Abstract
Translational stimulation of mRNAs during early development is often accompanied by increases in poly(A) tail length. Poly(A)-binding protein (PAB) is an evolutionarily conserved protein that binds to the poly(A) tails of eukaryotic mRNAs. We examined PAB’s role in living cells, using both Xenopus laevis oocytes and Saccharomyces cerevisiae, by tethering it to the 3′-untranslated region of reporter mRNAs. Tethered PAB stimulates translation in vivo. Neither a poly(A) tail nor PAB’s poly(A)-binding activity is required. Multiple domains of PAB act redundantly in oocytes to stimulate translation: the interaction of RNA recognition motifs (RRMs) 1 and 2 with eukaryotic initiation factor-4G correlates with translational stimulation. Interaction with Paip-1 is insufficient for stimulation. RRMs 3 and 4 also stimulate, but bind neither factor. The regions of tethered PAB required in yeast to stimulate translation and stabilize mRNAs differ, implying that the two functions are distinct. Our results establish that oocytes contain the machinery necessary to support PAB-mediated translation and suggest that PAB may be an important participant in translational regulation during early development.
Keywords: early development/mRNA turnover/poly(A)/translation/Xenopus oocytes
Introduction
During early development, changes in the pattern of protein synthesis are commonly due to the activation, repression or destruction of pre-existing mRNAs. Translational control of specific mRNAs is required for diverse processes in oocytes and embryos, including pattern formation in flies, regulation of cell fate in Caenorhabditis elegans and control of the cell cycle in vertebrates (reviewed in Curtis et al., 1995; Wickens et al., 2000). Translational control of many mRNAs correlates with changes in the length of their poly(A) tails: increases in length are generally associated with activation (reviewed in Richter, 1996; Gray and Wickens, 1998). Polyadenylation is required for the activation of several mRNAs with key roles in early development, including vertebrate c-mos and Drosophila bicoid (reviewed in Richter, 1996; Gray and Wickens, 1998). Yet the mechanism by which poly(A) tail lengthening enhances translation is unclear.
Popular models addressing the mechanism by which poly(A) tails stimulate translation focus on connections between it and the 5′ m7GpppX cap (reviewed in Jacobson, 1996; Richter, 1996; Sachs et al., 1997; Gray and Wickens, 1998). In oocytes in particular, two models have been proposed that link polyadenylation and the cap. One posits that polyadenylation induces ribose modification of the cap structure (Kuge and Richter, 1995). Ribose methylation plays a role in the translational activation of c-mos mRNA (Kuge et al., 1998), but does not appear to be required for poly(A)-stimulated translation of other mRNAs (Gillian-Daniel et al., 1998). A second class of models, based mainly on studies in yeast extracts, suggests that a protein that binds to poly(A) tails in vivo, poly(A)-binding protein (PAB), is important.
Pab1p is essential in yeast and may have roles in translation, stability (reviewed in Jacobson, 1996) and 3′ end processing (Amrani et al., 1997; Minvielle Sebastia et al., 1997). In yeast cell-free extracts, a Pab1p–poly(A) complex binds to the translation factor eukaryotic initiation factor (eIF)-4G, which in turn interacts with eIF-4E bound to the cap (Tarun and Sachs, 1996). Such a complex can be formed using purified components, effectively circularizing the mRNA (Wells et al., 1998) via an end-to-end complex (reviewed in Jacobson, 1996; Sachs et al., 1997), and stimulates translation in vitro (Tarun and Sachs, 1995, 1996; Tarun et al., 1997; Wells et al., 1998). In this model, translation is enhanced by the PAB–poly(A) complex helping eIF-4E bring eIF-4G to the mRNA; in turn, eIF-4G recruits the 40S ribosomal subunit. In mammalian cells, interaction between PAB and PAB-interacting protein-1 (Paip-1), which shares homology with eIF-4G, is also proposed to recruit ribosomal subunits (Craig et al., 1998).
Despite much progress in understanding PAB function in vitro, its role in living cells is less clear. Work on transcription amply demonstrates that the qualitative and quantitative effects of specific factors can differ substantially in vitro and in vivo (e.g. Moqtaderi et al., 1996; Walker et al., 1996; Lee and Struhl, 1997), and emphasizes the need for both approaches. Indeed, differences have been described between the effects of poly(A) on translation in nuclease-treated yeast extracts and in yeast spheroplasts; these may be a consequence of the heightened competition for translation factors in vivo (Preiss and Hentze, 1998). In vivo, genetic analysis of PAB function is complicated by the fact that it has multiple biological functions, and that other protein–protein and protein–RNA interactions may stabilize an end-to-end complex. Nevertheless, several lines of evidence suggest that PAB is important for translation in vivo: for example, in yeast pab1 mutants are synthetically lethal with specific alleles of cdc33 (eIF-4E) (Tarun et al., 1997), and polysomes are redistributed to free subunits in the absence of Pab1p (Sachs and Davis, 1989). Similarly, some viruses apparently interfere with PAB function to shut off host protein synthesis: a rotavirus protein, NSP3A, binds to eIF-4G and apparently displaces PAB from eIF-4F (a complex of eIF-4E, eIF-4G and eIF-4A) (Piron et al., 1998), while picornavirus proteases cleave PAB (Joachims et al., 1999; Kerekatte et al., 1999).
Although changes in poly(A) length are important for translational regulation during early development, the role of PAB in these processes is largely unexamined. The quantity of PAB in Xenopus oocytes is insufficient to occupy the poly(A) tails of all endogenous mRNAs (Zelus et al., 1989). Thus, PAB may be a central regulator for which mRNAs compete; alternatively, poly(A)-mediated translation in oocytes may be PAB independent. Although the injection of excess poly(A) into Xenopus oocytes perturbs translation of polyadenylated mRNAs (Drummond et al., 1985), overexpression of PAB does not result in global translational activation in these cells (Wormington, 1996).
In this report, we tethered PAB to the 3′-untranslated region (3′-UTR) of reporter mRNAs (Coller et al., 1998) to assess directly PAB’s role in translation in oocytes and yeast. Our results in oocytes demonstrate that multiple domains of PAB stimulate translation in vivo in a manner that is independent of binding to poly(A). Interaction with eIF-4G may be important for PAB’s stimulatory activity, but other mechanisms also appear sufficient. In addition, our results in yeast suggest that PAB’s effects on translation and stability are distinct. Our findings imply that, in oocytes, PAB is either missing or silenced on repressed mRNAs.
Results
PAB can stimulate translation in Xenopus oocytes
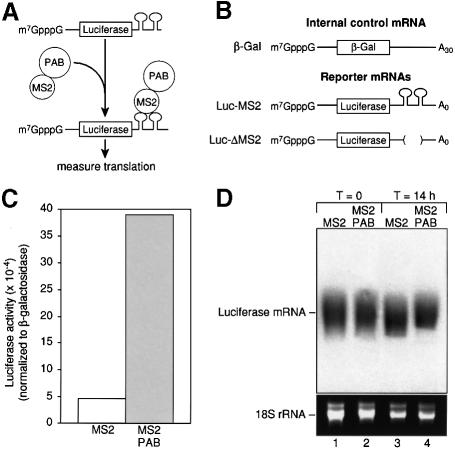
To determine whether PAB can stimulate translation in Xenopus laevis oocytes, we tethered PAB to the 3′-UTR of a reporter mRNA by adapting a ‘tethered function’ assay previously used to study mRNA stability in yeast (Coller et al., 1998; Figure 1A). This approach liberates studies of the role of PAB in translation from its other functions, and allows the presence of PAB on a specific mRNA to be modulated without the complications of PAB depletion in vivo. In vitro transcribed mRNAs encoding either MS2 or MS2 fused to PAB (MS2–PAB; Figure 1A) were injected into stage VI Xenopus oocytes. Oocytes were incubated for 6 h to allow protein production. Reporter mRNAs were then injected and the oocytes incubated overnight prior to harvesting. Two mRNAs were utilized and injected as a mixture (Figure 1B). Luc-MS2 mRNA encodes luciferase and contains MS2-binding sites; conversely, a control β-Gal mRNA encodes β-galactosidase but contains no MS2 sites (Figure 1B). Throughout this work, translational activity was quantified as luciferase activity normalized to β-galactosidase activity; variations in β-galactosidase levels were typically <10%, showing that the control mRNA was not responsive to the presence of the fusion proteins (see Figure 3 legend).
