Abstract
The class Ib ribonucleotide reductase of Escherichia coli can initiate reduction of nucleotides to deoxynucleotides with either a MnIII2-tyrosyl radical (Y•) or a FeIII2-Y• cofactor in the NrdF subunit. Whereas FeIII2-Y• can self-assemble from FeII2-NrdF and O2, activation of MnII2-NrdF requires a reduced flavoprotein, NrdI, proposed to form the oxidant for cofactor assembly by reduction of O2. The crystal structures reported here of E. coli MnII2-NrdF and FeII2-NrdF reveal different coordination environments, suggesting distinct initial binding sites for the oxidants during cofactor activation. In the structures of MnII2-NrdF in complex with reduced and oxidized NrdI, a continuous channel connects the NrdI flavin cofactor to the NrdF MnII2 active site. Crystallographic detection of a putative peroxide in this channel supports the proposed mechanism of MnIII2-Y• cofactor assembly.
Ribonucleotide reductases (RNRs) catalyze the reduction of nucleotides to deoxynucleotides in all organisms, providing the precursors required for DNA synthesis and repair (1). Class I RNRs are composed of two homodimeric subunits: α2, which contains the site of nucleotide reduction, and β2, which contains a metallocofactor required to initiate catalysis in α2. Two class I RNRs are found in Escherichia coli. The class Ia enzyme supplies deoxynucleotides during normal aerobic growth. In many prokaryotes, including several important human pathogens, the class Ib RNR plays a similar role. The function of the class Ib RNR in E. coli is not clear, but it is expressed under iron-limited and oxidative stress conditions (2–4). The class Ia RNRs require a stable diferric-tyrosyl radical (FeIII2-Y•) cofactor generated from diferrous (FeII2) β2 in the presence of O2 and a reductant. This process occurs in vitro by self-assembly (5) and in vivo by a biosynthetic pathway (6). By contrast, the identity of the metallocofactor in β2 of the class Ib RNRs (NrdF) has been controversial (7–10). Recent studies, however, have shown in vitro that both FeIII2-Y• and dimanganese(III)-Y• (MnIII2-Y•) cofactors are active in nucleotide reduction (7). Whereas the FeIII2-Y• cofactor in NrdF can be formed by self-assembly from FeII and O2 (8, 9, 11), the MnIII2-Y• cofactor can be generated from MnII2-NrdF only in the presence of O2 and the flavodoxin-like protein NrdI (7).
We have recently shown that E. coli MnII2-NrdF can form a complex with NrdI in vitro and in vivo and have proposed that reduced NrdI (NrdIhq) in this complex generates hydroperoxyl anion (HO2−), which channels to the metal site in NrdF (7). Two equivalents of HO2− would be required to generate the MnIII2-Y• cofactor. In vitro support for this proposal is based on the observations that NrdI, in contrast to most flavodoxins, behaves as a two-electron reductant (11) and that neither H2O2 nor O2•− activates cofactor assembly (7, 8). Support in vivo is provided by the conservation of NrdI in all organisms containing a class Ib RNR and by genetic studies showing that NrdI is essential for Streptococcus pyogenes class Ib RNR activity (12). Sequence alignments suggest that the E. coli system is representative of most class Ib RNRs, including those from Corynebacterium ammoniagenes and the human pathogens Mycobacterium tuberculosis and S. pyogenes.
The formation of two different active cofactors, FeIII2-Y• and MnIII2-Y•, in NrdF in vitro using the same protein ligands (two His, three Glu, and one Asp) to coordinate the metals (13), but different oxidants, raises the question of how this process is controlled in vivo. These same metal ligands are involved in Fe ligation in the class Ia RNR (14), which cannot form an active MnIII2-Y• cofactor. Here we present the crystal structures of E. coli NrdF in the MnII2 and FeII2 forms, providing structural insight into how NrdF generates Y• from two different metallocofactors using two different oxidants. We also present structures of NrdF in complex with NrdIhq and oxidized NrdI (NrdIox). Reaction of the NrdIox-NrdF complex in the crystal with 100 mM dithionite and O2 results in trapping of a small molecule best modeled as a peroxide in a channel linking the NrdI flavin to the NrdF metal site, supporting the proposed model of MnIII2-Y• cofactor activation by oxidant channeling.
