The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26
Spt6 and Iws1 are conserved and essential transcription regulators in eukaryotes. Here, we gain structural and functional insight into the Spt6-Iws1 complex itself and implications for the interactions of several other important factors involved in transcription.
Keywords: crystallography, mRNA export, RNA polymerase II, transcription elongation, yeast genetics
Abstract
Binding of elongation factor Spt6 to Iws1 provides an effective means for coupling eukaryotic mRNA synthesis, chromatin remodelling and mRNA export. We show that an N-terminal region of Spt6 (Spt6N) is responsible for interaction with Iws1. The crystallographic structures of Encephalitozoon cuniculi Iws1 and the Iws1/Spt6N complex reveal two conserved binding subdomains in Iws1. The first subdomain (one HEAT repeat; HEAT subdomain) is a putative phosphoprotein-binding site most likely involved in an Spt6-independent function of Iws1. The second subdomain (two ARM repeats; ARM subdomain) specifically recognizes a bipartite N-terminal region of Spt6. Mutations that alter this region of Spt6 cause severe phenotypes in vivo. Importantly, the ARM subdomain of Iws1 is conserved in several transcription factors, including TFIIS, Elongin A and Med26. We show that the homologous region in yeast TFIIS enables this factor to interact with SAGA and the Mediator subunits Spt8 and Med13, suggesting the molecular basis for TFIIS recruitment at promoters. Taken together, our results provide new structural information about the Iws1/Spt6 complex and reveal a novel interaction domain used for the formation of transcription networks.
Introduction
Eukaryotic RNA polymerase II (RNAPII) requires the timely and concerted recruitment of many factors to enhance pre-initiation complex (PIC) formation, promoter clearance, transcription elongation and to overcome the transcriptional barriers imposed by chromatin (Saunders et al, 2006; Li et al, 2007). Furthermore, recruitment by elongating RNAPII of factors involved in mRNA processing, mRNA surveillance and mRNA export enables a direct and tight coupling between transcription, mRNA maturation and mRNA export (Perales and Bentley, 2009).
The Iws1/Spt6 complex participates in this coupling by acting in transcription elongation, chromatin remodelling and mRNA export. Spt6 is a putative histone chaperone that interacts directly with histone H3, and appears to promote nucleosome reassembly at core promoters and within the body of genes in the wake of RNAPII (Bortvin and Winston, 1996; Winkler et al, 2000; Kaplan et al, 2003; Adkins and Tyler, 2006). Spt6 is also an elongation factor that enhances elongation rates of RNAPII both in vitro and in vivo (Endoh et al, 2004; Yoh et al, 2007; Ardehali et al, 2009). Yet, the role of Spt6 in elongation cannot be fully recapitulated by its histone chaperone activity as elongation enhancement also occurs on naked DNA templates (Endoh et al, 2004; Yoh et al, 2007). A recent study has shown that both Spt6 and Iws1 are associated genome-wide across transcribed regions (Mayer et al, 2010).
Interaction between Spt6 and the Iws1/Spn1 protein has been reported both in yeast and in mammals (Krogan et al, 2002; Lindstrom et al, 2003; Yoh et al, 2007). The involvement of Spt6 in mRNA export requires its association with Iws1, as Iws1 interacts directly with the mRNA export factor REF1/Aly (Yoh et al, 2007). Furthermore, either depletion of Iws1 or a mutation within the SH2 domain of Spt6, which impairs the ability of the Iws1/Spt6 complex to interact with elongating RNAPII, results in splicing defects and nuclear retention of bulk poly(A)+ mRNAs in mammalian cells (Yoh et al, 2007). Mammalian Iws1 is also required for the recruitment of the lysine methyltransferase, HYPB/Setd2, which trimethylates H3K36 across transcribed regions (Yoh et al, 2008).
Iws1 also functions independently of Spt6 by binding constitutively to the postassembly regulated CYC1 gene, thus preventing the recruitment of the Swi/Snf complex and repressing transcription of this gene (Fischbeck et al, 2002; Zhang et al, 2008). A single amino-acid change in yeast Iws1 (K192N) prevents its recruitment to the CYC1 gene, leading to the constitutive recruitment of Swi/Snf and increased transcription, even in non-inducing conditions. The direct interaction of Iws1 with the Ser5-phosphorylated RNAPII has been suggested for the constitutive recruitment of Iws1 (Zhang et al, 2008).
Despite the wealth of data on Spt6 and Iws1, their functional roles remain largely unknown at the molecular level. So far, only the structures of the Spt6 tandem SH2 domains (Diebold et al, 2010; Sun et al, 2010) and of the putative bacterial ancestor of Spt6, Tex (Johnson et al, 2008), have shed some light on the structural organization of Spt6. As for Iws1, part of the conserved region of Iws1 is homologous in sequence to the N-terminal domains of the elongation factors TFIIS and Elongin A, and of the co-activator Med26 (Wery et al, 2004; Ling et al, 2006). In TFIIS, this domain enables TFIIS recruitment to gene promoters by the co-activators SAGA and Mediator, in which it favours the formation of active PICs (Pan et al, 1997; Wery et al, 2004; Prather et al, 2005; Guglielmi et al, 2007; Kim et al, 2007). Yet, the NMR structures of the yeast and mouse TFIIS N-terminal domains adopt totally different folds (PDB code 1EO0; Booth et al, 2000; PDB code 1WJT, RIKEN Structural Genomics/Proteomics Initiative), precluding our understanding of the role of this domain.
Here, we present the biochemical, crystallographic and functional characterization of Iws1 and the Iws1/Spt6 complex from Encephalitozoon cuniculi and Saccharomyces cerevisiae. We show that Iws1 possesses two binding subdomains (HEAT and ARM subdomains) defined by the N- and C-termini of its conserved region. Another study has also recently identified these domains from the crystallographic analysis of S. cerevisiae Iws1 (Pujari et al, 2010). The HEAT subdomain has determinants for recognizing large negatively charged ions or small molecules such as phosphates, suggesting that it may serve to recognize phosphoproteins. This subdomain is specific to Iws1 and is most likely responsible for the Spt6-independent function of Iws1. The ARM subdomain of Iws1 specifically recognizes an N-terminal region of Spt6 (Spt6N). In yeast, we show that mutations that alter this region of Spt6 cause severe phenotypes, suggesting that the interaction between Spt6 and Iws1 is critical in vivo.
Importantly, the ARM subdomain of Iws1 encompasses the region of sequence homology with TFIIS, Elongin A and Med26. We show that the homologous subdomain from yeast TFIIS is able to form complexes with the Spt8 and Med13 subunits of SAGA and the Mediator, suggesting a molecular basis for TFIIS recruitment at promoters. These complexes combine interactions that are observed within the Iws1/Spt6N complex, along with interactions that are specific to the TFIIS/Spt8 and TFIIS/Med13 complexes. Taken together, our results provide a structural characterization of the Iws1/Spt6 complex and highlight a specific interaction domain shared by other transcriptional effectors.
Results
A small N-terminal region of Spt6 is sufficient to interact with Iws1
Yeast (S. cerevisiae; sc) and human (Homo sapiens; hs) Spt6 proteins are very large polypeptides that are not easily amenable to biochemical and structural studies. To overcome this problem, we use the fungi-related intracellular parasite, E. cuniculi (ec), as a model organism as its proteins are generally shorter than their eukaryotic orthologues (Katinka et al, 2001; Romier et al, 2007; Diebold et al, 2010). Spt6 and Iws1 are present in E. cuniculi and show strong sequence conservation with their yeast and human orthologues despite their shorter length (Figure 1A; Supplementary Figure 1).