Fig. 1. Tethered PAB causes translational stimulation. (A) The assay has two components: a luciferase reporter mRNA with binding sites for the MS2 coat protein within its 3′-UTR (Luc-MS2), and a fusion between MS2 coat protein and Xenopus PAB. Binding of coat protein to its sites tethers PAB to the mRNA. The effects of fusion proteins are measured by luciferase assay. (B) A luciferase reporter mRNA with MS2-binding sites within its 3′-UTR (Luc-MS2) was utilized. mRNAs encoding β-galactosidase (β-Gal) or luciferase (Luc-ΔMS2) that lacked MS2-binding sites were used as control mRNAs. Luciferase mRNAs lack poly(A). (C) Oocytes expressing MS2 or MS2–PAB mRNA were co-injected with Luc-MS2 and β-Gal mRNAs as a mixture. The translation of the reporter mRNAs was determined by luciferase and β-galactosidase assays. Luciferase activity (normalized for differences in the amount of β-galactosidase activity) was plotted. A representative experiment is shown. (D) The stability of 32P-radiolabeled luciferase mRNA after incubation for 14 h in oocytes expressing either MS2 or MS2–PAB from (C) is shown (lanes 3 and 4). RNA was also harvested from oocytes immediately following injection of luciferase mRNA (lanes 1 and 2). 18S rRNA (lower panel) is shown as a loading control.
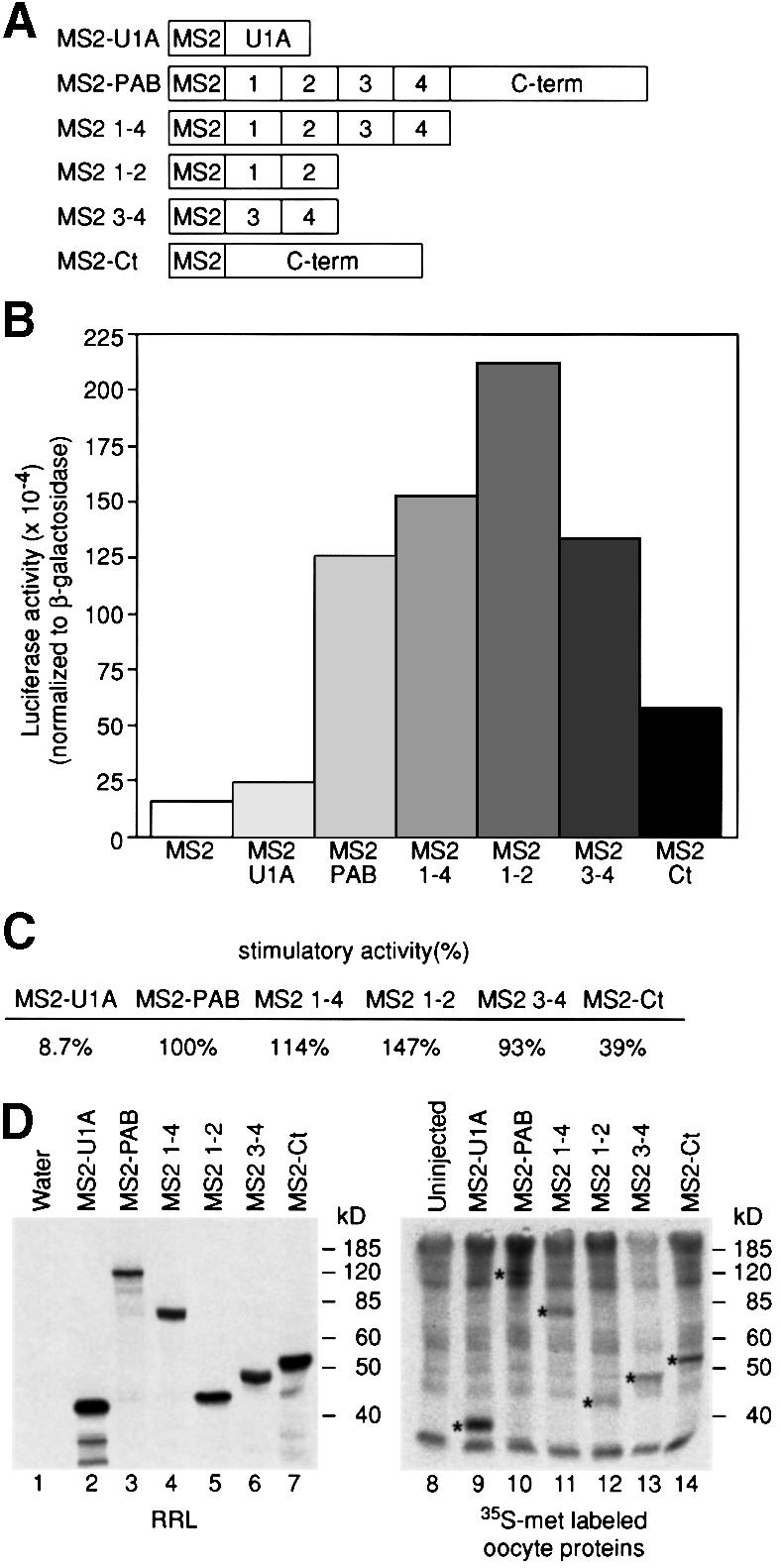
Fig. 3. Multiple regions of PAB stimulate translation. (A) Numbers 1–4 denote the RRM domains of PAB; Ct denotes the less conserved C-terminal region. (B) Oocytes expressing various MS2 fusions were injected with a mixture of β-Gal and Luc-MS2 mRNAs. Luciferase activities (corrected for differences in β-galactosidase activities) are plotted. A representative experiment is shown. Average β-galactosidase activity relative to oocytes expressing MS2 (set to 100%) was: MS2–U1A, 106%; MS2–PAB, 108%; MS2 1–4, 104%; MS2 1–2, 91%; MS2 3–4, 103%; MS2–Ct, 95%. (C) Data were derived from three experiments. The stimulatory activity of MS2–PAB protein, carrying full-length PAB compared with MS2, has been set to 100%. (D) mRNAs encoding fusion proteins were injected into oocytes (lanes 9–14) or translated in vitro (lanes 2–7). [35S]Methionine-labeled oocytes were subject to SDS–PAGE. Asterisks (lanes 9–14) mark proteins of the predicted size that are not present in uninjected oocytes (lane 8), and that co-migrate with in vitro translation products (lanes 2–7). The number of methionines in each protein is 21 in MS2–U1A, 34 in MS2–PAB, 21 in MS2 1–4, 11 in MS2 1–2, 16 in MS2 3–4 and 15 in MS2–Ct.
Expression of MS2–PAB stimulated luciferase activity from Luc-MS2 mRNA compared with MS2 protein alone (Figure 1C). The average stimulation observed was 7-fold; seldom being <4.5- or >10-fold. Reducing the amount of reporter mRNA by an order of magnitude did not increase stimulation by tethered PAB, suggesting that the ratio of reporter mRNA to PAB protein under standard conditions was optimal (data not shown). 32P-labeled Luc-MS2 mRNAs were equally stable in oocytes expressing MS2 or MS2–PAB over the course of the experiment (Figure 1D). This strongly suggests that the increase in luciferase activity was due to differences in translation and not reporter mRNA stability.
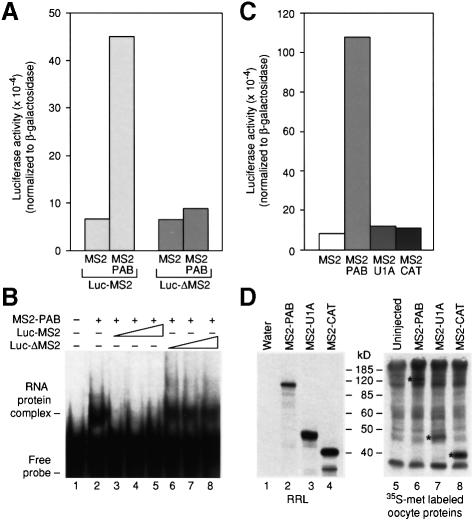
To determine whether stimulation by MS2–PAB occurred only in cis, we assayed the effects of MS2–PAB on Luc-ΔMS2 mRNA. This mRNA only differs from Luc-MS2 mRNA in that it lacks MS2-binding sites (Figure 1B). Translation of Luc-ΔMS2 was not stimulated by MS2–PAB (Figure 2A, compare bars 1 and 2 with 3 and 4). Gel mobility shift assays demonstrated that MS2–PAB interacted only with Luc-MS2 mRNA: unlabeled Luc-MS2 mRNA, but not Luc-ΔMS2 mRNA, competes for binding of MS2–PAB to a short radiolabeled RNA carrying MS2 sites (Figure 2B). The cis dependence of tethered PAB stimulation was corroborated by the lack of effect of MS2–PAB on the control, β-galactosidase mRNA (data not shown).