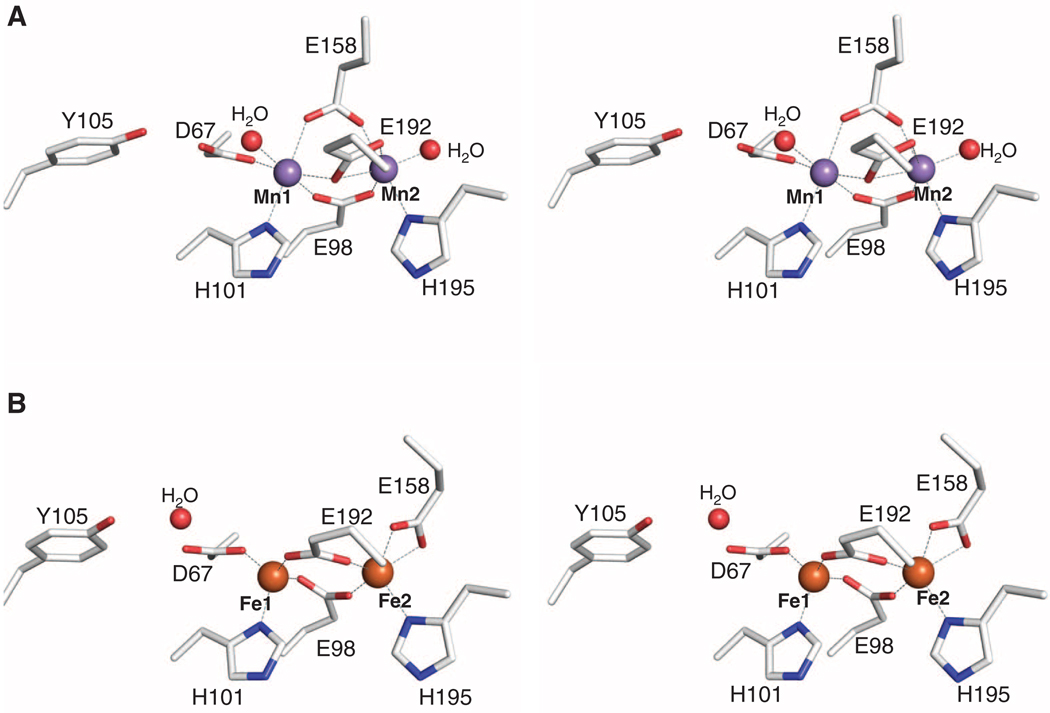
The 1.65 Å resolution (table S1) crystal structure (15) of MnII2-NrdF contains one monomer per asymmetric unit with the other half of the dimer related by crystallographic symmetry. The overall fold closely resembles that of other class I β2 subunits (16) (fig. S1). Anomalous diffraction data are consistent with the presence of two fully occupied MnII sites (Fig. 1A and fig. S2) with a Mn-Mn distance of 3.7 Å. Mn1 is coordinated by His101, Asp67, and a terminal water molecule, and Mn2 is coordinated by His195 and a terminal water molecule. Three glutamate residues (Glu98, Glu158, and Glu192) bridge the two metals in a manner previously not observed in RNRs and related carboxylate-bridged diiron enzymes (Fig. 1A and fig. S2) (17). Thus, each MnII is six-coordinate so ligand dissociation or reorganization, possibly via loss of the solvent molecule coordinated to Mn2 (17, 18), is necessary for reaction of the cluster with the oxidant. The non-coordinating side-chain oxygen atom of Asp67 is hydrogen bonded to the hydroxyl group of Tyr105, the site of the stable Y•. A similar interaction is observed in the E. coli class Ia FeII2 β2 structures (16). In the class Ib FeII2 β2 structures, however, the interaction between Asp67 and Tyr105 is mediated by a water molecule. As a result, the Mn1-Tyr OH distance is 5.8 Å as compared to the Fe1-Tyr OH distances of Salmonella typhimurium NrdF (6.4 to 7.0 Å) (18), C. ammoniagenes NrdF (6.2 to 6.7 Å) (13), and E. coli FeII2-NrdF (6.7 Å), which was determined to 1.9 Å resolution by soaking apo crystals of NrdF with FeII (table S1, Fig. 1B, and fig. S2) (19). The shorter Mn1-Tyr OH distance may be associated with the unusual electron paramagnetic resonance spectrum of the Mn-associated Y• relative to the Fe-associated Y• in NrdF (7).
