contained amino acids 1066–1394 of AEX-3, and pQE::CAB-1 contained amino acids 137–423 of CAB-1.
The plasmid pKI150, containing a truncated aex-3 gene, was constructed as follows. An aex-3 DNA fragment was generated by PCR using aex-3 cDNA as a template. The fragment was cloned into the pDONOR vector by recombinational cloning following the manufacturer’s protocol (Life Technologies). A 1.3 kb aex-3 promoter fragment was cloned into pPD97.77 (a gift from A.Fire) using the SphI and BamHI sites, resulting in paex-3p::gfp. The aex-3 cDNA fragment was transferred to the SmaI site of paex-3p::gfp using recombinational cloning. More details including primer sequences will be provided upon request. The resulting construct was named pKI150, which encoded amino acids 333–1409 of AEX-3 and also contained an insertion of 10 amino acids at the N-terminus as a cloning by-product (Gateway System of Life Technologies).
In vitro binding assay
For in vitro binding assays, the QIAexpressionist (Qiagen) and pMAL protein fusion and purification (New England BioLabs) systems were used. The M15[pREP4] Escherichia coli strain was transformed with pQE30-based constructs, and XL-1 Blue with pMAL-c2-based constructs. Column purification was performed following the manufacturer’s protocol (New England BioLabs and Qiagen). The concentration of MAL::AEX-3, MAL and H6::CAB-1 was calculated prior to mixing using the Bradford method (Frederick, 1987). The ratio between the main band and minor binds in SDS–PAGE was determined using densitometry (Bio-Rad Gel Doc) and the accurate concentration of the main-band proteins was calculated. MAL::AEX-3 and MAL proteins were applied to amylose-resin columns using column buffer [20 mM Tris–HCl pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF)]. After extensive washing with column buffer, the amylose-resin bound to proteins was mixed with purified H6::CAB-1 protein in binding buffer (20 mM Tris–HCl pH 7.4, 230 mM NaCl, 5 mM NaH2PO4 pH 8.0, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 25 mM imidazole). After incubation at 4°C for 18 h, the amylose-resin was washed with column buffer (10× volume of the resin bed), and bound proteins were eluted with column buffer containing 10 mM maltose. The eluted proteins were examined using SDS–PAGE followed by Coomassie Blue staining, and immunoblotting analysis using anti-Penta His (Qiagen) and anti-MBP (maltose binding protein) (New England BioLabs) antibodies as primary antibodies (Frederick, 1987). Approximately 1/100 of the eluted peak was applied to SDS–PAGE for analysis.
Acknowledgments
Acknowledgements
We thank H.Kouike and C.Li for technical assistance; T.Teramoto and T.Martin for advice about in vitro binding assays; E.Jorgensen for n2220; K.Matsumoto for jkk-1(km2); M.Nonet for rab-3 strains; A.Coulson for cosmids; R.Barstead for the C.elegans cDNA library; the C.elegans Genome Sequencing Consortium for C.elegans genome sequences; and A.Hart, E.Duncan, E.Jorgensen, K.Takimoto and E.Tisdale for helpful comments on the manuscript. This work was supported by Ministry of International Trading and Industry, Science and Technology Agency of Japan (K.I). Some strains were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institute of Health, National Center for Research Resources of the US.