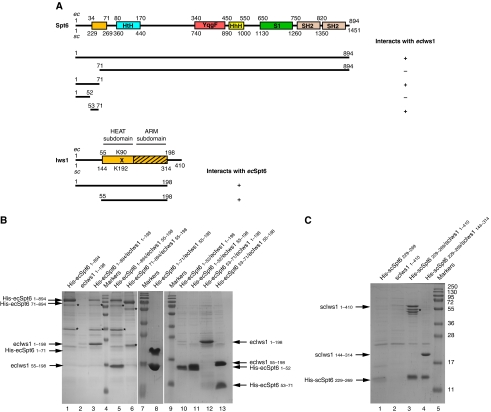
Figure 1.
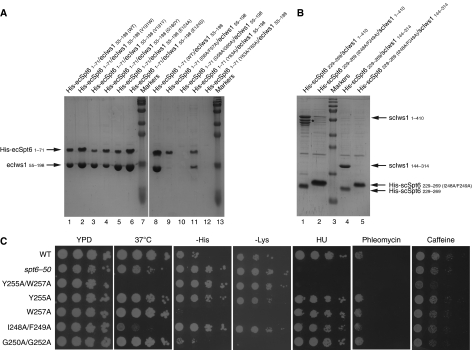
A small N-terminal region of Spt6 is sufficient to retain the full length or the conserved domain of Iws1. (A) Schematic view of the putative domain architecture of Spt6 and Iws1. The domains of Spt6 and Iws1 characterized in this study are shown as light orange boxes. This domain in Iws1 is composed of two subdomains (HEAT subdomain and ARM subdomain) and corresponds to the conserved region of Iws1 sufficient for Iws1 function in yeast (Fischbeck et al, 2002). The Iws1 invariant lysine (scK192/ecK90) involved in the Spt6-independent function of Iws1 is marked by an ‘X'. The Iws1 region homologous to TFIIS, Elongin A and Med26 N-terminal domains is hatched. The domains (HtH, YqgF, HhH and S1) that have been putatively assigned to the Spt6 core domain based on the structure of the bacterial Tex protein (Johnson et al, 2008) are shown. The two SH2 domains from the tandem SH2 domains at the C-terminus of Spt6 (Diebold et al, 2010; Sun et al, 2010) are indicated. The results of co-expression experiments shown in (B) are summarized below the proteins. E. cuniculi (ec) and S. cerevisiae (sc) numbering are shown. (B) Deciphering of E. cuniculi Iws1/Spt6 complex formation upon (co-) expression in E. coli of various constructs of both proteins and purification by affinity chromatography. All samples are analysed on SDS–PAGE. The fainter band for His-ecSpt6N53−71 in lane 12 compared with lane 13 is due to the lower amount of soluble complex obtained. Construct boundaries are indicated. Spt6 degradation products are marked with an ‘*'. (C) Characterization of the S. cerevisiae Iws1/Spt6N interaction based on the data obtained with the E. cuniculi proteins. The faint band for His-scSpt6N229−269 in lane 1 is most likely due to the poor solubility of this construct when expressed alone. Iws1 degradation products are marked with an ‘*'. Molecular weights are shown and are the same throughout the figures.
Our experiments show that full-length E. cuniculi Spt6 and Iws1 form a complex when co-expressed in Escherichia coli. Singly expressed ecSpt6, N-terminally tagged with a poly-histidine sequence, is retained on a cobalt affinity resin (Figure 1B, lane 1). In contrast, singly expressed, but untagged ecIws1 does not bind to the same resin, showing that no non-specific binding occurs (Figure 1B, lane 2). However, upon co-expression of his-tagged ecSpt6 with untagged ecIws1, both proteins are retained on the resin, revealing the formation of a complex between these two proteins (Figure 1B, lane 3).
Additional co-expression experiments defined regions in each protein sufficient for their interaction. First, the conserved region of Iws1 (ecIws1 residues 55–198), which is sufficient for the essential functions of Iws1 in yeast (Fischbeck et al, 2002), is also sufficient for binding to ecSpt6 (Figure 1B, lane 5). In the case of ecSpt6, as observed for its mammalian homologue (Yoh et al, 2007), the N-terminal region (ecSpt6 residues 1–71; ecSpt6N1−71), but not the remaining C-terminal region (ecSpt6 residues 71–894), is involved in Iws1 binding (Figure 1B, compare lanes 6 and 8).
Multiple alignment of Spt6 sequences shows that its N-terminal region contains a small conserved region (ecSpt6 residues 53–71; ecSpt6N53−71; Supplementary Figure 1A). Co-expression experiments showed that this small conserved region of Spt6 is sufficient for binding ecIws1 (Figure 1B, lanes 10–13). These results were confirmed by looking for a similar interaction between the yeast proteins: an scSpt6N229−269 construct, similar to ecSpt6N53−71, was able to form a complex with either full-length scIws1 or its conserved region (residues 144–314) (Figure 1C).
The conserved region of Iws1 is formed by HEAT and ARM repeats
To characterize precisely the interaction between Spt6 and Iws1, we solved the X-ray crystallographic structures of these proteins and their complex from E. cuniculi. Owing to rapid proteolytic N-terminal degradations occurring during purification, only the stable ecIws155−198 construct was considered and could be readily crystallized on its own in different space groups. For Spt6, due to yield and C-terminal proteolytic degradation problems with the full-length protein, only N-terminal constructs (encoding ecSpt6N residues 1–71, 34–71 and 53–71) were used in complex with ecIws155−198. Three different crystal forms were obtained: one form with the ecSpt6N34−71 construct and two forms with the ecSpt6N53−71 construct. All crystal forms were unrelated, providing an unbiased view of the interaction between Spt6N and Iws1.
The structure of ecIws155−198 was solved by multiple anomalous dispersion using selenomethionine derivatives. The model was refined against a 2.25 Å resolution native data set to an R-factor of 20.4% and an R-free of 26.5% (Supplementary Table I). The three structures of the complexes were then solved by molecular replacement using the structure of free Iws1. The Spt6N regions were built into the additional density and the structures of the ecSpt6N53−71/ecIws155−198 (forms 1 and 2) and ecSpt6N34−71/ecIws155−198 complexes were refined to 1.95, 2.10 and 1.75 Å resolution to R-factors of 21.0, 20.0 and 19.4% and R-frees of 23.6, 24.7 and 23.6%, respectively (Supplementary Table II). For Spt6N and Iws1, almost all residues could be seen in density, apart from a few N- and/or C-terminal residues of both proteins. A single exception was observed for the longer ecSpt6N34−71 construct in which the first 11 N-terminal residues could not be seen.