Fig. 1.
Structures of MnII2-NrdF and FeII2-NrdF. (A) Stereoview of the MnII2-NrdF active site. MnII ions are shown as purple spheres, water molecules are shown as red spheres, and NrdF side chains are represented in stick format and colored by atom type. (B) Stereoview of the FeII2-NrdF active site. FeII ions, modeled at 0.5 occupancy, are shown as orange spheres. Metal-ligand interactions are highlighted with dashed lines.
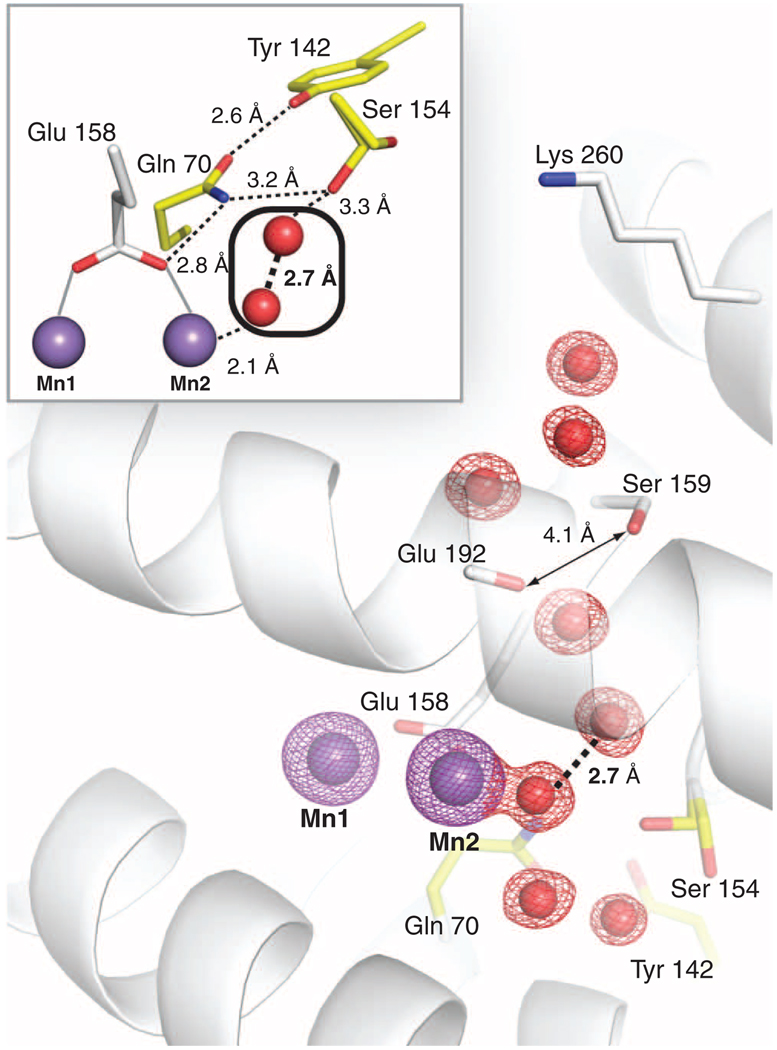
The two MnII ions are bridged by Glu98 in a µ-1,3 fashion and by Glu192 in a µ-(η1,η2) arrangement (Fig. 1A and fig. S2). The position of Glu192 is similar to that observed for the equivalent ligand in the E. coli class Ia β2 structure obtained by soaking FeII into apo crystals (14). The most notable difference between MnII2-NrdF and other β2 structures is the orientation of Glu158, which bridges the two MnII ions in a µ-1,3 mode, rather than coordinating Fe2 or Mn2 in a monodentate or bidentate fashion (Fig. 1 and fig. S2). Glu158 is in a short π-helical segment analogous to a conformationally flexible region observed in other β2s and diiron enzymes and hypothesized to dictate access to the active site (18, 20, 21). The space occupied by the Glu158 side chain in FeII2-NrdF is occupied by two solvent molecules in MnII2-NrdF: the coordinated water at Mn2 and a second water that links the coordinated water to the side chain of conserved residue Ser154 (Figs. 1 and 2). Ser154 is involved in a conserved hydrogen bonding network that connects Glu158 to a solvent channel from the protein surface (Fig. 2). The channel opening is located near conserved residue Lys260. This channel is better suited to allow access to the metal site by a peroxide oxidant than the channel present in class Ia β2 structures, which