Abstract
As a result of the recurring translocation t(11;16) (q23;p13.3), MLL (mixed-lineage leukemia) is fused in frame to CBP (CREB binding protein). This translocation has been documented almost exclusively in cases of acute leukemia or myelodysplasia secondary to therapy with drugs that target DNA topo isomerase II. The minimal chimeric protein that is produced fuses MLL to the bromodomain, histone acetyltransferase (HAT) domain, EIA-binding domain and steroid-receptor coactivator binding domains of CBP. We show that transplantation of bone marrow retrovirally transduced with MLL–CBP induces myeloid leukemias in mice that are preceded by a long preleukemic phase similar to the myelodysplastic syndrome (MDS) seen in many t(11;16) patients but unusual for other MLL translocations. Structure–function analysis demonstrated that fusion of both the bromodomain and HAT domain of CBP to the amino portion of MLL is required for full in vitro transformation and is sufficient to induce the leukemic phenotype in vivo. This suggests that the leukemic effect of MLL–CBP results from the fusion of the chromatin association and modifying activities of CBP with the DNA binding activities of MLL.
Keywords: chromosomal translocations/CREB binding protein/leukemia/MDS/MLL
Introduction
The MLL (mixed-lineage leukemia) gene (also called HRX, ALL-1 and Htrx), which is located on chromosomal band 11q23, is involved in translocations with up to 40 different partner genes (Rowley, 1993; Thirman et al., 1993; Bernard and Berger, 1995). We (Sobulo et al., 1997) and others (Satake et al., 1997; Taki et al., 1997) recently cloned this translocation from patients with a t(11;16) translocation, and determined that an MLL–CBP fusion was created as a result of this translocation. One unusual observation is that all of the t(11;16) patients had therapy-related acute leukemia or myelodysplasia after exposure to DNA topoisomerase II-targeting drugs (anthracyclines or epipodophyllotoxins) for treatment of a primary malignancy (Rowley et al., 1997). This is in contrast to other MLL translocations, such as the t(9;11), t(4;11) and the two types of t(11;19) involving the partner genes AF9, AF4, and ENL or ELL, respectively, where the majority are de novo leukemias and a small proportion, if any, are secondary leukemias that result from chemotherapy. More recently, it has been shown that MLL is also involved in translocations with p300, which is a functional homolog of CBP located on chromosome 22 (Ida et al., 1997).
MLL is a very large protein (431 kDa) with homology to the Drosophila trithorax (trx) protein in several domains (Djabali et al., 1992; Gu et al., 1992; Tkachuk et al., 1992). trx is required to maintain the proper expression of homeotic genes of the Bithorax and Antennapaedia complexes in Drosophila (Kennison, 1995). Mice with a single disrupted Mll allele display bidirectional homeotic transformations, similar to the changes observed in trx mutant Drosophila (Yu et al., 1995). It has also recently been shown that the expression of many HOX genes is not properly maintained in embryonic fibroblasts (MEFs) derived from the Mll null mouse embryos (Hanson et al., 1999). It is thought that trx regulates homeotic expression at the level of chromatin organization by maintaining an open chromatin structure, and it is likely that MLL regulates the HOX genes in an analogous manner, although the mechanism has not been defined.
CBP is the first MLL partner gene cloned for which there is much functional information. CBP is a transcriptional coactivator that interacts with many different proteins (reviewed in Mannervik et al., 1999). It was first shown to interact with the CREB transcription factor but has since been shown to be an essential coactivator of many different transcriptional activators, including the retinoic acid receptor, thyroid hormone receptor and the p65 subunit of NF-κB. It can act as an adapter to bring together specific transcription factors with members of the basal transcriptional apparatus. CBP possesses intrinsic histone acetyltransferase (HAT) activity that is thought to be important for decondensing chromatin and facilitating the binding of the RNA polymerase II (Pol II) transcription complex to the core promoter (Bannister and Kouzarides, 1996; Ogryzko et al., 1996). CBP is a component of large multiprotein complexes containing other HATs that include P/CAF (p300/CBP-associated factor) and the steroid receptor coactivators SRC-1 and TIF2. In addition, CBP can associate with other coactivator complexes such as ARC (Naar et al., 1998). Functional domains of CBP have been defined and include (from the N- to C-termini of the protein): a domain that binds nuclear hormone receptors; a transactivation domain; a domain that binds many proteins, including CREB; a bromodomain; a domain with HAT activity; a domain that binds proteins including E1A and P/CAF; and a domain that binds steroid receptor coactivators including SRC-1 and TIF2 (see Figure 4A).
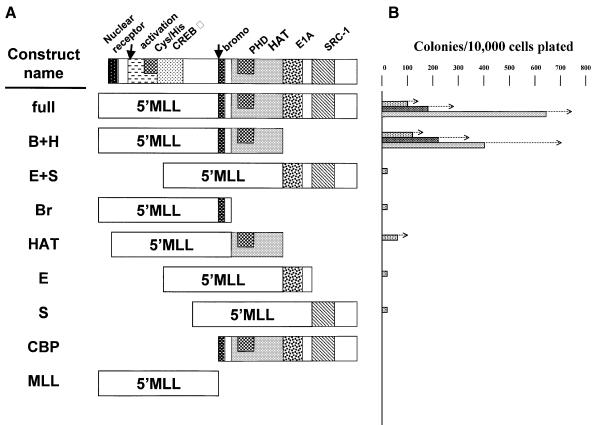
Fig. 4. Structure and in vitro proliferative effects of MLL–CBP mutants. (A) Structure of the constructs analyzed. B+H includes the bromodomain and the HAT domain; E+S includes the E1A-binding domain and the SRC-1-binding domain; Br includes the bromodomain; HAT includes the HAT domain; E includes the E1A-binding domain; and S includes the SRC-1-binding domain. (B) Number of secondary, tertiary and quaternary colonies (from top to bottom) generated per 104 input cells seeded following methylcellulose culture of Linlo cells transduced with the different MLL–CBP mutants. The values shown are the mean and the arrows indicate (+) standard error of the mean. Where no bars are indicated, too few colonies were generated to be shown on this graph.
Two different experimental systems have been exploited to generate mouse models of MLL-fusion leukemias. Homologous recombination targeted to the Mll locus was used to ‘knock-in’ the Mll–AF9 gene in embryonic stem cells, and the resulting chimeric mice developed acute myeloid leukemia (Corral et al., 1996). Retroviral transduction of MLL–ENL and MLL–ELL in murine bone marrow (BM) has also been used to transform myeloid progenitors in vitro and to generate myeloid leukemias in transplanted mice (Lavau et al., 1997, 2000). Furthermore, this approach was used to define the molecular requirements for transformation in vitro (Slany et al., 1998). Analysis of a series of MLL–ENL mutants demonstrated that domains within both MLL and ENL were indispensable for transformation. The critical features contributed by MLL were its DNA-binding properties, namely the AT-hooks and the methyltransferase homology motifs, while ENL’s contribution was concordant with its ability to transactivate transcription. Here we have applied the retroviral transduction/transplantation model to characterize the transforming properties of MLL–CBP in vivo and have investigated the molecular mechanisms of this activity.