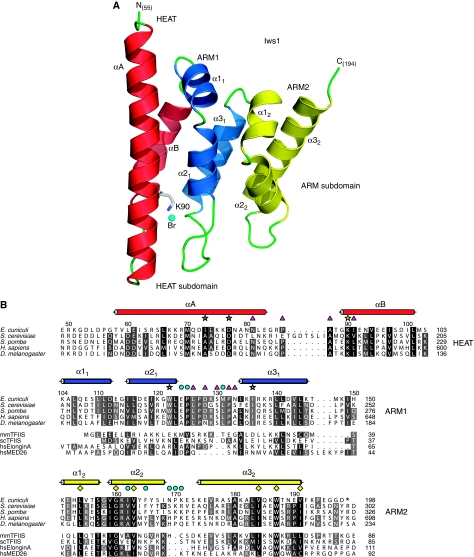
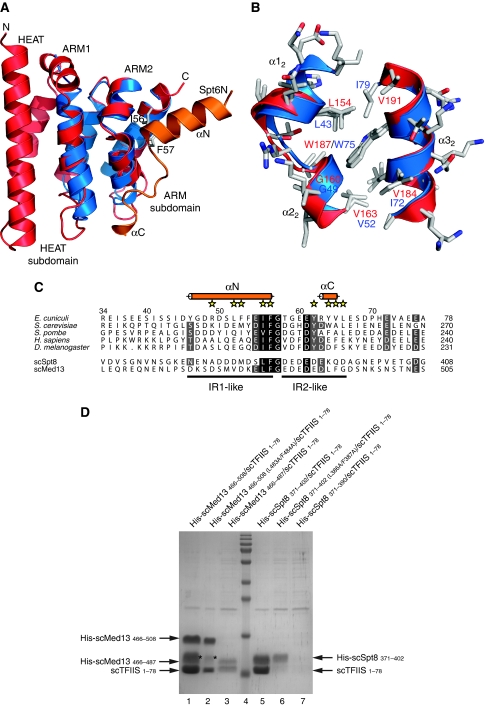
Structural analysis of ecIws155−198 in the different crystal forms reveals that the Iws1 conserved region is formed by a single HEAT repeat followed by two ARM repeats (ARM1 and ARM2) (Figure 2). HEAT and ARM motifs are related protein/protein interaction modules that are often found repeated several times in many proteins (Andrade et al, 2001). The HEAT motif is composed of an α-helical hairpin formed by two α-helices (αA and αB), whereas the ARM motif is composed of a short α-helix (α1), which is almost perpendicular to a helical hairpin formed by two α-helices (α2 and α3). Only minor structural differences are observed for Iws1 between all crystal forms, implying that binding of Spt6N does not induce large conformational changes.
Figure 2.
Structure of E. cuniculi Iws1. (A) Ribbon representation of ecIws155−198 crystal structure. α-helices of the HEAT, ARM1 and ARM2 motifs are coloured red, blue and yellow, respectively. Loops are coloured green. The two subdomains of Iws1 are indicated. The invariant K90 is displayed as sticks and coloured according to atom type. The anion bound to K90 Nɛ is shown as a cyan sphere. The first and last residues observed in the density are labelled. This colour scheme is used throughout the figures unless otherwise stated. The ribbon figures have been made with PYMOL (version 0.99; DeLano Scientific). (B) Multiple sequence alignment of the conserved region of Iws1 (top five rows) with the TFIIS, Elongin A and Med26 N-terminal domains (bottom three rows; not shown for the HEAT repeat, which is Iws1 specific). mm, Mus musculus; sc, S. cerevisiae; hs, H. sapiens. Sequence similarities are indicated by shading. Observed α-helices in the ecIws155−198 structure are shown above the sequences as cylinders coloured as in (A). Numbering above the sequences correspond to E. cuniculi, whereas the numbering at the end of each row relates to the different organisms. K90 and the residues involved in its packing at the interface of the HEAT and ARM1 repeats are labelled with yellow stars. Residues forming the highly conserved putative phosphate-binding domain of Iws1, together with K90, are labelled by magenta triangles. Residues involved in Spt6 IR1 and IR2 binding are labelled with yellow diamonds and cyan circles, respectively. Alignment features are identical in all figures unless otherwise stated. Alignments were created with ALINE (Bond and Schuttelkopf, 2009).
A conserved binding subdomain (HEAT subdomain) formed by the Iws1 HEAT repeat and part of the ARM1 repeat contains the invariant ecK90/scK192
In yeast Iws1, mutation of the invariant lysine K192 to asparagine leads to the rescue of TBP mutants that possess postrecruitment defects, and prevents constitutive recruitment of Iws1 to the CYC1 gene promoter in an Spt6-independent manner (Fischbeck et al, 2002; Zhang et al, 2008). The equivalent lysine of ecIws1, K90, is found at the start of helix αB in the HEAT repeat (Figures 2A and 3A). The K90 side chain is found at the interface of the tips of both HEAT and ARM1 repeats. The aliphatic part of the K90 side chain is at the centre of a hydrophobic core formed by the side chains of HEAT αA I73, ARM1 α21 W122 and ARM1 α31 I136. Furthermore, K90 Nɛ is fixed through a hydrogen bond with the carboxylate of invariant HEAT αA D77 that also hydrogen bonds with the K90 main chain amide (Figure 3A).
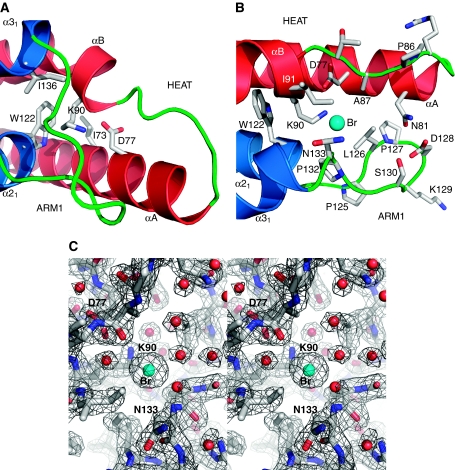
Figure 3.
The HEAT-subdomain Iws1. (A) Close-up view of K90 interactions at the interface of Iws1 HEAT and ARM1 repeats. The side chains involved in hydrophobic interactions and hydrogen bonding are shown. (B) Close-up view of the HEAT subdomain formed by Iws1 HEAT and ARM1 repeats. Residues forming the surface are displayed. The bromide anion (Br) is shown as a cyan sphere. (C) Stereo view of the structure at the bromide-binding site. The bromide (Br) and water molecules are shown as cyan and red spheres, respectively. The 2Fo–Fc electron density map of the refined structure is shown and contoured at 1.5 σ.
Importantly, the hydrogen bond formed between the carboxylate of D77 and the K90 Nɛ positions the latter atom precisely at the bottom of a pocket formed by both tips of the HEAT and ARM1 repeats, making it solvent accessible. Strikingly, most of the constituent residues of this pocket are highly conserved (Figures 2B and 3B). Inspection of this pocket reveals the direct binding of an ion to K90 Nɛ, which is further coordinated by the N133 main chain amide as well as one or two water molecules. In most structures, the bound ion is most likely a chloride ion provided by the protein buffer. In one structure, however, a much larger electron density was observed reflecting the replacement of the chloride ion by a negatively charged bromide ion provided by the crystallization conditions (Figure 3B and C). Interestingly, this ion-binding pocket would be large enough to accommodate a phosphate at the position of the bromide. Furthermore, the atoms coordinating the bromide ion (K90 Nɛ, N133 main chain amide and two water molecules) would be perfectly placed to coordinate an incoming phosphate. As one of the coordinating water molecules is located at the entrance of the pocket, it could also be replaced by the oxygen of a phosphorylated residue. Taken together, these results suggest that this conserved binding domain of Iws1 could be specific for phosphoproteins.