is more hydrophobic, smaller, and less solvent exposed, and therefore more appropriate for O2 passage (fig. S3). The ordered solvent and hydrogen bonding interactions may help constrain the unusual orientation of Glu158 in MnII2-NrdF. Moreover, the location of the two interacting solvent molecules at Mn2 could easily accommodate the proposed HO2− oxidant (22) (Fig. 2, inset). These waters may dissociate, allowing the oxidant to initially bind terminally to Mn2 in this position, by analogy to the proposal for H2O2 binding to the structurally related Mn catalases (23). By contrast, in both class Ia (fig. S3) and Ib FeII2 structures, including FeII2-NrdF (Fig. 1B),Glu158 and Phe162 create a hydrophobic pocket above the Fe2 site opposite His195 (E. coli NrdF numbering), an ideal destination for O2 before it reacts with the FeII2 site. This solvent channel likely represents the oxidant route to the metal cluster, and the environment near the active site may reflect different geometric or electrostatic requirements for correctly orienting O2 and the putative HO2− for reaction with their respective metal centers.
Fig. 2.
The MnII2-NrdF solvent-exposed active-site channel terminating at Mn2. A 2Fo − Fc electron density map (red mesh, contoured at 2σ) shows ordered waters in the channel. The Mn anomalous difference Fourier map (purple mesh, contoured at 12σ) is also shown. Residues implicated in channel access are shown as white sticks, and a conserved hydrogen bonding network (illustrated with dashed lines in inset) linking ordered solvent in the channel to Mn2 ligand Glu158 is shown as yellow sticks. Ser154 is modeled in two separate rotamer conformations in MnII2-NrdF, but in all NrdI-NrdF complex structures, it adopts the rotamer that points into the solvent channel.