Fig. 2. Stimulation of luciferase translation is cis dependent and specific. (A) Oocytes expressing MS2 or MS2–PAB were co-injected with mixtures of β-Gal mRNA and either Luc-MS2 or Luc-ΔMS2 mRNA. (B) Increasing amounts of unlabeled luciferase reporter mRNAs with (Luc-MS2) or without (Luc-ΔMS2) MS2 sites were added to mixtures of MS2–PAB fusion protein and a [32P]RNA containing an MS2-binding site. Complex formation was visualized by non-denaturing gel electrophoresis. (C) MS2 fusions with PAB, CAT and U1A were expressed in oocytes prior to the injection of the Luc-MS2 and β-Gal reporter mRNAs. (D) mRNAs encoding fusion proteins were injected into oocytes (lanes 6–8) or translated in vitro (lanes 2–4). [35S]Methionine-labeled oocytes were analyzed by SDS–PAGE. Asterisks (lanes 6–8) mark proteins of the predicted size that are not present in uninjected oocytes (lane 5), and that co-migrate with in vitro translation products (lanes 2–4). The number of methionines in each protein is 34 in MS2–PAB, 14 in MS2–CAT and 21 in MS2–U1A. In (A) and (C), representative experiments are shown; luciferase activities (corrected for differences in β-galactosidase activities) are plotted.
To determine whether stimulation required the PAB portion of the fusion protein, two other polypeptides were fused to MS2 coat protein (Figure 2C). MS2–CAT is an MS2 fusion with bacterial chloramphenicol acetyltransferase (CAT), a protein with no known function in RNA metabolism. MS2–U1A is a fusion of MS2 with U1A, a spliceosomal RNA-binding protein of the RNA recognition motif (RRM) family (Jovine et al., 1996 and references therein). Injection of Luc-MS2 mRNA into oocytes expressing these fusion proteins revealed that only MS2–PAB stimulated luciferase translation (Figure 2C). All fusion proteins were expressed in oocytes (Figure 2D). [MS2 protein could not be visualized easily due to the low number of methionines in this small protein (data not shown).] This suggests that tethered PAB stimulates translation in a specific manner that is not achieved by other fusion proteins, including other RRM-containing proteins. We therefore conclude that PAB stimulates translation in a specific, cis-dependent manner in Xenopus oocytes.
Multiple domains of PAB can stimulate translation
PAB is composed of four non-equivalent RNA-binding motifs of the RRM family and a C-terminal domain (Sachs et al., 1986; Burd et al., 1991; Figure 3A). RRM 2 provides most of PAB’s ability to bind poly(A), while RRM 4 provides most of its ‘non-specific’ RNA-binding activity (Nietfeld et al., 1990; Burd et al., 1991; Kühn and Pieler, 1996; Deardorff and Sachs, 1997). The C-terminus of PAB is the least conserved, and does not appear to bind RNA (Kühn and Pieler, 1996): it may promote intermolecular interactions between PAB molecules (Kühn and Pieler, 1996) and can contribute to mRNA stabilization in yeast (Coller et al., 1998).
To examine the contributions of separate PAB domains to translation in vivo, we tethered regions of PAB and assayed translation (Figure 3A). A representative experiment is shown in Figure 3B, and the average of several experiments in Figure 3C. A fusion protein containing RRMs 1–4 (MS2 1–4) had full stimulatory activity, suggesting that the C-terminus is not required. Stimulation by RRMs 1–4 is unlikely to be mediated by recruitment of endogenous full-length PAB via dimerization (data not shown; Kühn and Pieler, 1996). RRMs 1 and 2 (MS2 1–2) were sufficient to stimulate translation and appeared to be more active than full-length PAB; the significance of this reproducible difference is unclear. Surprisingly, a fusion protein containing only RRMs 3 and 4 (MS2 3–4) also stimulated Luc-MS2 translation to a degree similar to full-length MS2–PAB. A fusion containing only the C-terminal part of PAB (MS2–Ct) showed a smaller but reproducible and specific capacity to stimulate translation (Figure 3B and C). MS2–U1A, included as an additional control for any non-specific stimulatory activity, did not stimulate translation significantly. All fusion proteins were expressed in oocytes (Figure 3D), and differences in expression levels do not explain differences in their activity. Similarly, increasing the protein to mRNA ratio by injecting 10-fold less reporter mRNA had no effect on the magnitude of stimulation (data not shown), suggesting that sufficient levels of fusion proteins were present for maximal stimulation.
We conclude that multiple regions of PAB (RRMs 1–2, and RRMs 3–4) stimulate translation efficiently. Interest ingly, the activities of RRMs 1 and 2 and of RRMs 3 and 4 appear to be redundant: the activity of RRMs 1–4 is not greater than that of RRMs 1 and 2 or RRMs 3 and 4. The C-terminus has lower, but significant, stimulatory activity.
Factors that interact with Xenopus PAB
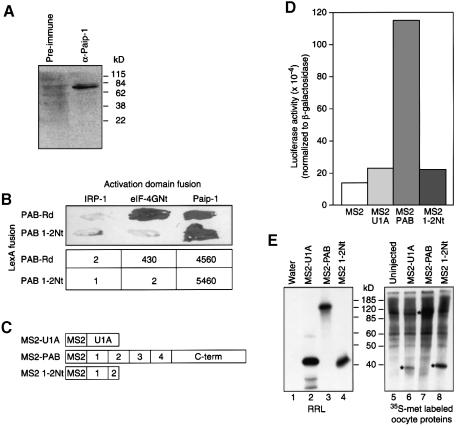
In other species (Saccharomyces cerevisiae, human and wheat), PAB interacts in vitro with eIF-4G, which forms part of a cap-binding complex with eIF-4E (Tarun and Sachs, 1996; Le et al., 1997; Imataka et al., 1998). In humans, Paip-1, which has significant homology to eIF-4G, also interacts with PAB (Craig et al., 1998). To understand further how Xenopus PAB stimulates translation in oocytes, we determined whether the regions of Xenopus PAB that elicit translational activation interact with eIF-4G or Paip-1. We performed two-hybrid tests using Xenopus PAB derivatives and the N-terminus of eIF-4G (eIF-4GNt) or full-length Paip-1 from humans; the Xenopus proteins are not yet cloned. Portions of PAB were presented as LexA fusions, and eIF-4G and Paip-1 as GAL4 activation domain fusions. PAB 1–2 (containing RRMs 1 and 2) interacted with both eIF-4G and Paip-1, but not with another RNA-binding protein (IRP1; Figure 4A). PAB’s interaction with Paip-1 appeared stronger than with eIF-4G; this is not due to higher levels of Paip-1 (data not shown). The C-terminus of PAB (PAB-Ct), which weakly stimulates translation in the tethered function assay, interacted only with Paip-1 (Figure 4A). PAB 3–4, which efficiently stimulates translation, interacted with neither factor (Figure 4A).
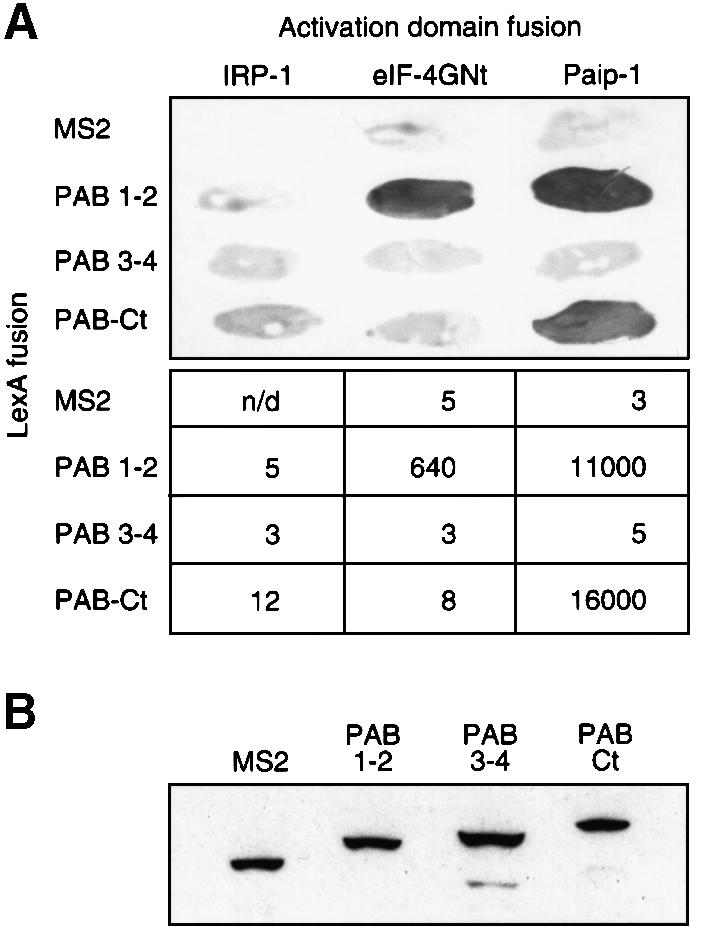
Fig. 4. Two-hybrid analyses with portions of PAB reveal interactions with eIF-4G and Paip-1. (A) Interactions of PAB 1–2 and PAB–Rd with eIF-4G and Paip-1 were tested by two-hybrid analysis. Top, qualitative filter assays. Bottom, specific activity of β-galactosidase, expressed as relative light units (RLU) × 10–3/µg protein. (B) Twelve-microgram aliquots of yeast extracts were analyzed by western blotting using an anti-LexA monoclonal antibody. Extracts were prepared from cells containing pACT-IRP and either LexA–MS2 (lane 1), LexA–PAB 1–2 (lane 2), LexA–PAB 3–4 (lane 3) or LexA–PAB-Ct (lane 4).