Results
MLL–CBP causes leukemia in mice preceded by a lengthy myeloproliferative phase
To analyze the transforming potential of MLL–CBP in an animal model, we used retroviral transduction to express the fusion gene in BM and to reconstitute lethally irradiated mice. For this study we used a cDNA encoding the MLL–CBP fusion protein, similar to the shortest version of the fusion that has been cloned from patient leukemia cells (Sobulo et al., 1997). This cDNA was subcloned upstream of the IRES–EGFP (internal ribosome entry site–enhanced green fluorescent protein) cassette of the MIE vector (Du et al., 1999), which is derived from the murine stem cell virus (MSCV) retrovirus (Hawley et al., 1994).
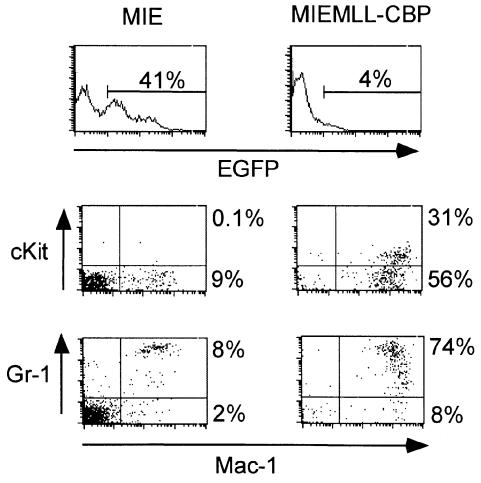
BM was harvested from BS/BA (Ly5.1) (see Materials and methods) donor mice 5 days after 5-fluorouracil treatment, and was further enriched for primitive hematopoietic cells by depletion of the population expressing markers of lineage differentiation. The resulting Linlo fraction was infected with the retroviral stocks by spinoculation as previously described (Slany et al., 1998). Transduction efficiency was determined by flow cytometry and indicated that 67% of Linlo cells infected with the MIE vector expressed EGFP, compared with <1% of the MIEMLL–CBP-infected cells. Ten irradiated BA.1 (Ly5.2) mice were transplanted each with 105 whole BM Ly5.2 cells, along with 15–16 × 103 Linlo Ly5.1 cells transduced with MIEMLL–CBP or the control MIE vector. Cell counts and immunostaining were performed on the peripheral blood (PB) of the mice to evaluate their hematopoietic reconstitution and monitor the development of disease. At 7 weeks post-reconstitution the MLL–CBP mice displayed a similar number of white blood cells (WBC), red blood cells (RBC) and platelets (PLT) compared with the MIE animals. However, the MLL–CBP mice had a much higher percentage of donor cells (64 versus 25% Ly5.1 positive cells in MLL–CBP and MIE mice, respectively, see experiment 1 in Table III). Moreover, the donor cells transduced with MLL–CBP preferentially yielded myeloid progeny as shown by a 3-fold increase in the percentage of Mac1/Gr1 positive compartment among the Ly5.1 population (24 versus 8% myeloid cells in MLL-CBP and MIE mice, respectively, see Table III). Despite this clear cut effect of the MLL–CBP transgene on the repopulating potential of the transplanted cells, only a small fraction of the engrafted leukocytes expressed EGFP in the MIEMLL–CBP mice (<10% of the Ly5.1 compartment were positive for EGFP whereas these represented 66–95% of the donor cells in the MIE cohort). Furthermore, the intensity of green fluorescence was very low in circulating cells transduced with MIEMLL–CBP (on average one order of magnitude lower than that seen in the MIE mice), often making it difficult to distinguish this population from the non-transduced leukocytes by flow cytometry analysis. The increase in the myeloid compartment of donor origin was not a transitory phenomenon as it could be seen with similar intensity at 12 weeks post-reconstitution (data not shown). To characterize better these myeloid cells in the PB we stained the leukocytes with an anti-Mac1 antibody together with an antibody to either Gr-1 or cKit. To compare MIE- and MIEMLL–CBP-transduced cells we restricted our analysis to the subfraction of cells expressing EGFP. We found consistent differences between the two cohorts of mice, which are illustrated in Figure 1. The fraction of EGFP-positive cells expressing Mac-1 was much larger in MIEMLL–CBP mice (ranging from 83 to 94%) than in MIE mice (ranging from 5 to 25%). Furthermore, the intensity of Mac-1 expression was also higher in the MIEMLL–CBP mice (the mean fluorescence intensity ranged between 103 and 2.2 × 103) compared with that seen in MIE mice (the mean fluorescence intensity ranged between 300 and 700). Interestingly, the circulating MIEMLL–CBP-transduced cells displayed features of incomplete myeloid maturation. First, a considerable fraction coexpressed Mac1 and cKit (the percentage of this double positive fraction ranged from 20 to 60% of the cells gated for EGFP expression in the MIEMLL–CBP mice, compared with <1% seen in the MIE cohort). Secondly, in most of the MIEMLL–CBP mice a significant proportion of the myeloid cells expressed moderate levels of Gr1.
Table III. The mice reconstituted with MLL–CBP (bromo + HAT) develop a preleukemic disease similar to that observed with full length MLL–CBP.
| Experimenta | Construct | Linlo cells injected per mouse (×103) | Proportion of donor cellsb (%) | Myeloid/donorb (%) | Mean survival (range) |
|---|---|---|---|---|---|
| 1 (n = 10) | MIE | 15 | 25 ± 3 | 8 ± 1 | >350 days |
| |
MLL–CBP |
16 |
64 ± 3 |
24 ± 3 |
164 days (126–188) |
| 2 (n = 9) | MIE | 12 | 15 ± 6 | 13 ± 2 | >180 days |
| MLL–CBP-bromo+HAT | 12 | 43 ± 9 | 39 ± 8 | NDc |
aThe number of mice reconstituted with each construct is shown in parentheses.
bThe mice were bled at 7 weeks (experiment 1) or 11 weeks (experiment 2) post-reconstitution. The values shown are the mean ± SEM.
cThree mice died of acute myeloid leukemia between 139 and 174 days after transplantation. Unfortunately, this cohort could not be studied further as the remaining mice succumbed to hyperthermia following an air conditioner failure.
Fig. 1. Characterization of the transduced leukocytes 10 weeks after reconstitution with MIE or MIEMLL–CBP (full). Following lysis of the red cells the peripheral blood from MIE and MIEMLL–CBP (full) mice collected 10 weeks post-reconstitution was simultaneously stained with a phycoerythrin (PE)-conjugated anti-Mac1 antibody and an allophycocyanin (APC)-conjugated antibody to either Gr-1 or cKit. Results from a representative MIE and MIEMLL–CBP (full) mouse are shown. The histograms represent the green fluorescence profiles; the limits of the EGFP-positive fraction are shown along with the percentage of cells it comprises. The plots represent the staining profiles of these EGFP-positive cells. The staining antibodies are indicated adjacent to each axis. Percentage values correspond to the content of the adjacent quadrants.