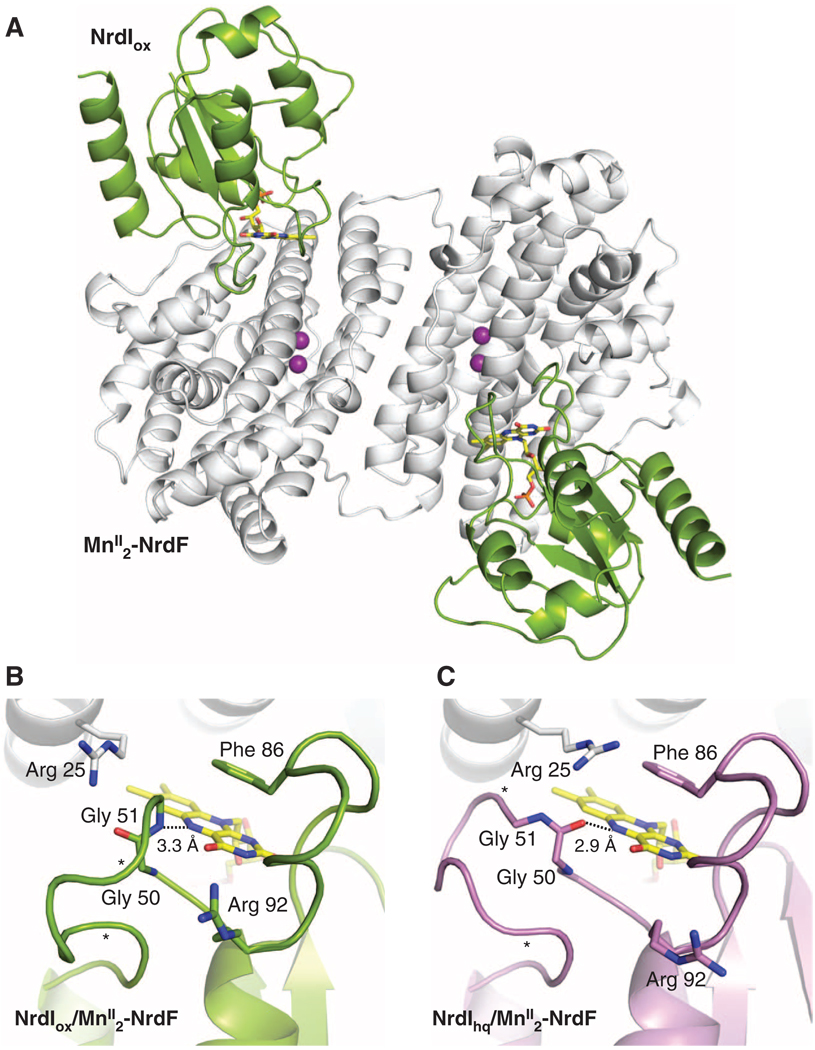
The complex between NrdIox and MnII2-NrdF (NrdIox-NrdF) was crystallized and its structure determined to 2.5 Å resolution (table S2). Two NrdI and two NrdF molecules are present in the asymmetric unit (Fig. 3A and fig. S4). One NrdI protein is bound to each NrdF opposite the dimer interface and directly over the solvent-exposed channel to the active site. The location of the C-terminal NrdF residue in this complex (fig. S4) suggests that the NrdI and α2 binding sites (24) are distinct. The NrdI-NrdF interface, which buries ~800 Å2 surface area per chain, is largely hydrophobic, with several interprotein hydrogen bonds (fig. S5). The overall fold of NrdF is the same as in the MnII2-NrdF structure, and the active site is nearly identical, including the unusual coordination mode of Glu158 (fig. S6). NrdI adopts a typical flavodoxin-like fold (25), with the isoalloxazine ring of the flavin mononucleotide (FMN) cofactor near the protein surface and enclosed by two loop regions (Fig. 3B). One of these loops provides the closest positive charge on NrdI to the reactive C4a position of the flavin (Arg92), whereas the other, the glycine-rich “50s loop,” interacts with the N5 position and displays pronounced redox-dependent conformational changes (see below). The 50s loop comprises residues 50 to 56 (Gly4-Thr-Ala-Gly), and as predicted (11), the amide nitrogen atom of Gly51 is within hydrogen bonding distance of the flavin N5 atom, similar to what is observed in oxidized long-chain flavodoxins (26). The electron density for the NrdI 50s loop is not completely continuous, suggesting conformational flexibility.
Fig. 3.
Structures of NrdI-NrdF protein-protein complexes. (A) A ribbon diagram of the NrdIox-NrdF structure. NrdI is shown in green and MnII2-NrdF is shown in white. The NrdI FMN cofactor is shown as yellow sticks. (B) The NrdI FMN environment in the NrdIox-NrdF structure (NrdI shown in green). (C) The NrdI FMN environment in the NrdIhq-NrdF structure (NrdI shown in purple). Hydrogen bonding interactions with the FMN N5 position are shown as dashed lines. The electron density for the 50s loop is not completely continuous between the asterisks.