All LexA fusion proteins were expressed at similar levels in yeast, as determined by western blotting using an anti-LexA monoclonal antibody (Figure 4B). Thus, lack of interaction between specific factors is not due to a lack of the proteins in yeast. Moreover, PAB 3–4 is capable of interacting with other proteins in the two-hybrid system, suggesting that it is nuclear and active (data not shown). We conclude that multiple interactions, involving eIF-4G, Paip-1 and other, as yet unidentified factors may mediate translational stimulation by Xenopus PAB.
Interaction with eIF-4G but not Paip-1 correlates with translational stimulation in vivo
RRMs 1 and 2 of PAB stimulate translation in oocytes and interact with both eIF-4G and Paip-1. Both factors are present in oocytes: eIF-4G has been detected previously (Keiper and Rhoads, 1999), and an anti-Paip-1 antibody detects a protein of the predicted molecular weight in oocyte extracts (Figure 5A). Thus, interactions with either eIF-4G or Paip-1 might plausibly underlie the stimulatory effect of RRMs 1 and 2 in oocytes.
Fig. 5. Interaction with eIF-4G correlates with translational stimulation by RRMs 1 and 2. (A) Oocyte extracts were probed with either pre-immune or anti-Paip-1 antiserum. Size markers in kilodaltons are shown on the right-hand side. (B) PAB 1–2Nt was tested for its ability to interact with eIF-4G and Paip-1 by two-hybrid analysis. PAB–Rd (see Figure 6) and IRP are used as positive and negative controls, respectively. Top, qualitative filter assays. Bottom, the specific activity of β-galactosidase, expressed as RLU × 10–3/µg protein. (C) MS2 1–2Nt contains a C-terminal deletion in RRM2. (D) Oocytes expressing MS2, MS2–U1A, MS2–PAB or MS2 1–2Nt were co-injected with Luc-MS2 and β-Gal mRNAs. Luciferase activities (corrected for differences in β-galactosidase activities) are plotted. A representative experiment is shown. (E) mRNAs encoding fusion proteins were injected into oocytes (lanes 6–8) or translated in vitro (lanes 2–4). [35S]Methionine-labeled oocytes were analyzed by SDS–PAGE. Asterisks (lanes 6–8) mark proteins of the predicted size that are not present in uninjected oocytes (lane 5), and that co-migrate with in vitro translation products (lanes 2–4). The number of methionines in each protein is 34 in MS2–PAB, 21 in MS2–U1A and 9 in MS2 1–2Nt.
To distinguish which interaction was important in vivo, we attempted to identify a mutant form of PAB that interacted with only one of the proteins. We constructed a C-terminal deletion of RRMs 1 and 2, in which the last 38 amino acids of RRM2 were removed (PAB 1–2Nt); this region of yeast PAB is required for the binding to eIF-4G (Otero et al., 1999). Two-hybrid analysis with PAB 1–2Nt revealed that it did not bind to eIF-4G, but did bind to Paip-1 (Figure 5B). A variant containing RRMs 1 and 2 in their entirety interacted with eIF-4G, as expected (Figure 5B).
To determine whether the deleted form of PAB stimulated translation, an MS2 fusion containing this region (MS2 1–2Nt; Figure 5C) was expressed in oocytes. MS2 1–2Nt did not stimulate Luc-MS2 translation (Figure 5D), although the protein was expressed efficiently (Figure 5E). Thus, deletion of the region of RRMs 1 and 2 that is required for binding to eIF-4G abolishes translational stimulation. These data provide evidence that interaction with eIF-4G is required for translational stimulation, and that interaction with Paip-1 is insufficient.
Translational activation requires neither poly(A) nor full RNA-binding activity
In yeast cell-free systems, a PAB–poly(A) tail complex has been suggested to be essential for PAB’s ability to interact with eIF-4G and stimulate translation (Tarun and Sachs, 1996; Tarun et al., 1997). Our data suggest that this is not the case in Xenopus oocytes: the reporter mRNAs in Figures 1–5 lack poly(A) yet were stimulated by tethered PAB. Similarly, translation was stimulated efficiently by RRMs 3 and 4 (Figure 3), despite significantly reduced ability to bind poly(A) (Burd et al., 1991; Kühn and Pieler, 1996; Deardorff and Sachs, 1997), and by RRMs 1 and 2, although Xenopus PAB lacking RRM4 and the C-terminus does not interact stably with poly(A) in oocytes (Wormington et al., 1996).
To test more decisively whether stimulation by tethered PAB required poly(A) binding, we introduced point mutations into the RNP1 sequences of PAB in MS2 1–2 (Figure 6A) previously shown to diminish poly(A) binding 100-fold in vitro (Deardorff and Sachs, 1997 and references therein). Stimulation by this mutant form of PAB (MS2–Rd) is comparable to that by full-length MS2–PAB, though reduced slightly relative to MS2 1–2 (Figure 6B and C). This result argues that tethered PAB does not contact poly(A) in trans on mRNAs other than the reporter, and that poly(A) binding is dispensible for stimulation.
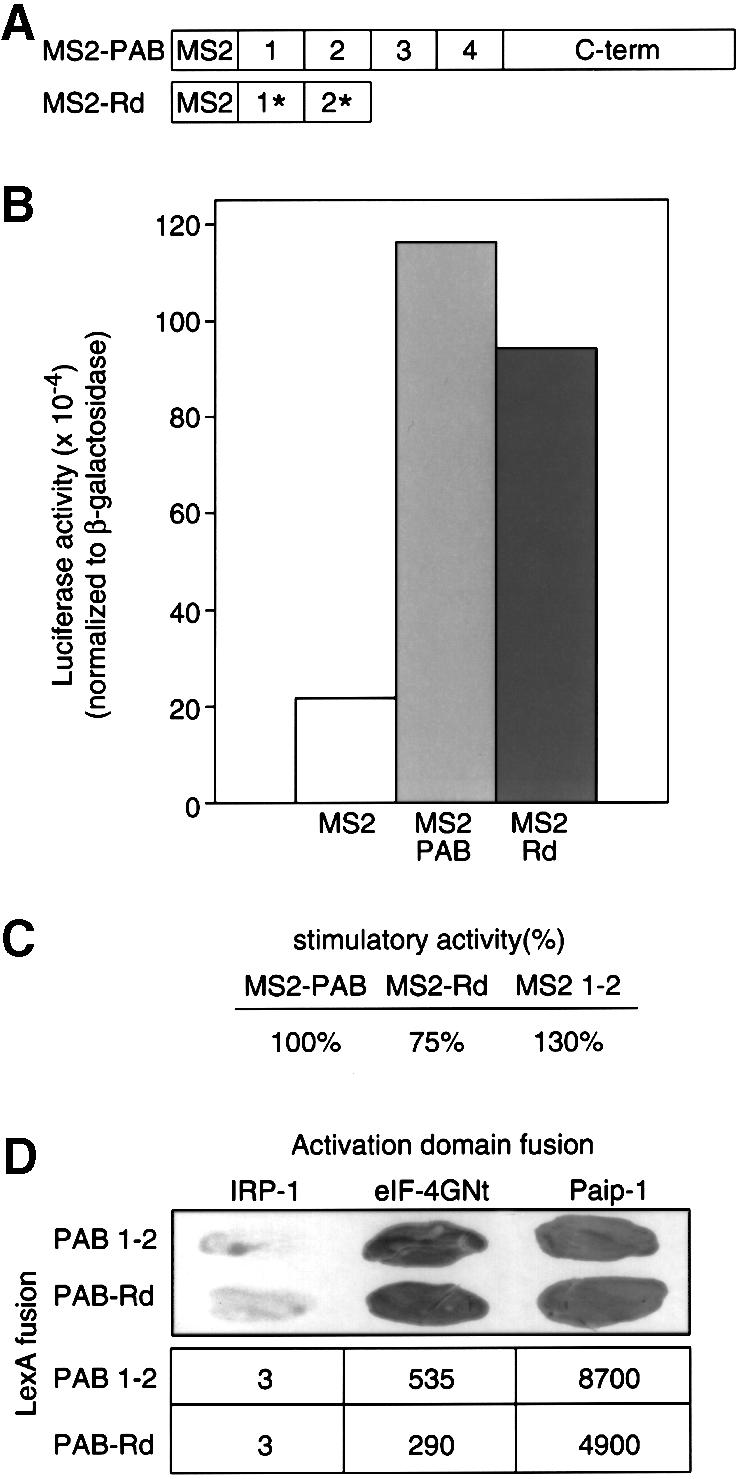
Fig. 6. A mutant form of PAB that does not bind poly(A) efficiently in vitro can still stimulate translation. (A) Asterisks indicate substitutions of aromatic residues within the RRMs. (B) Oocytes expressing MS2, MS2–PAB or MS2–Rd were co-injected with a mixture of Luc-MS2 and β-Gal mRNAs. Luciferase activities (corrected for differences in β-galactosidase activities) were plotted. A representative experiment is shown. (C) Data were derived from three experiments. The stimulatory activity of MS2–PAB protein, carrying full-length PAB compared with MS2, has been set to 100%. (D) Interactions of PAB 1–2 and PAB–Rd with eIF-4G and Paip-1 were tested by two-hybrid analysis. Top, qualitative filter assays. Bottom, specific activity of β-galactosidase, expressed as RLU × 10–3/µg protein.