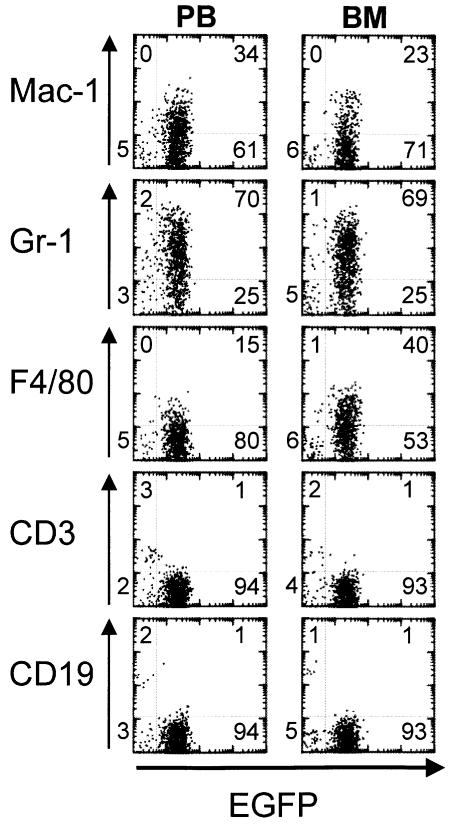
Eventually all the mice suffered from a rapidly evolving lethal disease between 126 and 188 days post-reconstitution. At this terminal stage, the mice exhibited a reduction in RBC and platelet PLT counts (Table I), and most showed a considerable increase in the number of WBC and the percentage of monocytes. Post-mortem examination showed consistent splenomegaly and pale femurs, and histological analysis confirmed that the spleen as well as the liver, kidney and thymus were infiltrated by leukemic cells. Morphological analysis of smears performed on the PB showed that generally a low percentage of myeloblasts and myelocytes were present, but that metamyelocytes, and band and segmented neutrophils were predominant (Figure 2). In contrast, a nearly homogeneous population of myeloblasts and myelocytes essentially replaced the BM (Figure 2). Flow cytometry analysis performed on the PB, BM and spleen revealed that at this stage the majority of the cells expressed EGFP (Figure 3). The intensity of green fluorescence was low and comparable to the intensity observed in the earlier phase described above. None of the EGFP-positive cells expressed the lymphoid markers CD3 or CD19. On the other hand, the myeloid nature of the leukemic cells was confirmed, as 65–95% of the EGFP-expressing cells in the spleen, PB or BM stained positively for Mac-1, Gr-1 or F4/80. All the signs exhibited by the animals are consistent with a diagnosis of acute myeloid leukemia. The clonality of the leukemias was assessed by Southern blot analysis, which revealed that all were mono- or pauci-clonal (data not shown).
Table I. Characteristics of the leukemias in recipients of MLL-CBP-transduced BM cells.
| Mouse | Latency (days) | WBC (103/µl) | Percentage monocytea | RBC (106/µl) | PLT (103/µl) | Spleen weight (g) |
|---|---|---|---|---|---|---|
| MIEMLL–CBP# 6 | 126 | ND | ND | ND | ND | 0.85 |
| MIEMLL–CBP# 4 | 129 | ND | ND | ND | ND | 0.36 |
| MIEMLL–CBP# 1 | 159 | 6.3 | 14 | 11 | 1870 | 0.65 |
| MIEMLL–CBP# 10 | 159 | 41 | 43 | 2.7 | 250 | 0.32 |
| MIEMLL–CBP# 5 | 165 | 53 | 6 | 5.5 | 369 | 0.53 |
| MIEMLL–CBP# 8 | 172 | 12 | 1 | 1.3 | 458 | 0.43 |
| MIEMLL–CBP# 9 | 172 | 120 | 40 | 2.5 | 248 | 0.70 |
| MIEMLL–CBP# 3 | 178 | ND | ND | ND | ND | ND |
| MIEMLL–CBP# 2 | 188 | 66 | 24 | 5.3 | 658 | 0.53 |
| MIEMLL–CBP# 7 |
188 |
7 |
24 |
4.4 |
469 |
0.36 |
| MIEMLL–CBP miceb | 164 ± 7 | 44 ± 14 | 22 ± 5 | 4.7 ± 1 | 342 ± 93 | 0.46 ± 0.05 |
| MIE micea | NA | 8 ± 0.9 | 7 ± 1 | 10 ± 0.3 | 1050 ± 95 | 0.090 ± 0.01c |
aDifferential counts determined by the Cell-Dyn counter.
bValues shown are the mean (± standard error of the mean) from the 10 MIEMLL–CBP mice at the time of sacrifice, or the 10 control MIE mice bled 158 days after reconstitution.
cSpleen weight from age-matched mice.
ND, not determined; NA, not applicable.
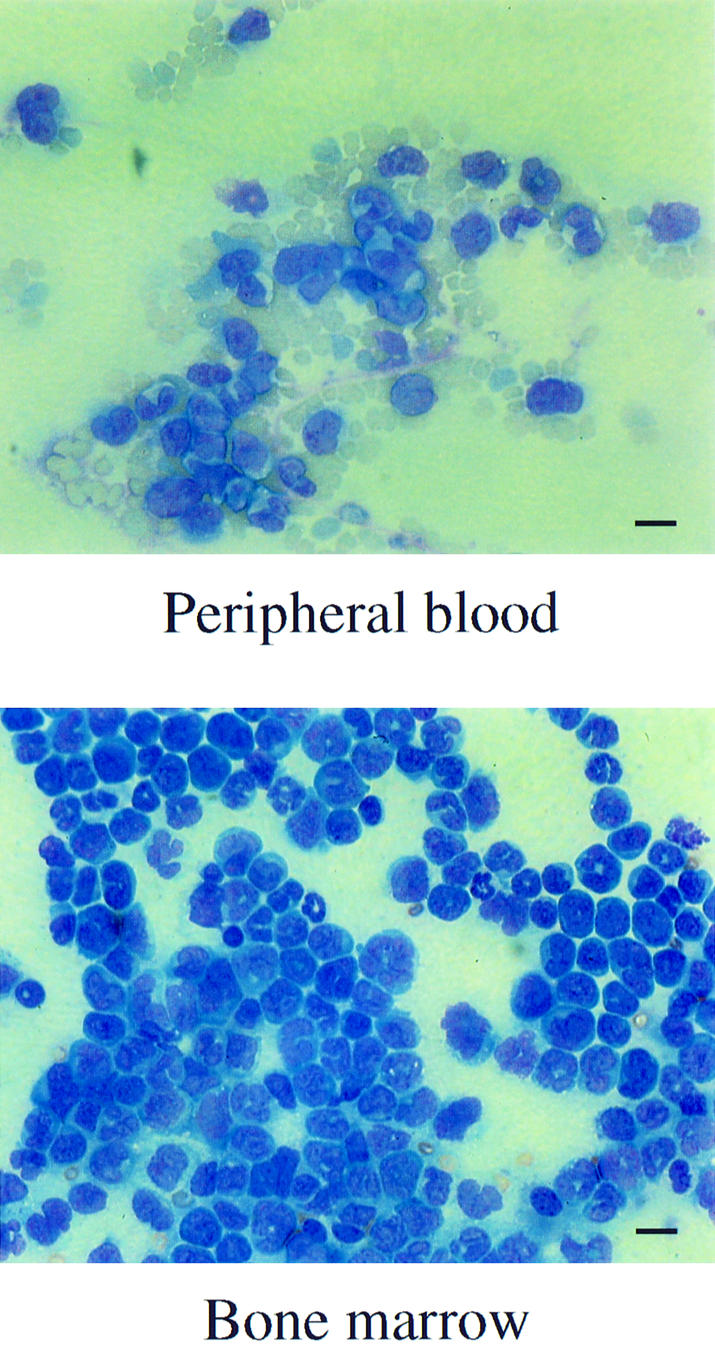
Fig. 2. Morphology of leukemic cells in the PB and BM of mice reconstituted with MIEMLL–CBP (full). Smears from PB and BM were performed at the time of sacrifice and stained with Wright–Giemsa (bar = 14 µM).