The structure of the NrdIox-NrdF complex reveals how NrdF contributes to the electrostatic environment of the FMN binding pocket. Typical flavodoxins carry out single electron transfers partly through destabilization of the reduced FMN, bound in the anionic form (FMNH−, protonated at N5 and deprotonated at N1), by acidic residues proximal to the isoalloxazine ring (27). The FMN environment in NrdI is more positively charged (Arg92 and Arg108), and in complex with NrdF, three additional charged residues from NrdF (Lys18, Arg25, and Arg190) are located within 12 Å of the FMN C4a position. The presence of positive charges near the O2-reactive C4a position is a conserved feature of flavoprotein oxidases, which reduce O2 to H2O2 as part of their catalytic cycle (28), suggesting that electrostatics may play a similar role in NrdI to favor two-electron reduction of O2 to a peroxide oxidant.
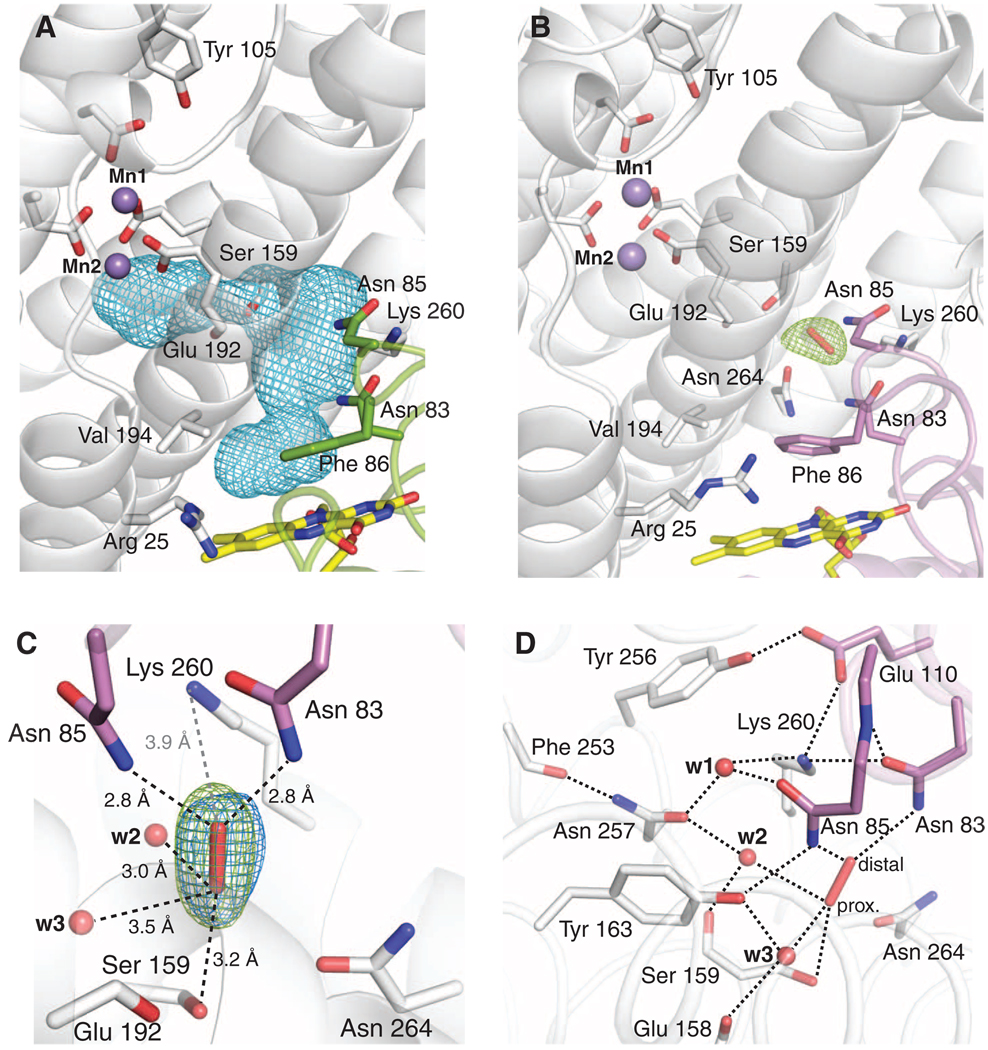
Complex formation between NrdI and NrdF results in a 10 Å extension of the NrdF active-site channel along the NrdI-NrdF interface toward the flavin ring (Fig. 4A). Solvent access to the channel is prevented by NrdI Phe86 and other hydrophobic and bulky residues near the flavin. Like the portion of the channel within NrdF, the interfacial region is lined with polar uncharged residues and backbone atoms. The side chains of NrdI residues Asn83 (completely conserved) and Asn85 (largely conserved) point into the channel. The position of the highly conserved Lys260 in NrdF enforces a sharp turn in the channel, leading directly to the MnII2 site. Lys260 is involved in a hydrogen bond network with the strictly conserved residues Tyr256 and NrdI Glu110 (figs. S5 and S7). The NrdF portion of the channel is lined by the side chains of Ser159, Tyr197, and Asn264 (figs. S7 and S8). Approximately 7 Å from the Mn2 site, the channel constricts to 4.1 Å (Figs. 2 and 4). This constriction is formed by the carbonyl oxygen of bridging ligand Glu192 and the side chain of Ser159, which follows in sequence space the unusual bridging ligand Glu158. Thus, the interactions that define this narrow point of the channel are intimately connected to theMnII2 coordination sphere.
Fig. 4.