References
- Castillo P.E., Janz,R., Sudhof,T.C., Tzounopoulos,T., Malenka,R.C. and Nicoll,R.A. (1997) Rab3A is essential for mossy fibre long-term potentiation in the hippocampus. Nature, 388, 590–593. [DOI] [PubMed] [Google Scholar]
- Christoforidis S., McBride,H.M., Burgoyne,R.D. and Zerial,M. (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature, 397, 621–625. [DOI] [PubMed] [Google Scholar]
- Debant A., Serra-Pages,C., Seipel,K., O’Brien,S., Tang,M., Park,S.H. and Streuli,M. (1996) The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain and has separate rac-specific and rho-specific guanine nucleotide exchange factor domains. Proc. Natl Acad. Sci. USA, 93, 5466–5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J., Zwartkruis,F.J., Verheijen,M.H., Cool,R.H., Nijman,S.M., Wittinghofer,A. and Bos,J.L. (1998) Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature, 396, 474–477. [DOI] [PubMed] [Google Scholar]
- Duerr J.S., Frisby,D.L., Gaskin,J., Duke,A., Asermely,K., Huddleston,D., Eiden,L.E. and Rand,J.B. (1999) The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J. Neurosci., 19, 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S. and Song,O. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Fleming J.T. et al. (1997) Caenorhabditis elegans levamisole resistance genes lev-1, unc-29 and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci., 17, 5843–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick A.M. (1987) Current Protocols in Molecular Biology. John Wiley and Sons Inc., New York, NY. [Google Scholar]
- Galiana E., Vernier,P., Dupont,E., Evrard,C. and Rouget,P. (1995) Identification of a neural-specific cDNA, NPDC-1, able to down-regulate cell proliferation and to suppress transformation. Proc. Natl Acad. Sci. USA, 92, 1560–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geppert M., Bolshakov,V.Y., Siegelbaum,S.A., Takei,K., De Camilli,P., Hammer,R.E. and Sudhof,T.C. (1994) The role of Rab3A in neurotransmitter release. Nature, 369, 493–497. [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth,R., Morisaki,H., Jahn,R. and Heuser,J.E. (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell, 90, 523–535. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. (1983) Male phenotypes and mating efficiency in C.elegans. Genetics, 103, 43–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl T.M., Parlati,F., Wimmer,C., Rothman,J.E., Sollner,T.H. and Engelhardt,H. (1998) Arrangement of subunits in 20 S particles consisting of NSF, SNAPs and SNARE complexes. Mol. Cell, 2, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H. et al. (1997) A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell, 90, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Horvitz H.R., Brenner,S., Hodgkin,J. and Herman,R.K. (1979) A uniform genetic nomenclature for the nematode Caenorhabditis elegans. Mol. Gen. Genet., 175, 129–133. [DOI] [PubMed] [Google Scholar]
- Huang L.S., Tzou,P. and Sternberg,P.W. (1994) The lin-15 locus encodes two negative regulators. Mol. Biol. Cell, 5, 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Staunton,J., Saifee,O., Nonet,M. and Thomas,J.H. (1997) aex-3 encodes a novel regulator of presynaptic activity in C.elegans. Neuron, 18, 613–622. [DOI] [PubMed] [Google Scholar]
- Jansen G., Hazendonk,E., Thijssen,K.L. and Plasterk,R.H. (1997) Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nature Genet., 17, 119–121. [DOI] [PubMed] [Google Scholar]
- Kawasaki M., Hisamoto,N., Iino,Y., Yamamoto,M., Ninomiya-Tsuji,J. and Matsumoto,K. (1999) A Caenorhabditis elegans JNK signal transduction pathway regulates coordinated movement via type-D GABAergic motor neurons. EMBO J., 18, 3604–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokkola T., Watson,M.A., White,J., Dowell,S., Foord,S.M. and Laitinen,J.T. (1998) Mutagenesis of human Mel1a melatonin receptor expressed in yeast reveals domains important for receptor function. Biochem. Biophys. Res. Commun., 249, 531–536. [DOI] [PubMed] [Google Scholar]
- Kornfeld K., Guan,K.L. and Horvitz,H.R. (1995) The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev., 9, 756–768. [DOI] [PubMed] [Google Scholar]
- Lackner M.R., Kornfeld,K., Miller,L.M., Horvitz,H.R. and Kim,S.K. (1994) A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev., 8, 160–173. [DOI] [PubMed] [Google Scholar]
- Lewis J.A., Wu,C.H., Levine,J.H. and Berg,H. (1980) Levamisole-resistant mutants of the nematode Caenorhabditis elegans appear to lack pharmacological acetylcholine receptors. Neuroscience, 5, 967–989. [DOI] [PubMed] [Google Scholar]
- Li C., Takei,K., Geppert,M., Daniell,L., Stenius,K., Chapman,E.R., Jahn,R., De Camilli,P. and Sudhof,T.C. (1994) Synaptic targeting of rabphilin-3A, a synaptic vesicle Ca2+/phospholipid-binding protein, depends on rab3A/3C. Neuron, 13, 885–898. [DOI] [PubMed] [Google Scholar]
- Lian J.P., Stone,S., Jiang,Y., Lyons,P. and Ferro-Novick,S. (1994) Ypt1p implicated in v-SNARE activation. Nature, 372, 698–701. [DOI] [PubMed] [Google Scholar]