To further investigate the role of K90, four mutants were created: K90R, K90A, K90D and K90N. All mutants could be expressed and purified. Despite a two- to four-fold decrease in yield for most of the mutants, none showed an increased tendency to aggregate as assessed by dynamic light scattering analyses. However, temperature-dependent unfolding of these proteins using the ThermoFluor technology showed that the K90D and K90N mutants have greater instability (Supplementary Figure 2A). We next tested the effect of these mutants on the formation of the complex with Spt6N. Upon co-expression of the mutants with ecSpt6N53−71, no complex was observed with the K90D mutant and the level of complex formation appeared to be reduced with the K90N mutant (Supplementary Figure 2B, lanes 1–5). Surprisingly, none of the mutants impaired the binding of the longer ecSpt6N1−71 construct (Supplementary Figure 2B, lanes 7–11). Taken together, our results suggest that the effects of the K90 mutants are local and suggest a more extensive interface between Iws1 and Spt6N.
Bipartite binding of Spt6N to Iws1 ARM repeats (ARM subdomain)
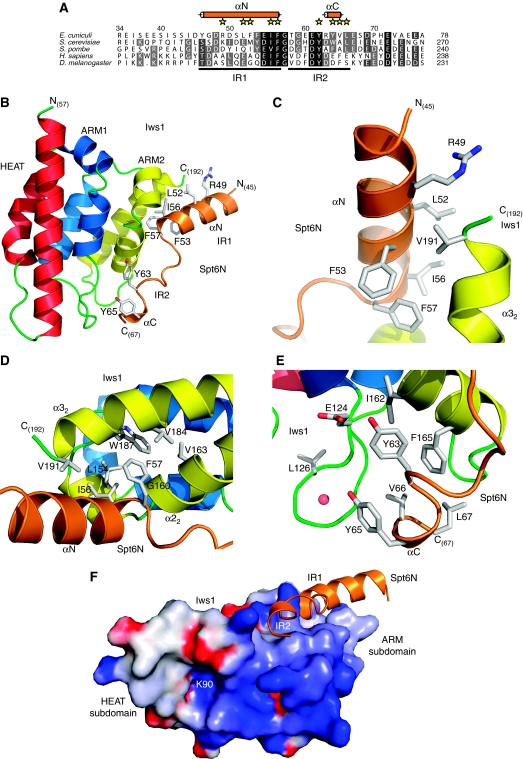
Analysis of the structures of the Iws1/Spt6N complex confirms the hypothesis of extended Iws1–Spt6N interactions, showing that the region of ecSpt6N interacting with Iws1 encompasses residues 45–67, forming contacts with both ARM repeats, but not with the HEAT repeat (Figure 4A and B). This region is composed of an N-terminal α-helix (αN) that interacts with Iws1 ARM2, pointing towards the ARM2 α22 helix. αN stops abruptly at the invariant glycine 58, which enables the peptide to go round the Iws1 α22 helix. The C-terminal part of the Spt6N peptide adopts a rather extended conformation followed by a short-helical turn (αC) and interacts with both ARM repeats (Figure 4B).
Figure 4.
Crystal structure of E. cuniculi Iws1/Spt6N complex. (A) Multiple sequence alignment of the Iws1-binding region of Spt6N. Both Spt6N IR1 and IR2 sub-regions are indicated below the sequences. Residues whose side chains are involved in Iws1 binding are labelled with yellow stars. Spt6N α-helices observed in the Iws1/Spt6N structures are shown as orange cylinders. (B) Ribbon representation of the Spt6N34−71/Iws155−198 structure. Spt6N is coloured orange. Most Spt6N side chains interacting with Iws1 are shown. (C) Close-up view of the interaction between Spt6N αN and Iws1 α32 helices. (D) Close-up view of Spt6N IF motif binding to Iws1 hydrophobic cavity. (E) Close-up view of Spt6N IR2 binding to Iws1. The red sphere represents a water molecule. (F) GRASP (Nicholls et al, 1991) representation of the electrostatic potential at the surface of E. cuniculi Iws1. The electrostatic potentials −8 and +8 kBT (kB, Boltzmann constant; T, temperature) are coloured red and blue, respectively. The Spt6N region binding to Iws1 is shown as orange ribbon. K90 Nɛ is located within a cavity and is labelled (K90).
As such, the interaction region (IR) of Spt6N with Iws1 can be divided into two sub-regions that we termed IR1 (Spt6N αN up to the invariant F57/G58 motif) and IR2 (C-terminal to G58) (Figure 4A). The interaction between the Spt6N IR1 region and Iws1 is dictated mostly by hydrophobic interactions. The Spt6N αN helix packs against the C-terminus of Iws1 ARM2 α32 helix (Figure 4C). This interaction is essentially hydrophobic and involves Iws1 V191 as well as several residues at the N-terminus of Spt6N αN. These latter residues are solvent exposed and their contribution to the hydrophobic core is mediated by their main chain Cβ and/or Cγ atoms. This most likely explains why the sequence of this helix is not evolutionarily conserved as only its α-helical propensity appears important. In agreement, secondary structure predictions of S. cerevisiae and H. sapiens Spt6N indicate an α-helix precisely at the position of ecSpt6N αN, ending at the invariant FG motif.
At the C-terminal end of Spt6N αN, I56 and F57 interact with a hydrophobic cavity of Iws1 formed by residues L154, G160, V163, V184, W187 and V191 of the ARM2 repeat (Figure 4D). The perfect conservation of the hydrophobic character of the I56/F57 motif, as well as the ability of the ecSpt6N53−71 construct to retain its interaction with Iws1 despite the large truncation of the αN helix, demonstrates the importance of the binding of the I56/F57 motif to the hydrophobic cavity of Iws1.
In contrast, the interaction between the Spt6N IR2 region and Iws1 is composed of both hydrophobic and hydrogen bond contacts. Following the I56/F57 motif, Spt6N G58 has the function of helix breaker, necessary to prevent steric clashes between Iws1 and an elongated αN helix of Spt6N. The four residues following G58 do not make extensive contacts with Iws1, but serve as a linker for the peptide to go around the α22 helix of ARM2. The next interaction between both proteins is mediated through the hydrogen bond formed between the hydroxyl of Spt6 Y63 and the carboxylate of Iws1 E124 (Figure 4E). This interaction is reinforced by hydrophobic contacts made between the Y63 side chain and Iws1 I162 and F165. Other interactions involve Spt6N tyrosine Y65 whose hydroxyl forms a water-mediated interaction with Iws1 L126 carboxyl, and Spt6 V66 and L67, which form hydrophobic contacts with residues of Iws1 ARM1 and ARM2 repeats.
Interestingly, both the ion-binding HEAT subdomain and the Spt6N-binding ARM subdomain of Iws1 are connected by a positively charged channel (Figure 4F). Specifically, the K90-binding pocket and the Spt6 IR2-binding surface of Iws1 are relatively close, suggesting that binding of a protein to the former pocket could prevent binding of Spt6 to the latter surface, or vice versa.
Point mutations within Spt6 Iws1-binding region cause Spt− and Ts− phenotypes in vivo
We next investigated the interaction between Spt6 and Iws1 by mutational analysis, using the ecSpt6N1−71 construct, which encompasses the full region of interaction of ecSpt6N with Iws1. Based on our structural data, single and double mutants of ecSpt6N and ecIws1 were made. At the Spt6N IR1/Iws1 interface, Iws1 residues G160 and V191 were changed to bulkier residues (single mutants G160Y, V191Y and V191W), whereas Spt6N residues I56 and F57 were changed to smaller alanine residues (double mutant I56A/F57A). At the IR1/IR2 transition, both Spt6N G58 and G60 were changed to alanines (double mutant G58A/G60A). At the Spt6N IR2/Iws1 interface, Iws1 E124 was changed to either serine or alanine (E124S and E124A), whereas Spt6N Y63 and Y65 were changed to alanines (single mutant Y63A and double mutant Y63A/Y65A).