Fig. 3. Immunophenotype of MLL–CBP leukemias. Two-parameter dot plots show green fluorescence (EGFP) versus expression of lineage-specific markers (Mac-1 and F4/80 for macrophages, Gr-1 for granulocytes, CD3 for T cells or CD19 for B cells) from the peripheral blood (PB) or bone marrow (BM) of a representative MIEMLL–CBP mouse. The numbers denote the percentage of cells that are present in each quadrant.
The phenotype of the leukemia seen in MLL–CBP reconstituted mice is similar to the one we have observed in mice transplanted with BM transduced with MLL–ENL (Lavau et al., 1997) or MLL–ELL (Lavau et al., 2000). ENL is a protein of unknown function containing a C-terminal transcriptional activation domain that is retained in the MLL–ENL fusion protein. ELL is an RNA polymerase II elongation factor that is fused almost in its entirety to MLL. Neither ENL nor ELL have sequence similarity to CBP. We wished to examine whether the long preleukemic myeloid proliferation seen with MLL–CBP was also a feature shared with other MLL fusion genes. This is not the case for MLL–ENL leukemias as these appear after a much shorter latency (48–82 days; our unpublished data). On the other hand, those induced by MLL–ELL evolve over a similar length of time to that observed with MLL–CBP. We therefore set up transduction/transplantation studies with these two genes in parallel to examine whether the early increase of myeloid cells could be seen with both MLL–CBP and MLL–ELL. As summarized in Table II, we did not see any preferential differentiation of the MLL–ELL-transplanted cells toward the myeloid lineage at early time points before the onset of leukemias. The long preleukemic phase therefore appears to be a specific characteristic of MLL–CBP in transplanted mice; this mirrors the human diseases in which translocations generating MLL–CBP are frequently associated with myelodysplastic syndrome (MDS).
Table II. The myeloproliferative phase that precedes acute leukemia is a specific trait of MLL–CBP reconstituted mice.
| Retroviral construct (number of mice) | Myeloid cells/donora |
Time before onset of overt leukemia (range) | |
|---|---|---|---|
| 7 weeks | 10 weeks | ||
| MIE (n = 10) | 12 ± 1% | 8 ± 1% | leukemia free >7months |
| MIEMLL–CBP (n = 9) | 23 ± 4, P = 0.012 | 17 ± 3%, P = 0.014 | >165 days (166–273) |
| MIEMLL–ELL (n = 10) | 17 ± 3%, P = 0.133 | 10 ± 1%, P = 0.370 | >146 days (147–229) |
aMean percentage (± SEM) of cells expressing Mac-1 and/or Gr-1 among the leukocytes of donor origin (Ly5.1 positive) measured at 7 and 10 weeks post-reconstitution. The statistical significance of the difference compared with the MIE mice was calculated using the unpaired t-test; the P value is shown (means are statistically significant if P <0.05).
The bromodomain and HAT domain contributed by CBP are sufficient for the transforming activity of MLL–CBP
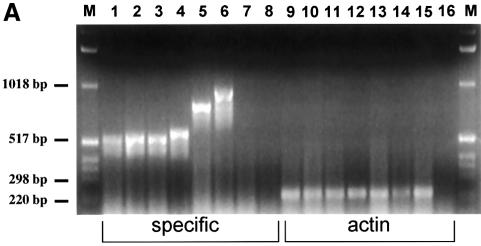
To unravel the molecular mechanism of transformation by MLL–CBP, we undertook a structure–function analysis of the fusion protein. For this we exploited an in vitro assay that measures the proliferative capacity of retrovirally transduced myeloid progenitors in methylcellulose culture. We have used this system to define which regions of the MLL moiety are critical to the transforming properties of MLL(HRX)–ENL (Slany et al., 1998). Here we focused on the various known functional domains of CBP. A series of deletion mutants of MLL–CBP was used to transduce Linlo cells, which were subsequently selected in methylcellulose culture in the presence of G418 (Figure 4A). The myeloid colonies that grew were then harvested, and the proliferative potential of the transduced cells was measured by sequentially reseeding the cells in methylcellulose culture to generate secondary, tertiary and quaternary colonies (Figure 4B). Expression of the various MLL–CBP constructs was confirmed in transfected 293 cells by reverse transcription PCR (RT–PCR) (Figure 5A).
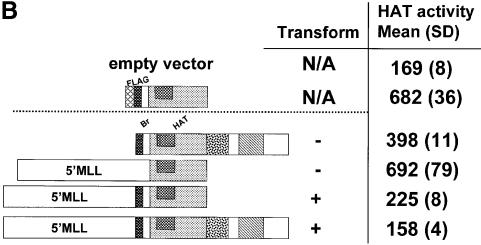
Fig. 5. Expression and in vitro HAT activity of MSCV-MLL–CBP constructs. (A) RT–PCR from 293 cells transiently transfected with MSCV constructs. The names of the constructs correspond to those described in Figure 4. MSCV-MLL–CBP full (lanes 1 and 9); B+H (lanes 2 and 10); Br (lanes 3 and 11); HAT (lanes 4 and 12); E+S (lanes 5, 8, 13 and 16); CBP (lanes 6 and 14); MIE vector (lanes 7 and 15); no reverse transcriptase (lanes 8 and 16). Lanes 1–8 used specific primers for MLL and CBP or CBP alone. Lanes 9–16 used actin primers. The 1 kb DNA ladder (Gibco-BRL) was used as marker (M). (B) HAT activity alone is not sufficient for transformation. Extracts from 293 cells transiently transfected with the indicated constructs were used to assay for in vitro HAT activity as assessed by measuring [3H]acetate incorporation with the filter binding assay on chicken histone substrate. Results of four independent experiments are shown as mean (standard deviation).