Two-hybrid analysis revealed that the RNA-binding-defective form of RRMs 1 and 2 still interacted with both eIF-4G and Paip-1, albeit with a slightly reduced affinity (Figure 6D). This result argues that the interaction between PAB and these factors in the two-hybrid system is via protein–protein interactions and not through an RNA bridge. Taken together, our data strongly suggest that translational stimulation is an intrinsic property of the Xenopus PAB protein, and that the role of poly(A) in PAB-mediated stimulation in oocytes is merely to recruit PAB.
Translational stimulation by yeast PAB in oocytes
Since translational stimulation by Xenopus PAB in oocytes did not exhibit the same requirement for poly(A) as does yeast PAB in vitro, we directly compared the effects of tethered yeast and Xenopus PAB in oocytes, using a reporter mRNA lacking poly(A). Surprisingly, tethered yeast PAB (MS2–yPAB) stimulated the translation of the non-adenylated mRNA even more efficiently than did the tethered Xenopus PAB (Figure 7). This suggests that differences between yeast and Xenopus PAB per se do not account for the differences in poly(A) dependence between Xenopus oocytes and yeast in vitro extracts. The higher activity of the yeast protein may reflect regulation of Xenopus PAB’s activity in oocytes (see Discussion).
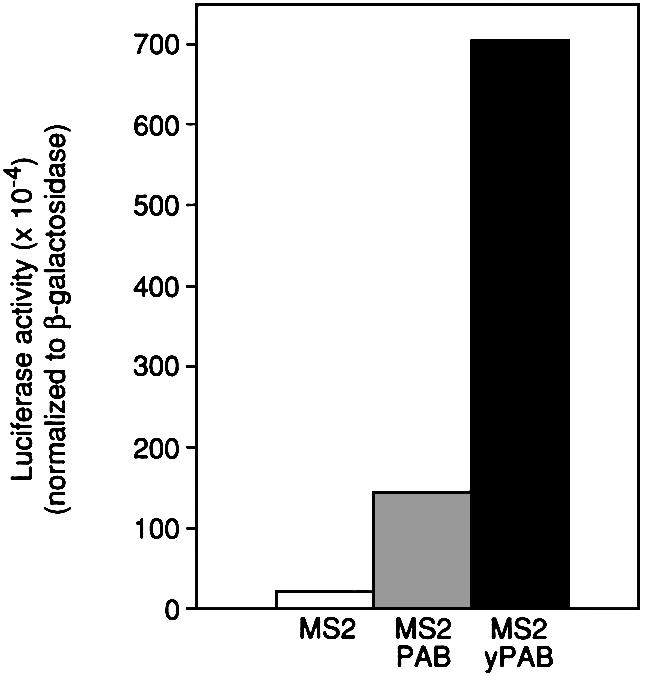
Fig. 7. Tethered yeast PAB stimulates translation in Xenopus oocytes. Oocytes expressing MS2, MS2–PAB or MS2–yPAB were co-injected with a mixture of Luc-MS2 and β-Gal mRNAs. Luciferase activities (corrected for differences in β-galactosidase activities) were plotted. A representative experiment is shown.
Translational stimulation by PAB in yeast
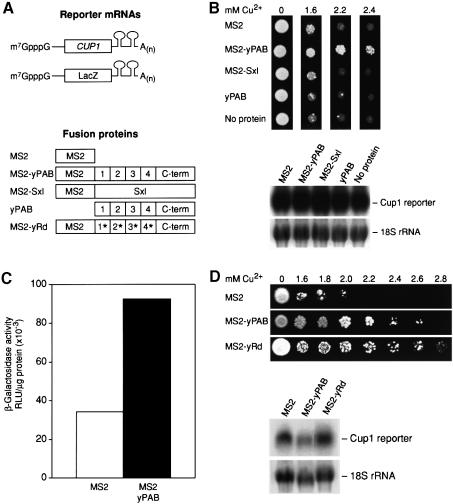
To examine the role of PAB in yeast, a phenotypic assay to measure PAB’s activity in intact yeast cells was devised. We used a relatively stable adenylated reporter mRNA to eliminate complications due to turnover (Coller et al., 1998). MS2 sites were inserted into the 3′-UTR of a reporter encoding the yeast copper metallothionein protein (CUP1) open reading frame (Figure 8A). The ability of yeast to survive in the presence of copper is proportional to Cup1p protein levels (Stutz and Rosbash, 1994). The CUP1/MS2 gene, carrying the MS2 sites, was functional in that it complemented a chromosomal disruption of the CUP1 gene (data not shown).
Fig. 8. PAB can stimulate translation in yeast in a poly(A)-independent manner. (A) Diagrams of polyadenylated reporter mRNAs with MS2 binding sites and either a CUP1 or lacZ open reading frame. ‘Sxl’ indicates Sex-lethal protein, a Drosophila protein, with multiple RRM domains. yPAB denotes yeast PAB, and yRD a poly(A)-binding-defective version of yeast PAB. (B) Upper panel: copper resistance assays of CUP1 levels using yeast transformed with several of the proteins depicted in (A). Lower panel: northern analysis of CUP1 reporter mRNAs from the assays above. 18S rRNA is used as a loading control. (C) β-galactosidase activity of yeast expressing either MS2 or MS2–yPAB and a lacZ reporter mRNA with MS2 sites. (D) Upper panel: copper resistance assays in yeast expressing MS2, MS2–yPAB or MS2–yRd. Lower panel: northern analysis of CUP1 reporter mRNAs. 18S rRNA is used as a loading control.
Expression of MS2–yPAB significantly increased survival on high levels of copper, allowing growth on concentrations up to 2.4 mM (Figure 8B, upper panel). These effects were specific, as neither MS2 alone, yPAB alone nor an MS2–Sex-lethal (MS2–Sxl) fusion (Crowder et al., 1999) enhanced cell growth at high copper concentrations (Figure 8B, upper panel). The steady-state mRNA level of the CUP1/MS2 reporter was not significantly affected by any of the fusion proteins (Figure 8B, lower panel), suggesting that the increased resistance to copper by MS2–yPAB is attributable to translation and not alterations of mRNA levels. Effects on mRNA transport cannot be formally eliminated, but polyadenylated reporter mRNAs were utilized to prevent nuclear retention.
To quantitate the translational effects of MS2–yPAB, we replaced CUP1 with LacZ (Figure 8A). β-galactosidase activity was elevated 2.7-fold by MS2–yPAB relative to cells expressing MS2 alone (Figure 8C). The magnitude of stimulation is consistent with the modest level of increase in copper resistance (Stutz and Rosbash, 1994), but considerably less than observed with MS2–yPAB in oocytes (Figure 7). This may be due to the presence of poly(A) on the reporter mRNAs in yeast cells; the tails could bind endogenous PAB and minimize MS2–yPAB’s effects. In support of this idea, polyadenylated reporter mRNAs in oocytes were stimulated only 2.4-fold by MS2–yPAB (data not shown).
To examine the role of poly(A) binding in yeast, we constructed a mutant form of MS2–yPAB (MS2–yRd), which contains point mutations in each of its RRMs that collectively depress poly(A) binding 300-fold (Deardorff and Sachs, 1997). Importantly, MS2–yRd stimulated CUP1/MS2 expression as well as MS2–yPAB (Figure 8D, upper panel), while not affecting steady-state mRNA levels (Figure 8D, lower panel).