Upon co-expression, none of the Iws1 mutants affected complex formation, showing that single mutations in Iws1 are not sufficient to destroy the complex with Spt6N (Figure 5A, lanes 1–6). In contrast, all Spt6N mutants, with the exception of the Y63A mutant, led to the loss of the complex (Figure 5A, lanes 8–12). Some of these Spt6N mutants appeared insoluble (Figure 5A, lanes 10 and 12), suggesting that the loss of the complex may be due to insolubility. However, the I56A/F57A mutant was still soluble, showing that, in this case, the loss of interaction with Iws1 was due to the mutations. In agreement, the same mutation introduced in the yeast Spt6N229−269 construct (I248A/F249A) leads to a drastic loss of interaction with scIws1 (Figure 5B).
Figure 5.
Mutational and in vivo analysis of the Iws1/Spt6 complex. (A) Analysis of the E. cuniculi Iws1/Spt6N complex by mutation of Iws1- and Spt6N-specific residues involved in complex formation. Stronger effects are observed by mutating the Spt6N protein. (B) Analysis of the full-length S. cerevisiae Iws1/Spt6 complex by mutation of the IF motif, confirming the result observed for the E. cuniculi Iws1/Spt6N complex. The mutant migrates more slowly than the wild-type construct. Degradation products are marked by an *. (C) Analysis of Spt6 mutants within the Iws1-binding region of full-length S. cerevisiae Spt6. Strains were grown to saturation in YPD, serially diluted 10-fold and spotted on the indicated media. Included for comparison was spt6-50, a previously characterized mutant.
As the Spt6N mutants appeared to have the strongest effect on the formation of the Iws1/Spt6N complex, we analysed the effect of these mutations in S. cerevisiae in vivo, comparing them with wild type and to a previously studied mutant, spt6–50. Initially, three mutants were constructed for the full-length yeast Spt6 protein: I248A/F249A, G250A/G252A and Y255A/W257A (equivalent to the I56A/F57A, G58A/G60A and Y63A/Y65A mutants of ecSpt6, respectively). When these mutant forms of full-length Spt6 were expressed in vivo as the only form of the protein, all of the yeast strains were viable, showing that all of the mutants still possess some Spt6 function (Figure 5C). The G250A/G252A mutant displayed a wild-type phenotype (Figure 5C), suggesting that either this form of Spt6 is functional in vivo or that it is redundant with some other function.
In contrast to the G250A/G252A mutant, the strains harbouring the I248A/F249A and Y255A/W257A mutants displayed several mutant phenotypes. Both mutants showed a strong temperature-sensitive phenotype at 37°C (Figure 5C). Furthermore, these two mutants caused a marked Spt− phenotype (suppression of auxotrophies caused by the insertion mutations his4–912δ and lys2–128δ), indicative of transcription defects. The Y255A/W257A mutant had the broadest range of phenotypes, as it was sensitive to both hydroxyurea and phleomycin, and modestly sensitive to caffeine (Figure 5C). To determine whether the phenotypes of Y255A/W257A required both amino-acid changes, the single mutants were constructed and tested for the same set of phenotypes. Compared with the double mutant, the effects of the Y255A and W257A single mutants were less drastic. Neither of them showed sensitivity to high temperature, hydroxyurea, phleomycin or caffeine. Although the Y255A mutant shows a partial Spt− phenotype (His+ phenotype), the W257A mutant displays a tighter His− phenotype than even the wild-type strain (Figure 5C). These results are in agreement with our co-expression tests in which we showed that the ecSpt6N Y63A mutant had less effect on the interaction with Iws1 than the Y63A/Y65A mutant. Taken together, these results confirm the importance of the Iws1-interacting region of Spt6 for the function of this protein in vivo.
The IR1-binding region of Iws1 is structurally conserved in TFIIS
Part of the conserved region of Iws1 has been shown to share sequence similarity with the N-terminal domains of TFIIS, Elongin A and Med26 (Wery et al, 2004; Ling et al, 2006); Figure 2B). Our structural data reveals that these domains correspond to the ARM subdomain of Iws1, with the HEAT subdomain of Iws1 being specific to this protein. We have tentatively superimposed our Iws1 structure on the mouse and yeast TFIIS Domain I structures (PDB code 1WJT, RIKEN Structural Genomics/Proteomics Initiative; and PDB code 1EO0; Booth et al, 2000). We observed a very good fit between ecIws1 and the mouse N-terminal domain of TFIIS (Figure 6A). Specifically, the upper parts of the ARM repeats superimpose extremely well, whereas the lower parts, notably the loop regions between helices α2 and α3 of the repeats, show much more structural divergence.
Figure 6.
Specific interaction of TFIIS N-terminal domain with Spt8 and Med13. (A) Superposition of E. cuniculi Iws1 (red) and mouse TFIIS Domain I (blue) structures. For comparison, the position of Spt6N (orange) bound to Iws1 is shown. (B) Superposition of the hydrophobic (IF-binding) pockets of E. cuniculi Iws1 (red) and mouse TFIIS domain I (blue). All side chains are shown. (C) Multiple sequence alignment of the Iws1-binding region of Spt6N (top five rows) and of the putative TFIIS-binding region of S. cerevisiae (sc) Spt8 and Med13 (bottom two rows). (D) Reconstitution by co-expression of the complexes formed between S. cerevisiae TFIIS (1–78) and Med13 (466–508), and S. cerevisiae TFIIS (1–78) and Spt8 (371–402). Importance of the IF motifs and acidic regions on complex formation is addressed by using specific mutants. Degradation products of Med13 are labelled with an ‘*'.
Surprisingly, no satisfying fit was found with the yeast protein (PDB code 1EO0). Careful inspection of the yeast TFIIS Domain I structure revealed the presence of several charged side chains in hydrogen bonding distance to hydrophobic side chains. To further investigate this discrepancy, we have established a model of the yeast TFIIS Domain I using the mouse structure and the alignment provided in Figure 2B (model available upon request). Inspection of the model reveals that the yeast TFIIS Domain I can perfectly adopt the fold observed for the mouse protein. Specifically, no steric clashes or any unconventional structural features are observed, suggesting that the current yeast TFIIS Domain I structure is most likely incorrect.
The most striking conservation between Iws1 and mouse TFIIS Domain I concerns the hydrophobic pocket binding the I56/F57 motif of Spt6 (Figure 6B). At the sequence level, this is reflected by the almost perfect conservation between the residues forming the pocket, with only a few conservative mutations. Therefore, both at the sequence and structural levels, the Spt6 IR1-binding region of Iws1 is almost completely conserved in TFIIS. In contrast, the IR2-binding region of Iws1 is different between these two proteins. Sequence analysis shows that the same pattern of conservation/divergence occurs in Elongin A and Med26 (Figure 2B).