As expected, the full length MLL–CBP cDNA, which induces myeloid proliferation and leukemias in transplanted mice, strongly increased the proliferative potential of myeloid colony-forming cells in methylcellulose. As can be seen in Figure 4B, the number of colonies generated per cells seeded was on average >100/104 on the second passage, and increased up to >600/104 at the fourth passage. Furthermore, most of the colonies were very large (made up of several thousands of cells after 7 days of culture) and the majority had a typical compact morphology very similar to the ones observed with MLL–ENL (Lavau et al., 1997). A construct expressing the CBP moiety alone had no transforming activity in the assay, as the number of colonies formed past the first round of plating was as low as that seen with the empty MSCV-neo vector. Among the various CBP deletion mutants, only the one containing both the bromodomain and the HAT domain fused to the amino portion of MLL had transforming activity equivalent to the full length MLL–CBP. These domains appeared sufficient while the other regions of the CBP protein were dispensable. Interestingly, while the CBP bromodomain alone fused to MLL had no transforming potential, the HAT domain alone did confer some growth advantage to the transduced cells, generating ∼50 secondary colonies. However, this effect was transient as the cells ceased to form colonies past the second round of plating.
We wished to determine whether HAT activity was indeed present in the MSCV-MLL–CBP constructs and whether the presence or absence of HAT activity correlated with transformation ability. We did not know whether HAT activity would be retained when CBP was expressed as a chimeric fusion protein with MLL. A liquid HAT assay was used to determine the relative ability of the MLL–CBP constructs to facilitate transfer of the [3H]acetyl group from [3H]acetylCoA to core chicken histones (Brownell and Allis, 1995). The results are shown in Figure 5B. A FLAG-tagged CBP (HAT domain alone) construct was used as a positive control for HAT activity in this assay. Whole extracts prepared from 293 cells transiently transfected with the various constructs were used to assay the HAT activity. As expected, the positive control FLAG-CBP (HAT domain) has a high level of HAT activity, as did the MSCV-CBP alone construct. For MSCV-MLL–CBP constructs, strong activity was present in the MLL–CBP (HAT alone) construct, whereas the MLL–CBP (bromo + HAT) construct had significant but much lower levels of HAT activity. The two full length MLL–CBP constructs tested (MSCV and MIE) both had levels of HAT activity that were indistinguishable from untransfected or vector-transfected cell extracts. This is most likely due to a very low level of expression of these full length constructs. Thus, the HAT activity of the particular constructs alone did not correlate with transformation ability. Specifically, MSCV-CBP alone and MSCV-MLL–CBP (HAT alone) had very good HAT activity but neither was fully transforming. The results from these experiments demonstrate that the HAT activity alone is not sufficient for transformation.
To examine whether the leukemogenic ability of MLL–CBP could be recapitulated by fusing the bromo- and HAT domains of CBP to the amino portion of MLL, we cloned the MLL–CBP (bromo + HAT) mutant in the MIE vector, and reconstituted mice with transduced BM. In this experiment the transduction efficiency in the Linlo cells was ∼60% with the MIE vector and <1% with the MIEMLL–CBP (bromo + HAT) mutant. Nine mice were inoculated with 12 × 103 infected cells and first analyzed for donor reconstitution and myeloid contribution at 11 weeks after transplantation. As seen in Table III, the MLL–CBP (bromo + HAT) mutant had a similar effect to the MLL–CBP (full length) on the production of myeloid cells of donor origin in the reconstituted mice, with a 3-fold increase compared with the MIE mice. Three of the mice reconstituted with the CBP (bromo + HAT) mutant developed a lethal myeloid leukemia very analogous to the one seen with MLL–CBP and within a comparable time frame (between 139 and 173 days post-transplantation). A technical breakdown fatal for the remaining mice precluded a comprehensive analysis of this cohort.
Discussion
Our in vivo study revealed that in addition to its leukemic effect MLL–CBP induces a myeloproliferative state characterized by an accumulation of immature forms, early on after transplantation. This seems to be specific of the CBP fusion partner as it was not observed with ENL or ELL fusion genes in the same experimental system. A similar pathology was observed with the Mll–AF9 knock-in mice with germ line transmission, which have a myeloproliferation as early as 5 days after birth (Dobson et al., 1999). However, with this gene the differentiation of the myeloid cells did not seem to be impaired. The preleukemic phase of the transplanted MLL–CBP mice, which precedes the overt myeloid leukemias by several months, is reminiscent of the MDS often associated with the (11;16) translocation in humans. It is tempting to speculate that this early myeloid proliferation generates a pool of transformed cells that are susceptible to further mutations that participate in the leukemic progression.
It has been shown recently in Cbp heterozygous knockout mice that highly penetrant multilineage defects in hematopoietic differentiation are observed (Kung et al., 2000). With advancing age (>1 year), these mice demonstrate an increased incidence of hematologic malignancies (39%), with loss of heterozygosity (LOH) at the Cbp locus observed in some cases. It has been argued that this demonstrates that Cbp has tumor-suppressing activity. The tumors were present only in animals >1 year of age, and the phenotypes were histiocytic sarcomas, lymphocytic leukemia or myelogenous leukemia. This is very different from what we observe in the MLL–CBP infected mice where the leukemias develop in 100% of the animals by 6.5 months of age, and are all myelomonocytic leukemia. Both Cbp alleles should be intact in these mice; however, this has not been specifically analyzed. In humans with the MLL–CBP translocation, it is certainly possible that the inactivation of one normal CBP allele and one MLL allele, in addition to the novel functions provided by the MLL–CBP fusion protein, contribute to leukemogenesis. There has been no observed LOH at the remaining CBP allele in the t(11;16) patients; however, a finer analysis of the seemingly intact allele has not yet been pursued.
In our analysis of leukemias with the t(11;16) that contain the MLL–CBP fusion, RT–PCR from one of our patients revealed that the contribution from CBP to the fusion was limited to the region from the bromodomain to the C-terminus of the protein (Sobulo et al., 1997). In contrast, the majority of patients for which the breakpoints have been identified demonstrated that almost all of CBP was brought into the MLL–CBP chimeric protein (Satake et al., 1997; Sobulo et al., 1997; Taki et al., 1997). Therefore, although our transformation studies demonstrate that only the bromodomain and the HAT domain are required from CBP in the fusion protein, the predominant fusion in vivo includes much more of CBP. This could be due to the structure of the CBP gene itself. The intron in which the break usually occurs is quite large (40 kb) compared with the rest of the CBP gene (total ∼150 kb), so one would expect more breaks to occur in this large intron just by chance. It is also possible, however, that there are some structural elements present in this intron that render it more susceptible to breakage and the possibility of translocation once patients are treated with drugs that target DNA topoisomerase II. This is particularly intriguing because of the association of this particular type of MLL translocation almost exclusively with therapy-related leukemia (Rowley et al., 1997). We have previously shown that certain structural elements are present within the MLL gene as well as within AF9, another partner gene of MLL that is associated with therapy-related leukemia (Strissel et al., 1998, 2000). These include scaffold attachment regions, DNA topoisomerase II cleavage sites and DNase hypersensitive sites. It will be interesting to determine whether the region in CBP where the breaks primarily occur also contains these same structural elements.