- Liu D.W. and Thomas,J.H. (1994) Regulation of a periodic motor program in C.elegans. J. Neurosci., 14, 1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonart G., Janz,R., Johnson,K.M. and Sudhof,T.C. (1998) Mechanism of action of rab3A in mossy fiber LTP. Neuron, 21, 1141–1150. [DOI] [PubMed] [Google Scholar]
- Lopez-Barahona M., Bustelo,X.R. and Barbacid,M. (1996) The TC21 oncoprotein interacts with the Ral guanosine nucleotide dissociation factor. Oncogene, 12, 463–470. [PubMed] [Google Scholar]
- Lupashin V.V. and Waters,M.G. (1997) t-SNARE activation through transient interaction with a rab-like guanosine triphosphatase. Science, 276, 1255–1258. [DOI] [PubMed] [Google Scholar]
- McBride H.M., Rybin,V., Murphy,C., Giner,A., Teasdale,R. and Zerial,M. (1999) Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell, 98, 377–386. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K., Alfonso,A., Nguyen,M., Crowell,J., Johnson,C. and Rand,J. (1996) A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl Acad. Sci. USA, 93, 12593–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M.L., Grundahl,K., Meyer,B.J. and Rand,J.B. (1993) Synaptic function is impaired but not eliminated in C.elegans mutants lacking synaptotagmin. Cell, 73, 1291–1305. [DOI] [PubMed] [Google Scholar]
- Nonet M.L., Staunton,J.E., Kilgard,M.P., Fergestad,T., Hartwieg,E., Horvitz,H.R., Jorgensen,E.M. and Meyer,B.J. (1997) Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J. Neurosci., 17, 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P. and Zerial,M. (1997) The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol., 9, 496–504. [DOI] [PubMed] [Google Scholar]
- Oishi H., Sasaki,T., Nagano,F., Ikeda,W., Ohya,T., Wada,M., Ide,N., Nakanishi,H. and Takai,Y. (1998) Localization of the Rab3 small G protein regulators in nerve terminals and their involvement in Ca2+-dependent exocytosis. J. Biol. Chem., 273, 34580–34585. [DOI] [PubMed] [Google Scholar]
- Poirier M.A., Xiao,W., Macosko,J.C., Chan,C., Shin,Y.K. and Bennett,M.K. (1998) The synaptic SNARE complex is a parallel four-stranded helical bundle. Nature Struct. Biol., 5, 765–769. [DOI] [PubMed] [Google Scholar]
- Riddle D.L. (1988) The dauer larva. In Wood,W. (ed.), The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 393–412. [Google Scholar]
- Ross L.H., Freedman,J.H. and Rubin,C.S. (1995) Structure and expression of novel spliced leader RNA genes in Caenorhabditis elegans. J. Biol. Chem., 270, 22066–22075. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning, A Laboraotry Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schievella A.R., Chen,J.H., Graham,J.R. and Lin,L.L. (1997) MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase. J. Biol. Chem., 272, 12069–12075. [DOI] [PubMed] [Google Scholar]
- Sogaard M., Tani,K., Ye,R.R., Geromanos,S., Tempst,P., Kirchhausen,T., Rothman,J.E. and Sollner,T. (1994) A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell, 78, 937–948. [DOI] [PubMed] [Google Scholar]
- Sudhof T.C. (1995) The synaptic vesicle cycle: a cascade of protein–protein interactions. Nature, 375, 645–653. [DOI] [PubMed] [Google Scholar]
- Sudhof T.C. (1997) Function of Rab3 GDP-GTP exchange. Neuron, 18, 519–522. [DOI] [PubMed] [Google Scholar]
- Sulston J. and Hodgkin,J. (1988) Methods. In Wood,W. (ed.), The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sutton R.B., Fasshauer,D., Jahn,R. and Brunger,A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature, 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Thomas J.H. (1990) Genetic analysis of defecation in Caenorhabditis elegans. Genetics, 124, 855–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Nakanishi,H., Satoh,A., Hirano,H., Obaishi,H., Matsuura,Y. and Takai,Y. (1997) Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J. Biol. Chem., 272, 3875–3878. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C., Collins,R.N. and Novick,P.J. (1997) Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol., 137, 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Okamoto,M., Schmitz,F., Hofmann,K. and Sudhof,T.C. (1997) Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature, 388, 593–598. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman,B.V., McNew,J.A., Westermann,B., Gmachl,M., Parlati,F., Söllner,T.H. and Rothman,J.E. (1998) SNAREpins: minimal machinery for membrane fusion. Cell, 92, 759–772. [DOI] [PubMed] [Google Scholar]
- Xu X.Z., Wes,P.D., Chen,H., Li,H.S., Yu,M., Morgan,S., Liu,Y. and Montell,C. (1998) Retinal targets for calmodulin include proteins implicated in synaptic transmission. J. Biol. Chem., 273, 31297–31307. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhou,L. and Miller,C.A. (1998) A splicing variant of a death domain protein that is regulated by a mitogen-activated kinase is a substrate for c-Jun N-terminal kinase in the human central nervous system. Proc. Natl Acad. Sci. USA, 95, 2586–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]