TFIIS has been shown to be recruited to promoters in a SAGA- and Mediator-dependent manner (Prather et al, 2005; Kim et al, 2007) and two-hybrid analyses have shown that the yeast TFIIS N-terminal domain interacts with SAGA subunit Spt8 and Mediator subunit Med13 (Wery et al, 2004). In this latter study, the TFIIS IRs of Spt8 and Med13 have been delimited to 100 and 200 residues, respectively. Alignment of these two regions revealed a single patch of conservation composed of a Leucine–Phenylanine–Glycine sequence (LFG motif) followed by acidic residues. Furthermore, despite poor sequence conservation, both sequences upstream of the LFG motif are predicted to be α-helical. Therefore, these two proteins contain a region highly reminiscent of the Spt6 IR1 region (Figure 6C). In contrast, the IR2 region is not conserved in these proteins and is replaced in both proteins by a stretch of aspartate and glutamate residues.
We investigated the possibility of an interaction between Spt8 and Med13 with TFIIS by co-expressing the scSpt8371−402 and scMed13466−508 (IR1/IR2)-like motifs with the yeast TFIIS N-terminal domain (residues 1–78). Formation of a complex was observed in both cases (Figure 6D, lanes 1 and 5; Supplementary Figure 3). Interestingly, upon mutation of the LF motifs, the interaction with scTFIIS1−78 was apparently weakened with scMed13466−508 and abolished with scSpt8371−402 (Figure 6D, lanes 2 and 6), suggesting that residues of scMed13466−508, which are not conserved in scSpt8371−402, strengthen the interaction. Furthermore, removal of the IR2-like regions of these two proteins by using shorter constructs encompassing only two acidic residues after the LFG motif (scSpt8371−390 and scMed13466−487) abolishes interaction in the case of Spt8, but only weaken the interaction in the case of Med13 (Figure 6D, lanes 3 and 7), demonstrating the importance of the IR2-like region in Spt8 and Med13 and suggesting that N-terminal residues in Med13 interact more extensively with scTFIIS1−78. Taken together, these results indicate that Spt8 and Med13 form specific interactions with TFIIS N-terminal domain. These complexes rely on features observed not only in the formation of the Iws1/Spt6 complex (recognition of the IR1 region), but also on TFIIS-specific features.
Discussion
Despite the progress made in the functional characterization of Spt6 and Iws1, the roles of these proteins in molecular terms are not well understood. We have started addressing this issue by studying the Spt6 and Iws1 proteins from the fungi-related parasite E. cuniculi. We show here that the interaction between Spt6 and Iws1 only requires a small N-terminal region of Spt6 (E. cuniculi residues 45–67), which agrees with and refines previous results obtained with the mammalian versions of these proteins (Yoh et al, 2007). Furthermore, the crystallographic structures of Iws1 and Iws1 bound to this N-terminal region of Spt6 reveal the presence of two distinct subdomains in Iws1. An independent determination of the Iws1 crystallographic structure (Pujari et al, 2010) fits well with the structure that we determined.
The first subdomain (HEAT subdomain) is specific to Iws1 and is formed by its N-terminal HEAT repeat and part of the following ARM repeat. At the heart of this domain is an invariant lysine (E. cuniculi K90, S. cerevisiae K192), which is instrumental in the Spt6-independent constitutive recruitment of Iws1 to promoters in yeast (Fischbeck et al, 2002; Zhang et al, 2008). The Nɛ atom of this lysine is located at the bottom of a highly conserved pocket that has the ability to preferentially bind negatively charged ions as large as phosphates. In agreement, it has been proposed that Ser5-phosphorylated RNAPII CTD repeats directly recruit yeast Iws1 in a K192-dependent manner (Zhang et al, 2008). Yet, our preliminary investigations in vitro have failed to characterize a strong interaction between Iws1 and the phosphorylated CTD of RNAPII. This suggests that such an interaction requires another partner that could either stabilize a direct interaction between Iws1 and the RNAPII CTD or, alternatively, bridge both molecules.
The second subdomain (ARM subdomain) of Iws1 is defined by its two ARM repeats and forms a specific complex with a small N-terminal region of Spt6 (Spt6N). Biochemical and structural analysis of Spt6 recognition by Iws1 revealed two anchoring regions in Spt6N, IR1 and IR2, which are both important for proper complex formation. Furthermore, introduction of amino-acid changes in the context of the full-length yeast Spt6 protein in vivo showed that two mutants of the IR1 and IR2 regions, I248A/F249A and Y255A/W257A, respectively, display strong Spt− and Ts− phenotypes, thus highlighting the importance of the full Iws1-binding region of Spt6 in vivo.
Interestingly, the HEAT subdomain of Iws1, which most likely modulates the Spt6-independent role of Iws1, and the IR2-binding region of the Iws1 ARM subdomain are relatively close to each other (Figure 4F). Furthermore, almost all residues of the highly conserved loop, which connects helices α21 and α31 of the Iws1 ARM1 repeat, participate either in the HEAT subdomain or in the IR2-binding region of Iws1 ARM subdomain (Figure 2B). Thus, the two Iws1 subdomains appear strongly linked, which suggests that the switch between the Spt6-independent and Spt6-dependent functions of Iws1 occurs through steric hindrance as well as by possible structural rearrangements. It cannot be excluded that a longer C-terminal construct of Spt6 might interact directly with the HEAT subdomain of Iws1. Our co-expression experiments suggest, however, that this interaction is highly unlikely.
Another striking result of our studies is the almost perfect structural conservation between the IR1-binding region of the Iws1 ARM subdomain and the mouse TFIIS N-terminal domain (Domain I). In yeast, the TFIIS Domain I has recently been associated with the newly discovered function of TFIIS in PIC formation and stabilization through its recruitment by the SAGA and Mediator complexes (Prather et al, 2005; Guglielmi et al, 2007; Kim et al, 2007). In agreement, a direct interaction has been observed between yeast TFIIS Domain I and SAGA and Mediator respective subunits, Spt8 and Med13 (Wery et al, 2004).
The strong structural homology observed between the IR1-binding region of Iws1 ARM subdomain and TFIIS Domain I suggests that both domains bind their targets in an analogous manner. We show here that Spt8 and Med13 contain a single patch of conserved sequence that is composed of a highly conserved IR1 motif followed by a stretch of acidic residues. Co-expression of these conserved regions from yeast Spt8 and Med13 with yeast TFIIS Domain I results in complex formation, and requires their IR1-like regions as well as their specific acidic regions. Altogether, these results suggest a specific interaction between TFIIS and Spt8, and TFIIS and Med13, thereby suggesting a molecular basis for TFIIS recruitment at promoters, in order to participate in active PIC formation, possibly by recruitment of RNAPII through TFIIS Domain II.
Importantly, in yeast, only TFIIS Domain I shares sequence homology with the Iws1 IR1-binding region. In contrast, in humans, this homology extends not only to the different isoforms of TFIIS, but also to the isoforms of Elongin A, to Med26 and to two other nuclear proteins, PNUTS and PIBP, whose functions are less well characterized (Wery et al, 2004; Ling et al, 2006). Surprisingly, in humans, no homologue of yeast Spt8 has been found to date and the TFIIS-binding domain of yeast Med13 does not seem to be conserved in its human homologue. However, in agreement with what has been observed in yeast, both human TFIIS and Elongin A IR1-binding regions are sufficient to interact with the RNAPII holoenzyme containing various PIC components as well as the Mediator subunit Cdk8 (Pan et al, 1997). This suggests that one or several subunits of the human holoenzyme have evolved specific sequence for the binding of TFIIS and Elongin A N-terminal domains, possibly recapitulating the role of Spt8 and Med13 in yeast.