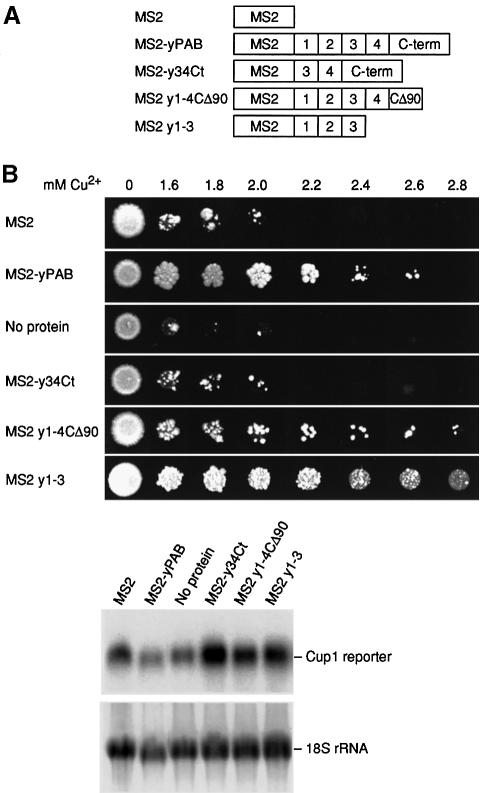
To identify specific regions of MS2–yPAB that are required for translational stimulation in yeast, we analyzed a series of PAB deletions fused to MS2 coat protein (Figure 9A). Removal of the last 90 amino acids of yeast PAB (MS2 y1–4CΔ90) prevented the ability of PAB to stabilize otherwise unstable reporter mRNAs (data not shown) but not its ability to promote translation (Figure 9B). A larger deletion revealed that RRMs 1–3 are sufficient to support growth on high copper concentrations (MS2 y1–3; Figure 9B). Interestingly, in contrast to Xenopus PAB, RRMs 3 and 4 and the C-terminus of yeast PAB do not promote translation (Figure 9B). This fusion protein is active, since it can stabilize mRNAs (Coller et al., 1998). These results suggest that RRMs 1–3 are sufficient for translational stimulation in yeast and are consistent with the observation that RRM2 is critical for the in vitro association of Pab1p with eIF-4G (Kessler and Sachs, 1998).
Fig. 9. RRMs 1–3 of yeast PAB are sufficient to stimulate translation. (A) MS2 y1–4CΔ90 indicates an MS2 fusion with yeast PAB that carries all but the last 90 amino acids of yeast PAB. MS2 y1–3 contains RRMs 1–3, and MS2–y34Ct contains RRMs 3 and 4 and the C-terminus of yeast PAB. (B) Upper panel: copper resistance assays with tethered yeast PAB, using strains carrying the forms of PAB indicated in (A). Lower panel: northern analysis of CUP1 reporter mRNAs. 18S rRNA is used as a loading control.
Discussion
An important and central conclusion of our work is that PAB stimulates translation of reporter mRNAs in vivo in otherwise unperturbed cells, extending previous studies. In addition, our work leads to the following main conclusions. (i) The stimulatory activity is an intrinsic property of both Xenopus and yeast PAB, as it requires neither PAB’s RNA-binding activity nor a poly(A) tail. However, PAB must be bound to the mRNA as the effect is observed only in cis. (ii) Multiple regions of PAB possess stimulatory activity in Xenopus oocytes. (iii) Analysis of PAB mutants suggests that an interaction of RRMs 1 and 2 with eIF-4G is important for stimulation, but that binding to Paip-1 is insufficient. (iv) Interactions between RRMs 3 and 4 and novel factors may also stimulate. (v) Tethered yeast PAB stimulates translation substantially more in Xenopus oocytes than does Xenopus PAB. (vi) In intact yeast, the regions required for stability and translation functions differ.
PAB stimulates translation in oocytes
The effect of poly(A) on translation in oocytes and embryos is pronounced, yet the small amount of PAB present in these cells appears to be insufficient to occupy all available binding sites on endogenous mRNAs (Zelus et al., 1989). Our results demonstrate that oocytes contain, in an active state, the machinery necessary to respond to PAB. Thus the stimulatory effects of poly(A) in oocytes may involve the recruitment of endogenous PAB. Consistent with this view, the magnitude of translational stimulation in oocytes by poly(A) (10-fold at the mRNA concentrations used here) is comparable to that of tethered PAB (7-fold).
Repressed mRNAs in oocytes carry short but significant poly(A) tails, sufficiently long to bind PAB. Our observation that tethered PAB stimulates translation suggests that, in the absence of other influences, PAB bound to those tails would enhance translation. Overexpression of PAB in oocytes does not stimulate translation of endogenous mRNAs, though it does prevent deadenylation during maturation (Wormington et al., 1996). It is possible that a threshold number of PAB molecules must be bound to activate translation, and that number is not reached on repressed mRNAs, even with excess PAB in trans. At the extreme, PAB may be occluded from repressed mRNAs. Alternatively, PAB’s stimulatory activity may be prevented by mRNA-bound repressors. However, at least some mRNAs can be derepressed without a change in poly(A) length (reviewed in Gray and Wickens, 1998; Wickens, 2000).
Tethered PAB stimulates translation of reporter mRNAs only in cis (Figures 2 and 8). Although trans effects of PAB were observed in yeast extracts (Otero et al., 1999), differences in experimental design make it difficult to compare results directly; similar interactions may underlie the two phenomena.
Our results suggest that translational stimulation is an intrinsic property of Xenopus and yeast PAB, and are consistent with in vitro studies of mammalian PAB (Imataka et al., 1998). Although it is formally possible that PAB mutants that are defective in RNA binding still interact with the mRNAs to which they are tethered, our results show that a specific PAB–poly(A) complex is not required for stimulation. Trans stimulation in vitro by PAB mutants with reduced RNA-binding activity has been reported (Otero et al., 1999). Our results suggest that the role of poly(A) in PAB-mediated stimulation is merely to form a scaffold for PAB binding; our data do not exclude the possibility that the complex is required for other aspects of mRNA function, or that other poly(A)-binding proteins contribute to poly(A)’s translational effects.
In Xenopus oocytes, translation is stimulated by PAB from yeast more than by that from Xenopus (Figure 7). The activity of Xenopus PAB may be down-regulated in oocytes by proteins that do not recognize yeast PAB. Alternatively, full activity of Xenopus but not yeast PAB could require modifications of one or more initiation factors, as occurs during maturation (Pain, 1996).
Multiple regions of PAB stimulate translation
In oocytes, RRMs 1 and 2 and RRMs 3 and 4 act redundantly, not additively, to stimulate translation: each portion stimulates to an extent similar to the full-length protein. In contrast, in yeast, RRMs 34Ct lacked detectable activity. Thus, Xenopus and yeast PAB may differ in the precise manner in which they facilitate translation, or in the balance between different protein–protein interactions. Trans activation is not detected in vivo (Figures 2 and 8), but in vitro appears to require RRM 4 of yeast PAB (Otero et al., 1999), perhaps mimicking the effects of tethered Xenopus RRMs 3 and 4. The C-terminal portion of PAB modestly stimulates translation in oocytes (Figure 3) and interacts with Paip-1 (Figure 4); however, the interpretation of this result is complicated because the C-terminus may recruit endogenous PAB (Kühn and Pieler, 1996; data not shown).
Factors involved in translational stimulation
The ability of multiple domains of PAB to stimulate translation redundantly raises the question of how many interactions a single PAB molecule directs. RRMs 1 and 2 of Xenopus PAB presumably contact at least two proteins, eIF-4G and Paip-1 (Figure 4), and other factors probably contact RRMs 3 and 4. It is unclear whether these different contacts co-exist in a single PAB molecule.
eIF-4G interacts in vitro with PAB from several species (Tarun and Sachs, 1996; Le et al., 1997; Tarun et al., 1997; Imataka et al., 1998); our data extend this observation to Xenopus, and are consistent with the interaction between the Xenopus eIF-4F and PAB (Fraser et al., 1999; Keiper and Rhoads, 1999). In each species, including Xenopus, the binding site for eIF-4G lies in RRMs 1 and 2. The Paip-1 interaction site has not been identified in other species. We find two Paip-1 sites in Xenopus PAB: one in RRMs 1 and 2 and one in the C-terminus.
Our data suggest that interaction with eIF-4G, and not Paip-1, is critical for PAB-mediated stimulation in oocytes. The key observation is that removal of a portion of RRM2 does not affect interaction with Paip-1, but eliminates both translational stimulation and binding to eIF-4G. This provides strong in vivo support for the model, based on in vitro studies, that eIF-4G is important for PAB-mediated stimulation. Paip-1 interacts with RRM 1–2Nt, and with the C-terminus of PAB, neither of which significantly stimulates translation in oocytes. Thus, although a putative Paip-1 protein can be detected immunologically in oocytes, our data provide no evidence of its having a major role in PAB-stimulated translation at this developmental stage.
Our data with RRMs 3 and 4 raise the notion that interactions other than with eIF-4G or Paip-1 may also be sufficient for PAB-mediated stimulation (Figure 3). These may be novel factors, or known components of the translation machinery. The translation of capped, polyadenylated mRNAs in yeast is relatively resistant to perturbations in the eIF-4G–PAB interaction, and RRM 4 may be required for trans stimulation (Tarun et al., 1997; Kessler and Sachs, 1998; Otero et al., 1999). Taken together, these findings suggest that other factors may also play a role in yeast. However, in contrast to Xenopus PAB, RRMs 3 and 4 of yeast PAB are not sufficient to stimulate in yeast. The quantitative effect of different interactions may vary during development: for instance, the impact on translation of cleaving eIF-4G with viral protease changes during oocyte maturation (Keiper and Rhoads, 1999).