The NrdI-NrdF channel. (A) NrdI-NrdF complex formation extends the NrdF active-site channel to the FMN cofactor. The complex channel is depicted as a light blue mesh and was calculated using a 1.4 Å probe radius. Selected NrdI (green) and NrdF (white) residues lining the channel are shown as sticks. (B) Observation of a trapped species, best modeled as peroxide, in the NrdI-NrdF channel in a crystal reduced by dithionite in the presence of oxygen (NrdIhq-NrdFperox). Strong Fo − Fc electron density (green mesh, contoured at 3.3σ) is present in the channel after the first refinement cycle. The FMN cofactor (yellow), NrdI side chains lining the channel (purple), NrdF residues in the channel and at the active site (white), and the peroxide (red) are all shown as sticks. (C) A magnified view of the modeled peroxide shown in Fig. 3B and hydrogen bonding interactions with residues and solvent in the channel. The final 2Fo − Fc electron density (blue mesh, contoured at 1.8σ) is superimposed on the initial Fo − Fc electron density map from Fig. 3A. Water molecules are shown as red spheres. Dashed black lines indicate potential hydrogen bonding interactions. The gray dashed line represents the distance between the modeled peroxide and the nearest charged residue, conserved NrdF residue Lys260. The Glu192 backbone carbonyl group and the side chain of Ser159 constitute the narrowest point of the active-site channel. The oxygen atom distal to the MnII2 site interacts strongly with the side chains of NrdI residues Asn85 (2.8 Å) and conserved Asn83 (2.8 Å). (D) The extended hydrogen bonding network near the putative peroxide binding site. The side-chain orientations of Asn83 and Asn257 can be assigned unequivocally based on their interactions with Lys260 and the backbone amide nitrogen of Asn85, and the carbonyl oxygen of Phe253, respectively. The interactions of w2 with Asn257 (2.8 Å) and the backbone carbonyl of Ser159 (2.7 Å) constrain w2 to act as a hydrogen bond acceptor for the proximal oxygen atom of the modeled peroxide (2.9 Å), suggesting that this oxygen is protonated. The distal oxygen accepts two hydrogen bonds from Asn83 and Asn85. Because no other potential hydrogen bond donor or acceptor exists for this oxygen atom, its protonation state cannot be determined from this analysis.