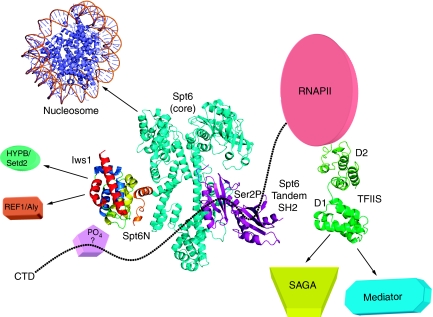
The results presented here provide a detailed view of the Iws1/Spt6 complex. Specifically, we show the presence of two specific conserved subdomains within Iws1, one that may recognize a phosphoprotein and a second subdomain specific for Spt6 recognition. Our structural, mutational and in vivo functional data demonstrate a bipartite binding of Spt6 to this second subdomain of Iws1. Importantly, part of the recognition determinants within Iws1 for Spt6 are conserved in yeast TFIIS and suggest the molecular basis for recruitment of TFIIS by SAGA Spt8 and Mediator Med13 to promoters. Therefore, our results deepen our understanding of the transcription networks made by the Iws1/Spt6 complex and TFIIS (Figure 7). Furthermore, the broad distribution of the Iws1 (Spt6 binding) ARM subdomain in proteins of higher eukaryotes reveals the larger functional implication of this domain in these organisms. Our study paves the way for deciphering these additional interaction networks, which will require further structural, biochemical and in vivo investigations in order to better understand the role of these networks in transcriptional regulation.
Figure 7.
Model of the transcription networks made by Iws1/Spt6 and TFIIS. Based on the results presented in this article as well as previously published data, the figure provides a current model of the Iws1/Spt6 complex and recapitulates our current knowledge of the transcription networks affected by Iws1/Spt6 and TFIIS, including homologous domains (ARM subdomain for Iws1 and Domain I for TFIIS). The model for Spt6 has been assembled from (i) the structure of the bacterial Tex protein that shares sequence homology with the core domain of Spt6 (cyan; Johnson et al, 2008), (ii) the recently discovered C-terminal tandem SH2 domains of Spt6 (purple; Diebold et al, 2010; Sun et al, 2010) and (iii) the Iws1/Spt6N complex (coloured as in previous figures; this manuscript). The N-terminal acidic domain of Spt6 is not included. Interactions of Iws1/Spt6 and TFIIS with different transcriptional effectors are shown with arrows. The C-terminal domain of the RNA polymerase II (CTD) is shown as dotted line. Binding of the Spt6 tandem SH2 repeats to Ser2-phosphorylated CTD repeats has been labelled with ‘Ser2P'. The putative CTD binding to the HEAT subdomain of Iws1, possibly through a phosphorylated residue and the requirement of an unknown factor, has been labelled with ‘?'. The PDB codes for the structures used to make the figure are 1AOI, 1WJT, 2XP1, 3BZC, 3GTM and 3GXW.
Materials and methods
Expression and purification
The constructs and mutants used were created by standard PCR procedures and inserted in the pET-MCN series of multi-expression vectors (Romier et al, 2006), using NdeI and BamHI restriction sites. All proteins were expressed or co-expressed in E. coli BL21[DE3] cells containing the pRARE vector (Novagen). For single expression, proteins were fused with an N-terminal (His)6 tag. For co-expression, one protein was his-tagged and the other untagged and complex formation was monitored by retention of the untagged protein on affinity resin (Talon Metal affinity resin, Clontech) through its interaction with the tagged protein. Following expression, cells were resuspended in lysis buffer (10 mM Tris pH 8.0 with either 50 mM NaCl for the different complexes or 400 mM NaCl for Iws1 alone) and then lysed by sonication.
For small-scale (2 ml) interaction analysis, soluble fractions were incubated for 1 h with 25 μl Talon Metal affinity resin (Clontech) and washed with lysis buffer. Laemmli buffer (30 μl) was then added to the resin for analysis on SDS–PAGE. For large-scale purification, to produce the protein and complexes for crystallization trials, the soluble fraction was recovered by high-speed centrifugation and incubated with Talon Metal affinity resin for 1 h. The resin was then extensively washed with lysis buffer and resuspended in 2 ml of this buffer, and bovine thrombin was added overnight for cleaving off the histidine tag. The supernatant was recovered and applied onto a gel filtration column Hiload 16/60 Superdex 75 (Amersham Pharmacia) equilibrated with lysis buffer supplemented with 2 mM dithiotreitol. The purified proteins/complexes were concentrated on an Amicon 10K system (Millipore). For production of selenomethionylated Iws1, protein expression was carried out in minimal medium containing selenomethionine.
Biophysical characterization
Dynamic Light Scattering experiments were performed using a Dynapro-MS (Protein Solutions). Thermal Shift Assay experiments (Thermofluor) were performed using the Mini Opticon Real Time PCR Detection System from Bio-Rad. The fluorescent reporter used was Sypro Orange (Invitrogen; final concentration 25 ×). The temperature was raised from 20 to 90°C in increments of 1°C every minute. Wild type and mutant Iws1 proteins were used at a concentration of 1 mg/ml.
Crystallization
Crystallization trials were performed using the sitting and hanging drop vapour diffusion method. The different crystallization conditions for the ecIws155−198 construct are reported elsewhere (Koch et al, 2010). The ecSpt653–71/ecIws155−198 complex (10 mg/ml) gave two crystal forms. The first form was obtained at 24°C in the presence of 22% PEG 3350 (Fluka) and 0.2 M NaBr. The second crystal form was grown at 4°C in the presence of 0.1 M sodium acetate pH 5, 18% PEG 1500 (Fluka) and 0.05 M MgCl2. The ecSpt634–71/ecIws155−198 complex (10 mg/ml) was crystallized at 17°C in the presence of 33% PEG 1500 (Fluka) and 0.1 M of the succinic acid/sodium dihydrogen phosphate/glycine system, pH 7 (Hampton). For data collection, the crystals were frozen into liquid nitrogen, either directly (ecSpt634–71/ecIws155−198) or after cryoprotection within the crystallization solution supplemented with 20% PEG 200 (ecIws155−198 and ecSpt653–71/ecIws155−198 form 2) or 20% glycerol (ecSpt653–71/ecIws155−198 form 1).
Data collection, structure determination, model building and refinement
All data obtained in this project were collected at 100 K on ESRF beamlines ID14-1, ID14-2, ID14-4, ID23-1, ID29 and SOLEIL beamline PROXIMA1. All data were processed and scaled using HKL2000 (Otwinowski and Minor, 1997). The ecIws155−198 structure was solved by performing three-wavelength MAD experiments on selenomethionylated protein-containing crystals (Supplementary Table I). Phasing was performed with SHARP (La Fortelle and Bricogne, 1997). Model building was carried out using TURBO-FRODO and COOT (Emsley and Cowtan, 2004), and refinement with REFMAC (CCP4, 1994). The structures of the complexes were solved by molecular replacement with PHASER (McCoy et al, 2007) using the ecIws155−198 structure as a search model. The Spt6 protein constructs were built into the additional electron density and the structures refined using identical procedures as for ecIws155−198. All structures showed good deviations from ideal geometry (Supplementary Tables I and II), with no Ramachandran outliers. The structures described in this project have been deposited under the PDB codes 2xpl, 2xpn, 2xpo and 2xpp.