The studies reported here demonstrate that a deletion mutant of MLL–CBP, which only retains the bromodomain and the HAT domain of CBP fused to MLL, has equivalent transforming activity in vitro and in vivo to the full length fusion protein. This indicates that the critical functions provided by CBP to the fusion protein are contained within these two domains. The HAT domain of CBP alone fused to N-terminal MLL caused some increase in myeloid proliferation but was not fully transforming in vitro. The fact that this particular mutant displayed high HAT activity indicates that the full oncogenicity of MLL–CBP requires additional properties contributed by the bromodomain.
CBP has been shown to acetylate several different proteins to date. CBP can acetylate all four core histones, H2A, H2B, H3 and H4, and thus resembles a type-A HAT (Bannister and Kouzarides, 1996; Brownell and Allis, 1996). These HATs are generally nuclear proteins that modify chromatin-associated histones at distinct sites. Almost all of the type-A HATs also contain a bromodomain that has been postulated to tether these HATs to specific active chromatin domains (Winston and Allis, 1999). It has been suggested that this could be how specificity is generated to link histone acetylation to gene activation. Genes of the HOX cluster are known to be downstream targets of MLL function (Yu et al., 1995, 1998; Hanson et al., 1999). It is possible that aberrant constitutive histone acetylation at this cluster of target genes caused by the MLL–CBP fusion could be critical to the leukemogenesis pathway. A mechanism of aberrant transcriptional activation of MLL target genes has been proposed for other 11q23 leukemias in which MLL is fused to proteins with transactivation properties, such as AF4 or ENL (Prasad et al., 1995; Slany et al., 1998). The structure–function analysis of MLL–ENL, indicating that the transactivating domain of ENL was necessary and sufficient to confer transforming activity to the chimeric protein, gave weight to this hypothesis.
The HAT activity of CBP is not limited to histone substrates and it is likely to influence other cellular events in addition to its role in transcriptional co-activation. CBP and its homolog p300 have indeed been shown to acetylate non-histone proteins including p53 (Gu and Roeder, 1997), dTCF (Waltzer and Bienz, 1998), HMG-I(Y) (Munshi et al., 1998), GATA-1 (Boyes et al., 1998), EKLF (Zhang and Bieker, 1998), NF-Y (Li et al., 1998), TFIIE and TFIIF (Imhof et al., 1997), HIV-1 Tat (Kiernan et al., 1999) and c-Myb (Tomita et al., 2000). As a result of acetylation, the functional activities of these proteins are altered. For example, acetylation of p53 increases its sequence-specific DNA binding activity, providing a potential mechanism for transcriptional control of p53 target genes (Gu and Roeder, 1997). When T-cell factor (TCF) is activated by Wnt/Wingless signaling, it binds its coactivator β-catenin/Armadillo, and stimulates target genes. Drosophila CBP acetylates a conserved lysine in the Armadillo-binding domain of Drosophila TCF, which lowers the affinity of Armadillo binding to dTCF (Waltzer and Bienz, 1998). HMG-I(Y) is an essential architectural component required for IFNB gene enhanceosome assembly. In the context of the enhanceosome, acetylation of HMG-I by CBP leads to enhanceosome destabilization and disassembly, resulting in post-induction turn-off of the IFNB gene (Munshi et al., 1998). Any of these, or other as yet unidentified targets of CBP acetyltransferase activity, may be important in leukemogenesis.
The specific function of the CBP bromodomain is unknown. However, by analogy to the bromodomains of other proteins some educated guesses can be made as to potential roles of this domain. The NMR structure of the P/CAF bromodomain has recently been solved and demonstrated to interact with acetylated lysine residues (Dhalluin et al., 1999). It is interesting that the interaction between the bromodomain and acetyl lysine is similar to that between acetyl-CoA and HAT. Acetyl lysine residues may be present in very many different contexts, either as specifically modified lysines in the various histone tails or as residues from transcription factors that are acetylated. The residues of the bromodomain that are critical for acetyl lysine binding are highly conserved throughout bromodomain family members (Dhalluin et al., 1999). It may be that specificity for the particular acetylated target protein is determined by the other variable residues within the bromodomain. This could contribute to specificity of histone acetylation by tethering HATs to specific chromosomal sites, or it could play a role in the acetylation of other proteins. For example, the bromodomain was shown to be required, in addition to the HAT domain of p300, to acetylate effectively the c-Myb protein involved in hematopoietic neoplasms (Tomita et al., 2000). It is also possible that the regulated acetylation of activator proteins could be used to signal or enhance the binding of bromodomain-containing coactivator complexes that are important for transcriptional control, such as SAGA, RSC and Snf/Swi. Very recently it has been shown that the Gcn5 bromodomain is important in nucleosome remodeling of a downstream target gene (Syntichaki et al., 2000). Mutation of specific bromodomain residues that are critical for acetyl-lysine binding demonstrated that these residues are not required in vivo for Gcn5-mediated histone acetylation, but rather are necessary for Swi2-dependent nucleosome remodeling that follows acetylation (Syntichaki et al., 2000).
Both the CBP bromodomain and the HAT domain, which are necessary for leukemic transformation, are functionally linked to recognition and/or modification of lysine residues in proteins that bind to CBP. Because these proteins are numerous and diverse, it remains to be determined which of these interactions are relevant for leukemia. Certainly, identification of these critical pathways will aid our understanding of this particular type of leukemia and potentially provide specific targets for intervention. Whether or not the same mechanisms will be relevant to the very many other MLL translocations in leukemia will also need to be elucidated.
Materials and methods
Plasmid cloning and production of retroviral constructs
MSCV-MLL–CBP plasmids were constructed by three-way ligations using the following fragments. The MSCV-neo vector (Hawley et al., 1994) (kindly provided by R.Hawley) was digested with EcoRI and BglII, and dephosphorylated with CIP (Boehringer Mannheim). N-terminal MLL was obtained by digestion of MSCV-MLL–SalI–ELL (R.Luo and M.Thirman) with EcoRI and SalI, followed by gel purification of the 4.2 kb MLL fragment. CBP fragments were obtained by first creating a silent site mutant of CBP, CBPmut1 (CBP2 plasmid kindly provided by J.Borrow), which destroyed an internal SalI site using the QuikChange site-directed mutagenesis kit (Stratagene) as per the manufacturer’s recommendations. The relevant domains of CBP were PCR amplified using pfu polymerase and a low number of cycles from either the wild-type CBP2 or the CBPmut1 plasmid, digested with SalI and BglII, followed by gel purification. The three fragments were ligated, and resultant clones were analyzed by restriction digest and sequencing. The regions of CBP that were included in each construct (numbers refer to CBP amino acids as per DDBJ/EMBL/GenBank accession No. U47741) were: (1) full (1021–2442); (2) B+H (1021–1850); (3) E+S (1715–2442); (4) Br (1021–1170); (5) HAT (1174–1850); (6) E (1715–1960); (7) S (1960–2442); (8) CBP (1021–2442). For in vivo leukemogenesis studies, we used the MIE vector (Du et al., 1999) that encodes EGFP downstream of an IRES. The MIE-MLL–CBP constructs were prepared similarly to the MSCV-MLL–CBP constructs except that the CBP portions were excised from the MSCV-MLL–CBP plasmids used for in vitro transformation, and used directly for subcloning without further PCR amplification. Retroviral supernatants were obtained by transiently transfecting the different retroviral constructs in the Bosc23 packaging cell line as previously described (Lavau et al., 1997).