The translation and stability functions of yeast PAB differ
In principle, the translation and turnover functions of yeast PAB might be manifestations of the same underlying event: formation of an end-to-end complex via eIF-4G, for example. Tethered yeast RRMs 1–3 stimulate translation (Figure 9), but do not stabilize (Coller et al., 1998). Tethered yeast PAB lacking the last 90 amino acids behaves comparably. In addition, although two Arabidopsis PAB isoforms can provide PAB’s translation function in yeast, only the form with extensive homology to the C-terminal portion of yeast PAB fully couples deadenylation and decapping (Belostotsky and Meagher, 1996; R.Palaniverlu and R.Meagher, personal communication). The C-terminal region of PAB promotes PAB oligomerization (Kühn and Pieler, 1996; Mangus and Jacobson, 1998), which may be required for stabilization. While our results separate the translation and stabilization functions of tethered PAB, it is also true that translation and turnover are intimately linked (e.g. Schwartz and Parker, 1999); for example, the ability of tethered PAB to stabilize requires translation of the mRNA (Coller et al., 1998).
Lengthening of poly(A) in oocytes and embryos is commonly associated with translational activation, and removal of the tail with repression. Our results demonstrate that PAB can stimulate translation in the oocyte, and that all the requisite machinery is present and active. Thus, it appears that the function of poly(A) tail lengthening during development may involve recruitment of PAB to the mRNA, or relief of repression of PAB activity. Dissecting the interplay of PAB with 3′-UTR-bound repressors and the translational machinery is a clear challenge.
Materials and methods
Plasmids
Plasmids are summarized in Table I. Details of their construction can be found in our supplementary Materials and methods (the supplementary data are available at The EMBO Journal Online).
Table I. Plasmids used.
| Plasmid | Use | Comment |
|---|---|---|
| MSP | tether protein | MS2 coat protein ORF |
| MS2–PAB | fusion protein (Xenopus) | PAB amino acids 3 (Pro)–633 (Ala) |
| MS2 1–4 | fusion protein (Xenopus) | PAB amino acids 3 (Pro)–390 (Ile) |
| MS2–Ct | fusion protein (Xenopus) | PAB amino acids 395 (Val)–633 (Ala) |
| MS2 1–2 | fusion protein (Xenopus) | PAB amino acids 3 (Pro)–182 (Glu) |
| MS2 3–4 | fusion protein (Xenopus) | PAB amino acids 182 (Glu)–396 (Ile) |
| MS2 1–2Nt | fusion protein (Xenopus) | PAB amino acids 3 (Pro)–137 (Ser) |
| MS2–Rd | fusion protein (Xenopus) | PAB amino acids 3 (Pro)–182 (Glu)* |
| MS2–yPAB | fusion protein (Xenopus) | yeast PAB ORF |
| MS2–CAT | fusion protein (Xenopus) | CAT ORF |
| MS2–U1A | fusion protein (Xenopus) | U1A ORF |
| LexA–MS2 | two-hybrid | MS2 coat protein ORF |
| PAB 1–2 | two-hybrid | PAB amino acids 1 (Met)–182 (Glu) |
| PAB 3–4 | two-hybrid | PAB amino acids 182 (Glu)–396 (Ile) |
| PAB–Ct | two-hybrid | PAB amino acids 396 (Ile)–633 (Ala) |
| PAB 1–2Nt | two-hybrid | PAB amino acids 3 (Pro)–137 (Ser) |
| PAB–Rd | two-hybrid | PAB amino acids 3 (Pro)–182 (Glu)* |
| pACT-IRP | two-hybrid | Iron Regulatory Protein ORF |
| pACT-4GNt | two-hybrid | eIF-4GI amino acids 1 (Met)–641 (Arg) |
| pACT-Paip | two-hybrid | Paip-1 ORF |
| pJK350 | reporter | β-galactosidase ORF, no MS2 sites |
| pLGENB1 | reporter | luciferase ORF, no MS2 sites |
| pLGMS2 | reporter | luciferase ORF, contains MS2 sites |
| Cup1/MS2 | reporter (yeast) | CUP1 ORF, contains MS2 sites |
| LacZ/MS2 | reporter (yeast) | CUP1 ORF, contains MS2 sites |
| MS2 | tether protein (yeast) | MS2 coat protein ORF |
| PAB1p | non-tethered yeast PAB (yeast) | yeast PAB ORF |
| MS2–yPAB | fusion protein (yeast) | yeast PAB amino acids 2 (Ala)–576 (Ala) |
| MS2–Sxl | fusion protein (yeast) | Sex-lethal ORF |
| MS2–yRd | fusion protein (yeast) | yeast PAB amino acids 16 (Ile)–576 (Ala)* |
| MS2 y1–4CΔ90 | fusion protein (yeast) | yeast PAB amino acids 2 (Ala)–486 (Gln) |
| MS2 y1–3 | fusion protein (yeast) | yeast PAB amino acids 2 (Ala)–286 (Glu) |
| MS2–y34Ct | fusion protein (yeast) | yeast PAB amino acids 180 (Lys)–576 (Ala) |
ORF, open reading frame.
An asterisk denotes changes in aromatic residues in the RRMs that reduce poly(A)–RNA binding: in MS2–Rd and PAB–Rd, these are Y56V and F142V; in MS2–yPAB, these are Y83V, F170V, F263V and F366V.
(yeast) identifies plasmids used in tethered function assays in S.cerevisiae.
In vitro transcription and translation
Plasmids were digested with BglII (luciferase reporters) or HindIII (β-Gal reporter and fusion proteins). T7 and SP6 transcription were performed as described (Gray et al., 1993) except that SP6 enzyme was purchased from Promega (WI, USA) and 50 mM exogenous dithiothreitol (DTT) was added. 32P-labeled RNAs were prepared as described (Gray and Hentze, 1994). In vitro translation reactions (25 µl) in rabbit reticulocyte lysate were programmed with 1 µg of mRNA and were performed and analyzed as described (Gray et al., 1993).
Electrophoretic mobility shift assays
N-terminal His6-tagged proteins were purified by nickel NTA–agarose chromatography as described (Gray et al., 1993) except that overnight cultures were diluted to OD 0.4 and grown for 1 h with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 25°C prior to lysis. Protein preparations were pre-treated on ice for 5 min with 1 mM acetic acid and 1 mM DTT. Binding was performed on ice in 200 mM Tris pH 8.5, 160 mM KCl, 20 mM magnesium acetate and 160 µg/ml bovine serum albumin (BSA) for 1 h. Unlabeled competitor RNAs were added prior to 32P-labeled probes. Heparin (final concentration 5 mg/ml) was added 1 h after the addition of 32P-labeled probes. RNA–protein complex formation was analyzed by non-denaturing gel electrophoresis.
Oocyte injections and in vivo labeling of oocytes
Oocyte micromanipulation and microinjection were performed as described (Gillian-Daniel et al., 1998). Fifty nanoliters of a 1 µg/µl solution of mRNAs encoding fusion proteins was injected 6 h prior to injection of 50 nl of a solution containing 24 fmol of luciferase reporter mRNAs and 12 fmol of β-Gal control mRNA. Incubation was continued overnight before harvesting.
Isotopic labeling was achieved by incubating oocytes in MMR (100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM HEPES pH 7.4, 1 mg/ml penicillin and streptomycin) (Gillian-Daniel et al., 1998) containing 100 µCi/ml [35S]methionine for 6 h. Oocytes were washed and homogenized in 10 µl/cell 10 mM Tris–HCl, 1 mM EDTA containing a protease inhibitor cocktail (Boehringer Mannheim, Germany). Homogenates were centrifuged for 10 min and supernatant collected and subjected to SDS–PAGE.
Luciferase assays and RNA isolation from oocytes
A minimum of three pools of five oocytes was collected and assayed per experimental point. Oocytes were homogenized in lysis buffer (40 µl/oocyte; Tropix, MA). A 5 µl aliquot was assayed for luciferase activity using luciferase assay reagent (Promega, WI) and 2.5 µl for galactosidase activity using Galacto-light Plus (Tropix). Relative light determinations were measured in a Monolight 2010 (PharMingen, CA). Luciferase activities were adjusted for variations in β-galactosidase activities among pools. 32P-labeled luciferase RNAs were recovered and analyzed as described (Gillian-Daniel et al., 1998).
Western analysis
Cells were pelleted from 50 ml cultures, washed in S buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3 pH 7.6), repelleted and resuspended in 300 µl of RIPA buffer [150 mM NaCl, 1% (v/v) NP-40, 0.5% (w/v) deoxycholate, 0.1% (w/v) SDS, 0.05 mM Tris–HCl pH 8.0] containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and yeast protease inhibitors (Sigma). Cells were disrupted by two, 1 min pulses in a bead beater using 200 µl of glass beads. A 12 µg aliquot of cleared lysate was subjected to western analysis using a LexA monoclonal antibody (Clontech) according to the manufacturer’s instructions, with the exception that the horse serum blocking step was omitted. Xenopus extracts were prepared and analyzed as described (Dickson et al., 1999) using a 1:1000 dilution of anti-Paip-1 or pre-immune serum (gift of Nahum Sonenberg).