Yeast genetics
Mutations encoding the amino-acid changes I248A/F249A, G250A/G252A, Y255A/W257A, Y255A and W257A in S. cerevisiae SPT6 were each individually introduced into a copy of SPT6 on a pRS414-based CEN plasmid. Mutagenesis was performed using the Quikchange Lightning Site-directed Mutagenesis kit (Agilent). The mutated plasmids were used to transform S. cerevisiae strain FY857 (MATα his4-912δ lys2-128Δ leu2Δ1 ura3-52 trp1Δ63 spt6Δ::LEU2 containing pCC11 (SPT6 in a CEN URA3-marked plasmid)) using standard protocols. A 5-FOA plasmid shuffle (Boeke et al, 1984) was performed to obtain a strain that contained only the plasmid with the spt6 mutant allele. Spot tests were performed on the following media, all incubated at 30°C unless otherwise indicated: YPD, YPD at 37°C, SC-his, SC-lys, YPD+150 mM hydroxyurea, YPD + 13 μg/ml phleomycin and YPD+15 mM caffeine. YPD, SC-his and SC-lys were made as previously described (Rose et al, 1990). All strains are listed in Supplementary Table III. The plasmids they contain are indicated in parentheses.
Note added in proof
An independent study (McDonald SM, Close D, Xin H, Formosa T and Hill CP. Mol Cell, 10.1016/j.molcel.2010.11.014) has also determined a very similar structure of an Iws1/Spt6 complex; the results of the two studies are highly complementary.
Supplementary Material
Acknowledgments
We thank Michel Werner for providing the yeast TFIIS cDNA, Jean-Marie Wurtz and Jean-Paul Renaud for technical discussions, Irwin Davidson for critical reading of the manuscript and members of the Structural Genomics Platform of IGBMC for setting up automated procedures. We thank members of the ESRF-EMBL joint structural biology groups and members of SOLEIL for the use of their beamline facilities and for help during data collection. MLD is supported by a PhD grant from the French Research Ministry. This work was supported by institutional funds from the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Université de Strasbourg (UDS), the European Commission SPINE2-Complexes project (contract n° LSHG-CT-2006-031220) and NIH grant GM32967.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adkins MW, Tyler JK (2006) Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 [DOI] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P (2001) Comparison of ARM and HEAT protein repeats. J Mol Biol 309: 1–18 [DOI] [PubMed] [Google Scholar]
- Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT (2009) Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J 28: 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197: 345–346 [DOI] [PubMed] [Google Scholar]
- Bond CS, Schuttelkopf AW (2009) ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr 65(Part 5): 510–512 [DOI] [PubMed] [Google Scholar]
- Booth V, Koth CM, Edwards AM, Arrowsmith CH (2000) Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70. J Biol Chem 275: 31266–31268 [DOI] [PubMed] [Google Scholar]
- Bortvin A, Winston F (1996) Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50(Part 5): 760–763 [DOI] [PubMed] [Google Scholar]
- Diebold ML, Loeliger E, Koch M, Winston F, Cavarelli J, Romier C (2010) A non-canonical tandem SH2 enables interaction of elongation factor SPT6 with RNA polymerase II. J Biol Chem (doi:10.1074/jbc.M110.146696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 (Part 12 Part 1): 2126–2132 [DOI] [PubMed] [Google Scholar]
- Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, Sato H, Yamaguchi Y, Mandal SS, Reinberg D, Wada T, Handa H (2004) Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol 24: 3324–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbeck JA, Kraemer SM, Stargell LA (2002) SPN1, a conserved gene identified by suppression of a postrecruitment-defective yeast TATA-binding protein mutant. Genetics 162: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B, Soutourina J, Esnault C, Werner M (2007) TFIIS elongation factor and Mediator act in conjunction during transcription initiation in vivo. Proc Natl Acad Sci USA 104: 16062–16067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SJ, Close D, Robinson H, Vallet-Gely I, Dove SL, Hill CP (2008) Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J Mol Biol 377: 1460–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099 [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453 [DOI] [PubMed] [Google Scholar]
- Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA (2007) The transcription elongation factor TFIIS is a component of RNA polymerase II preinitiation complexes. Proc Natl Acad Sci USA 104: 16068–16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M, Diebold ML, Cavarelli J, Romier C (2010) Crystallization and preliminary crystallographic analysis of eukaryotic transcription and mRNA export factor Iws1 from Encephalitozoon cuniculi. Acta Crystallogr Sect F Struct Biol Cryst Commun 66(Part 2): 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. In: Methods in Enzymology, Carter JCW, Sweet RM (eds), Vol. 276, pp 472–494. New-York: Academic Press [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR III, Hartzog GA (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol 23: 1368–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Smith AJ, Morgan GT (2006) A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS. Nucleic Acids Res 34: 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P (2010) Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40(Part 4): 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing X-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Carter JCW, Sweet RM (eds), Vol. 276, pp 307–326. New-York: Academic Press [DOI] [PubMed] [Google Scholar]
- Pan G, Aso T, Greenblatt J (1997) Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem 272: 24563–24571 [DOI] [PubMed] [Google Scholar]
- Perales R, Bentley D (2009) ‘Cotranscriptionality': the transcription elongation complex as a nexus for nuclear transactions. Mol Cell 36: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather DM, Larschan E, Winston F (2005) Evidence that the elongation factor TFIIS plays a role in transcription initiation at GAL1 in Saccharomyces cerevisiae. Mol Cell Biol 25: 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujari V, Radebaugh CA, Chodaparambil JV, Muthurajan UM, Almeida AR, Fischbeck JA, Luger K, Stargell LA (2010) The transcription factor Spn1 regulates gene expression via a highly conserved novel structural motif. J Mol Biol (doi:10.1016/j.jmb.2010.09.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier C, Ben Jelloul M, Albeck S, Buchwald G, Busso D, Celie PH, Christodoulou E, De Marco V, van Gerwen S, Knipscheer P, Lebbink JH, Notenboom V, Poterszman A, Rochel N, Cohen SX, Unger T, Sussman JL, Moras D, Sixma TK, Perrakis A (2006) Co-expression of protein complexes in prokaryotic and eukaryotic hosts: experimental procedures, database tracking and case studies. Acta Crystallogr D Biol Crystallogr 62(Part 10): 1232–1242 [DOI] [PubMed] [Google Scholar]
- Romier C, James N, Birck C, Cavarelli J, Vivares C, Collart MA, Moras D (2007) Crystal structure, biochemical and genetic characterization of yeast and E. cuniculi TAF(II)5 N-terminal domain: implications for TFIID assembly. J Mol Biol 368: 1292–1306 [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Laboratory Course Manual for Methods in Yeast Genetics. New York: Cold Spring Harbor [Google Scholar]
- Saunders A, Core LJ, Lis JT (2006) Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7: 557–567 [DOI] [PubMed] [Google Scholar]
- Sun M, Lariviere L, Dengl S, Mayer A, Cramer P (2010) A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II CTD. J Biol Chem (doi:10.1074/jbc.M110.144568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wery M, Shematorova E, Van Driessche B, Vandenhaute J, Thuriaux P, Van Mullem V (2004) Members of the SAGA and Mediator complexes are partners of the transcription elongation factor TFIIS. EMBO J 23: 4232–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M, aus Dem Siepen T, Stamminger T (2000) Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J Virol 74: 8053–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Cho H, Pickle L, Evans RM, Jones KA (2007) The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev 21: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA (2008) The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 22: 3422–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA (2008) Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol 28: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.