Infection of progenitors and methylcellulose colony forming assays
Infection of lineage-depleted (Lin–) BM from 5-fluorouracil-treated C57BL/Ka-Ly5.1, Thy1.1 (known as BS/BA) mice and culture of the transduced progenitors in methylcellulose were conducted as previously described (Slany et al., 1998).
Liquid HAT assays
Extracts prepared from 293 cells transiently transfected with vector controls or with various CBP contructs were used to assay for liquid HAT activity essentially as described (Brownell and Allis, 1995). Approximately 48 h after calcium phosphate transfection, cells were harvested, washed with TEN (40 mM Tris–HCl pH 7.6, 1 mM EDTA, 150 mM NaCl) plus protease inhibitors (Sigma). After removal of TEN, cells were extracted with NET-N (100 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl pH 8.0, 0.2% NP-40) plus protease inhibitors, incubated on ice for 10 min and then microfuged for 30 min at 4°C. The supernatant was retained and a fraction electrophoresed for Coomassie staining and/or western blotting to assess the amount of protein expressed. Crude chicken core histones (10 µg; kindly provided by Craig Mizzen and David Allis) and 21 µl of cell extract or control proteins were incubated for 15 min at 30°C in a final volume of 50 µl of buffer A [50 mM Tris–HCl pH 8.0, 10% (v/v) glycerol, 10 mM sodium butyrate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride]. Reactions were initiated by the addition of [3H]acetyl-CoA (100 nCi, 6.1 Ci/mmol) to a final concentration of 0.328 µM. HAT activity was determined by liquid scintillation counting of aliquots of reactions spotted on P-81 filters (Whatman), and processed as described (Brownell and Allis, 1995).
RT–PCR analysis
Total RNA was isolated from transiently transfected 293 or Bosc cells using TriReagent (Sigma, St Louis, MO) as recommended by the manufacturer. Residual DNA was removed from the samples by digestion with RNase-free DNase, and cDNA prepared using a cDNA cycle kit (Invitrogen) according to the manufacturer’s instructions. The RT–PCR reaction was performed essentially as previously described (Sobulo et al., 1997), except that the MLL primer used was 5′-CTCCACCACCAGAATCAGGTC-3′ and the CBP primers used were: 5′-GGGATTCTTTACGATGTCAAATGCGTCTGGAATTCCGAGG-3′ for most of the MLL–CBP fusions, except 5′-GAGCTTGTGTTTGATGTTGAGGCAGAAGG-3′ for MLL–CBP(1715–2442), 5′-GCAGAACGCAAATCTGTGCCATCTTCCGGCCACA-3′ for MLL-CBP HAT alone, 1497B 5′-CCACTCCTGCAGTCGTGCTGGCTTGGGTATTTTTTGATCAGG-3′ and 1174T 5′-CGCAAGACATCCCGAGCATATAAGTTTTGCAGTAAGCTTGC-3′ for CBP alone. The actin primers used were: forward, 5′-CTTCCAGCCTTCCTTCCTGG-3′; and reverse, 5′-GTACTTGCGCTCAGGAGGAG-3′.
Reconstitution assay and characterization of leukemias
Reconstitution of lethally irradiated C57BL/Ka-Ly5.2, Thy1.1 (known as BA.1) mice with transduced progenitors was performed as described (Lavau et al., 1997) with the following modifications. Each mouse was inoculated by tail vein injection with 1–2 × 104 Lin– BM cells transduced with MLL–CBP, together with 105 normal BS/BA BM cells to ensure radioprotection. PB was collected from the retro-orbital plexus of anesthetized mice at regular intervals after transplantation. Circulating leukocytes, erythrocytes and PT were counted by analysis of 20 µl of blood using a Cell Dyn 3500R (Abbott Diagnostics, Abbott Park, IL). The degree of engraftment and the proportion of myeloid cells among the circulating donor leukocytes were assessed by flow cytometric analysis of samples co-stained with phycoerythrin (PE)-conjugated anti-Ly5.1 antibody (Pharmingen Inc., San Diego, CA) and a mix of allophycocyanin (APC)-conjugated antibodies against CD11b/Mac-1 and Gr-1.
More detailed immunophenotypic characterization of the leukemic cells was done by co-staining the circulating leukocytes with a biotin-conjugated anti-Ly5.1 antibody and phycoerythrin (PE)-conjugated antibodies against F4/80 (Caltag, Burlingame, CA), CD11b/Mac-1, Gr-1, CD19, CD3, or with the IgG2a or IgG2b isotype controls (Pharmingen Inc., San Diego, CA) followed by a secondary stain with streptavidin-APC (Caltag, Burlingame, CA). Stained cells were washed, resuspended with propidium iodide (PI) and examined with a FACSCalibur instrument (Becton-Dickinson, Mountain View, CA) following exclusion of dead cells by high PI staining and forward light scatter. The sick animals were euthanized by CO2 inhalation and their tissues fixed in formalin, sectioned, and stained with hematoxylin and eosin for histological analysis. Blood smears and cytospin preparations of the BM were stained with Wright–Giemsa for morphological analysis.
Acknowledgments
Acknowledgements
We thank Janet Rowley for helpful discussions, Robert Slany for advice on western blotting, Roger Luo for 5′MLL(Sal1) construct, David Allis and Craig Mizzen for chicken core histones and advice on the liquid HAT assay, Ronald Tomek and Alanna Harden for technical assistance. This work was supported by grants from the National Institutes of Health (CA40046) (N.Z.-L.) and by National Institutes of Health grant CA78431, the Burroughs Wellcome Fund and the American Society of Hematology (M.T.)