Yeast methods
Two-hybrid assays and β-galactosidase assays were performed as described (Zhang et al., 1999). Copper sensitivity assays were performed by transforming yRP1209 (MATa ura3, trp1, his3, leu2, cup1::URA3; gift from Roy Parker) with appropriate plasmids on minimal media. Transformants were grown overnight in nutrient-rich media supplemented with 2% galactose/2% raffinose. Cells were washed twice in water and plated on synthetic media supplemented with 2% galactose/2% raffinose, at varying concentrations of CuSO4. Growth was scored after 5 days of incubation at 30°C. RNA purification and northern analysis from yeast were performed as described (Coller et al., 1998).
Supplementary data
Supplementary data to this paper (Materials and methods) are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Diane Lawson and Nicole Benkers for invaluable technical assistance, the Wickens laboratory for their expertise, and Drs R.Meagher, R.Parker and N.Sonenberg for discussing results prior to publication. We thank Drs M.Hentze, R.Moon, R.Parker, A.Sachs, D.Schoenberg and N.Sonenberg for reagents and strains. Work in the Wickens laboratory is supported by the NIH (GM31892). N.K.G. is supported by a Wellcome Trust Prize International Travelling Fellowship.
References
- Amrani N., Minet,M., Le Gouar,M., Lacroute,F. and Wyers,F. (1997) Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro.Mol. Cell. Biol., 17, 3694–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belostotsky D.A. and Meagher,R.B. (1996) A pollen-, ovule- and early embryo-specific poly(A) binding protein from Arabidopsis com plements essential functions in yeast. Plant Cell, 8, 1261–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C.G., Matunis,E.L. and Dreyfuss,G. (1991) The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell. Biol., 11, 3419–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J.M., Gray,N.K. and Wickens,M.P. (1998) mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev., 12, 3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig A.W.B., Haghighat,A., Yu,A.T.K. and Sonenberg,N. (1998) Interaction of polyadenylate-binding protein with the eIF-4G homologue PAIP enhances translation. Nature, 392, 520–523. [DOI] [PubMed] [Google Scholar]
- Crowder S.M., Kanaar,R., Rio,D.C. and Alber,T. (1999) Absence of interdomain contacts in the crystal structure of the RNA recognition motifs of Sex-lethal. Proc. Natl Acad. Sci. USA, 96, 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis D., Lehmann,R. and Zamore,P.D. (1995) Translational regulation in development. Cell, 81, 171–178. [DOI] [PubMed] [Google Scholar]
- Deardorff J.A. and Sachs,A.B. (1997) Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J. Mol. Biol., 269, 67–81. [DOI] [PubMed] [Google Scholar]
- Dickson K.S., Bilger,A., Ballantyne,S. and Wickens,M.P. (1999) The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol. Cell. Biol., 19, 5707–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond D.R., Armstrong,J. and Colman,A. (1985) The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res., 13, 7375–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C.S., Pain,V.M. and Morley,S. (1999) The association of initiation factor 4F with poly(A) binding protein is enhanced in serum-stimulated Xenopus kidney cells. J. Biol. Chem., 274, 196–204. [DOI] [PubMed] [Google Scholar]
- Gillian-Daniel D.L., Gray,N.K., Åstrom,J., Barkoff,A.F. and Wickens,M. (1998) Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol. Cell. Biol., 18, 6152–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.K. and Hentze,M.W. (1994) Iron regulatory protein prevents binding of the 43S translation pre-initiation complex to ferritin and eALAS mRNAs. EMBO J., 13, 3882–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N.K. and Wickens,M.P. (1998) Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol., 14, 399–457. [DOI] [PubMed] [Google Scholar]
- Gray N.K., Quick,S., Goossen,B., Constable,A., Hirling,H., Kühn,L. and Hentze,M.W. (1993) Recombinant iron regulatory factor as an iron-responsive-element-binding protein, a translational repressor and an aconitase. A functional assay for translational repression and direct demonstration of the iron switch. Eur. J. Biochem., 218, 657–667. [DOI] [PubMed] [Google Scholar]
- Imataka H., Gradi,A. and Sonenberg,N. (1998) A newly identified N-terminal amino acid sequence of human eIF-4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. (1996) Poly(A) metabolism and translation: the closed loop model. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–480. [Google Scholar]
- Joachims M., Van Breugel,P.C. and Lloyd,R.E. (1999) Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol., 73, 718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovine L., Oubridge,C., Avis,J.M. and Nagai,K. (1996) Two structurally different RNA molecules are bound by the spliceosomal protein U1A using the same recognition strategy. Structure, 4, 621–631. [DOI] [PubMed] [Google Scholar]
- Keiper B.D. and Rhoads,R.E. (1999)Translational recruitment of Xenopus maternal mRNAs in response to poly(A) elongation requires initiation factor eIF4G-1. Dev. Biol., 206, 1–14. [DOI] [PubMed] [Google Scholar]
- Kerekatte V., Keiper,B.D. Badorff,C., Cai,A., Knowlton,K.U. and Rhoads,R.E. (1999) Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechansim of host protein synthesis shutoff? J. Virol., 73, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S.H. and Sachs,A.B. (1998) RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol., 18, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge H. and Richter,J.D. (1995) Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J., 14, 6301–6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge H., Brownlee,G.G., Gershon,P.D. and Richter,J.D. (1998) Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis.Nucleic Acids Res., 26, 3208–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U. and Pieler,T. (1996) Xenopus poly(A) binding protein: functional domains in RNA-binding and protein–protein interaction. J. Mol. Biol., 256, 20–30. [DOI] [PubMed] [Google Scholar]
- Le H., Tanguay,R.L., Balasta,M.L., Wei,C.C., Browning,K.S., Metz,A.M., Goss,D.J. and Gallie,D.R. (1997) Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem., 272, 16247–16255. [DOI] [PubMed] [Google Scholar]
- Lee M. and Struhl,K. (1997) A severely defective TATA-binding protein–TFIIB interaction does not preclude transcriptional activation in vivo. Mol. Cell. Biol., 17, 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus D. and Jacobson,A. (1998) Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol., 18, 7383–7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L., Preker,P.J., Wiederkehr,T., Strahm,Y. and Keller,W. (1997) The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl Acad. Sci. USA, 94, 7897–7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Bai,Y., Poon,D., Weil,P.A. and Struhl,K. (1996) TBP-associated factors are not generally required for transcriptional activation in yeast. Nature, 383, 188–191. [DOI] [PubMed] [Google Scholar]
- Nietfeld W., Mentzel,H. and Pieler,T. (1990) The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J., 9, 3699–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero L.J., Ashe,M.P. and Sachs,A.B. (1999) The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J., 18, 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain V.M. (1996) Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem., 236, 747–771. [DOI] [PubMed] [Google Scholar]
- Piron M., Vende,P., Cohen,J. and Poncet,D. (1998) Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts poly(A) binding protein from eIF4F. EMBO J., 17, 5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T. and Hentze,M.W. (1998) Dual function of the messenger RNA cap structure in poly(A)-tail promoted translation in yeast. Nature, 392, 516–520. [DOI] [PubMed] [Google Scholar]
- Richter J.D. (1996) Dynamics of poly(A) addition and removal during development. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 481–503. [Google Scholar]
- Sachs A.B. and Davis,R.W. (1989) The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell, 58, 857–867. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Bond,M.W. and Kornberg,R.D. (1986) A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell, 45, 827–835. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- Schwartz D.C. and Parker,R. (1999) Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz F. and Rosbash,M. (1994) A functional interaction between Rev and yeast pre-mRNA is related to splicing complex formation. EMBO J., 13, 4096–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs,A.B. (1995) A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev., 9, 2997–3007. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z., Wells,S.E., Deardorff,J.A. and Sachs,A.B. (1997) Translation initiation factor eIF-4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl Acad. Sci. USA, 94, 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.S., Reese,J.C., Apone,L.M. and Green,M.R. (1996) Tran scription activation in cells lacking TAFIIs. Nature, 382, 185–188. [DOI] [PubMed] [Google Scholar]
- Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Circulariz ation of mRNA by eukaryotic translation initiation factors. Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- Wickens M., Goodwin,E., Kimble,J., Strickland,S. and Hentze,M. (2000) Translational control of developmental decisions. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 295–370. [Google Scholar]
- Wormington M., Searfoss,A. and Hurney,C.A. (1996) Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J., 15, 900–909. [PMC free article] [PubMed] [Google Scholar]
- Zelus B.D., Giebelhaus,D.H., Eib,D.W., Kenner,K.A. and Moon,R.T. (1989) Expression of poly(A)-binding protein during development of Xenopus laevis. Mol. Cell. Biol., 9, 2756–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Kraemer,B., SenGupta,D., Fields,S. and Wickens,M. (1999) A three-hybrid system to detect and analyze RNA–protein interactions in vivo. Methods Enzymol., 306, 93–113. [DOI] [PubMed] [Google Scholar]