References
- Allen R.J. et al. (1998) Establishment and characterization of a megakaryoblast cell line with amplification of MLL. Leukemia, 12, 1119–1127. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bernard O.A. and Berger,R. (1995) Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer, 13, 75–85. [DOI] [PubMed] [Google Scholar]
- Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- Brownell J.E. and Allis,C.D. (1995) An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc. Natl Acad. Sci. USA, 92, 6364–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E. and Allis,C.D. (1996) Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev., 6, 176–184. [DOI] [PubMed] [Google Scholar]
- Corral J. et al. (1996) An M11–AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell, 85, 853–861. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.-M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Djabali M. et al. (1992) A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukaemias. Nature Genet., 2, 113–118. [DOI] [PubMed] [Google Scholar]
- Dobson C.L., Warren,A.J., Pannell,R., Forster,A., Lavenir,I., Corral,J., Smith,A.J.H. and Rabbitts,T.H. (1999) The Mll–AF9 gene fusion in mice controls myeloproliferation and specifies acute myeloid leukaemogenesis. EMBO J., 18, 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Redner,R.L., Cooke,M.P. and Lavau,C. (1999) Overexpression of wild-type retinoic acid receptor α (RARα) recapitulates retinoic acid-sensitive transformation of primary myeloid progenitors by acute promyelocytic leukemia RARα-fusion genes. Blood, 94, 793–802. [PubMed] [Google Scholar]
- Fidanza V., Melotti,P., Yano,T., Nakamura,T., Bradley,A., Canaani,E., Calabretta,B. and Croce,C.M. (1996) Double knockout of the ALL-1 gene blocks hematopoietic differentiation in vitro. Cancer Res., 56, 1179–1183. [PubMed] [Google Scholar]
- Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Gu Y. et al. (1992) The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell, 71, 701–708. [DOI] [PubMed] [Google Scholar]
- Hanson R.D. et al. (1999) Mammalian Trithorax and Polycomb-group homologues are antagonistic regulators of homeotic development. Proc. Natl Acad. Sci. USA, 96, 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley R.G., Lieu,F.H.L., Fong,A.Z.C. and Hawley,T.S. (1994) Versatile retroviral vectors for potential use in gene therapy. Gene Ther., 1, 136–138. [PubMed] [Google Scholar]
- Haynes S.R., Dollard,C., Winston,F., Beck,S., Trowsdale,J. and Dawid,I.B. (1992) The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res., 20, 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida K., Kitabayashi,I., Taki,T., Taniwaki,M., Noro,K., Yamamoto,M., Ohki,M. and Hayashi,Y. (1997) Adenoviral E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13). Blood, 90, 4699–4704. [PubMed] [Google Scholar]
- Imhof A., Yang,X.J., Ogryzko,V.V., Nakatani,Y., Wolffe,A.P. and Ge,H. (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol., 7, 689–692. [DOI] [PubMed] [Google Scholar]
- Kennison J.A. (1995) The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet., 29, 289–303. [DOI] [PubMed] [Google Scholar]
- Kiernan R.E. et al. (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J., 18, 6106–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung A.L., Rebel,V.I., Bronson,R.T., Ch’ng,L.-E., Sieff,C.A., Livingston,D.M. and Yao,T.-P. (2000) Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev., 14, 272–277. [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Szilvassy,S.J., Slany,R. and Cleary,M.L. (1997) Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J., 16, 4226–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavau C., Du,C., Luo,R. and Thirman,M.J. (2000) Retrovirus mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemia in mice. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Herrler,M., Landsberger,N., Kaludov,N., Ogryzko,V.V., Nakatani,Y. and Wolffe,A.P. (1998) Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive tran scription from the Xenopus hsp70 promoter in vivo. EMBO J., 17, 6300–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik M., Nibu,Y., Zhang,H. and Levine,M. (1999) Transcriptional coregulators in development. Science, 284, 606–609. [DOI] [PubMed] [Google Scholar]
- Munshi N., Merika,M., Yie,J., Senger,K., Chen,G. and Thanos,D. (1998) Acetylation of HMG I(Y) by CBP turns off IFNB expression by disrupting the enhanceosome. Mol. Cell, 2, 457–467. [DOI] [PubMed] [Google Scholar]
- Naar A.M., Beaurang,P.A., Robinson,K.M., Oliner,J.D., Avizonis,D., Scheek,S., Zwicker,J., Kadonaga,J.T. and Tjian,R. (1998) Chromatin, TAFs and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev., 12, 3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Yoshihiro,N. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Prasad R., Yano,T., Sorio,C., Nakamura,T., Rallapalli,R., Gu,Y., Leshkowitz,D., Croce,C.M. and Canaani,E. (1995) Domains with transcriptional regulatory activity within the ALL1 and AF4 proteins involved in acute leukemia. Proc. Natl Acad. Sci. USA, 92, 12160–12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley J.D. (1993) Rearrangements involving chromosome band 11q23 in acute leukaemia. Semin. Cancer Biol., 4, 377–385. [PubMed] [Google Scholar]
- Rowley J.D. et al. (1997) All patients with the T(11;16)(q23;p13.3) that involves MLL and CBP have treatment-related hematologic disorders. Blood, 90, 535–541. [PubMed] [Google Scholar]
- Satake N., Ishida,Y., Otoh,Y., Hinohara,S., Kobayashi,H., Sakashita,A., Maseki,N. and Kaneko,Y. (1997) Novel MLL–CBP fusion transcript in therapy-related chronic myelomonocytic leukemia with a t(11;16)(q23;p13) chromosome translocation. Genes Chromosom. Cancer, 20, 60–63. [PubMed] [Google Scholar]
- Slany R.K., Lavau,C. and Cleary,M.L. (1998) The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol. Cell. Biol., 18, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobulo O.M. et al. (1997) MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3). Proc. Natl Acad. Sci. USA, 94, 8732–8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strissel P.L., Strick,R., Rowley,J.D. and Zeleznik-Le,N.J. (1998) An in vivo topoisomerase II cleavage site and a Dnase I hypersensitive site colocalize near exon 9 in the MLL breakpoint cluster region. Blood, 92, 3793–3803. [PubMed] [Google Scholar]
- Strissel P.L., Strick,R., Tomek,R.J., Roe,B.A., Rowley,J.D. and Zeleznik-Le,N.J. (2000) DNA structural properties of AF9 are similar to MLL and could act as recombination ‘hot spots’ resulting in MLL/AF9 translocations and leukemogenesis. Hum. Mol. Genet., 9, 1671–1679. [DOI] [PubMed] [Google Scholar]
- Syntichaki P., Topalidou,I. and Thireos,G. (2000) The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature, 404, 414–417. [DOI] [PubMed] [Google Scholar]
- Taki T., Sako,M., Tsuchida,M. and Hayashi,Y. (1997) The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood, 89, 3945–3950. [PubMed] [Google Scholar]
- Thirman M.J. et al. (1993) Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N. Engl. J. Med., 329, 909–914. [DOI] [PubMed] [Google Scholar]
- Tkachuk D.C., Kohler,S. and Cleary,M.L. (1992) Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell, 71, 691–700. [DOI] [PubMed] [Google Scholar]
- Tomita A., Towatari,M., Tsuzuki,S., Hayakawa,F., Kosugi,H., Tamai,K., Miyazaki,T., Kinoshita,T. and Saito,H. (2000) c-Myb acetylation at the carboxyl-terminal conserved domain by transcriptional co-activator p300. Oncogene, 19, 444–451. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Winston F. and Allis,C.D. (1999) The bromodomain: a chromatin-targeting module? Nature Struct. Biol., 6, 601–604. [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hess,J.L., Horning,S.E., Brown,G.A.J. and Korsmeyer,S.J. (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature, 378, 505–508. [DOI] [PubMed] [Google Scholar]
- Yu B.D., Hanson,R., Hess,J.L., Norning,S.E. and Korsmeyer,S.J. (1998) MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl Acad. Sci. USA, 95, 10632–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeleznik-Le N.J., Harden,A.M. and Rowley,J.D. (1994) 11q23 translocations split the ‘AT-hook’ cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc. Natl Acad. Sci. USA, 91, 10610–10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. and Bieker,J.J. (1998) Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]