As a first step toward investigating the NrdI-mediated activation of the NrdF metallocofactor on the molecular level, we determined two structures of reduced NrdI in complex with NrdF (table S2). For the first structure (NrdIhq-NrdF, 2.0 Å resolution), crystals were grown in the presence of 3 mM dithionite in an anaerobic chamber. These crystals were colorless, indicating that the flavin was reduced. For the second structure, which reveals a trapped species best modeled as peroxide (NrdIhq-NrdFperox, 2.35 Å resolution), NrdIox-NrdF crystals were soaked in 100 mM dithionite outside the anaerobic chamber until the bright yellow color bleached (~2 min). Both structures reveal conformational changes near the FMNH− cofactor suggesting reduction of the flavin (Fig. 3C and fig. S9). The most pronounced change involves the NrdI 50s loop, which adopts a more open conformation exposing the isoalloxazine ring to solvent (Fig. 3C). Protonation of the flavin N5 concomitant with reduction breaks the N5 hydrogen bond with the amide nitrogen of Gly51, and the carbonyl oxygen of Gly50 is positioned to accept a hydrogen bond from the FMNH− N5H. In addition, NrdF Arg25 is oriented closer to C4a (6 Å) compared with the NrdIox-NrdF complex, and in one NrdF monomer, it is hydrogen bonded to the backbone carbonyl of NrdI Gly50, perhaps locking in place the orientation of the 50s loop. The proximity of a positively charged residue to the reduced FMN may be important in its reaction with O2 and is consistent with the unusual redox properties of NrdI (29). As in the NrdIox-NrdF structure (Fig. 4A), there is no clear solvent access route to the channel. The NrdIhq-NrdF structure reveals little conformational change within the complex when compared with NrdIox-NrdF. However, a highly ordered water network is evident within the channel (fig. S8) that extends along the NrdI-NrdF interface to the NrdI FMN cofactor.
Surprisingly, strong (~7σ) unexplained electron density (Fig. 4B) was observed in both active-site channels of the structure obtained from aerobically dithionite-soaked crystals (NrdIhq-NrdFperox). This density is oblong in shape (Fig. 4C), and a number of small molecules have been modeled in an effort to determine its identity (fig. S10). Modeling as a single water molecule results in strong difference electron density (4.5σ), consistent with a diatomic species. Modeling as dioxygen yields difference density at the edges, suggesting that the O-O bond distance is longer than 1.2 Å. Less difference density was evident with superoxide (O-O bond distance 1.34 Å), with the best fit being a fully occupied peroxide species with an O-O bond distance of 1.47 Å. Given that the crystals were exposed to 100 mM dithionite, we also considered the possibility that bisulfite, a dithionite breakdown product, could account for the density. Modeling with bisulfite (fig. S10E) yields Fo − Fc difference density and high B factors, however. Therefore, we modeled the electron density as a peroxide. Peroxide could have been produced by reaction of O2 with NrdIhq or with the dithionite used to reduce NrdIox (30, 31).
The putative peroxide species is lodged at the bend in the active-site channel ~10 Å from the FMN isoalloxazine ring and ~10 Å from the Mn2 site (Fig. 4B). It is within hydrogen bonding distance of residues from both NrdF and NrdI, as well as solvent molecules in the channel (Fig. 4, C and D). The side chain of the single charged residue in the channel, NrdF Lys260, is 3.9 Å from the distal peroxide oxygen atom, consistent with a role in electrostatic attraction, but not close enough for protonation. It is also protected from protonation by Tyr163 by an intervening solvent molecule. The modeled peroxide is involved in an extensive hydrogen bonding network in which the proton donors and acceptors can be inferred (Fig. 4D and figs. S11 and S12). The structure suggests that the proximal oxygen atom is protonated, consistent with either H2O2 or HO2− (32). The presence of the trapped species supports the relevance of the channel for oxidant transport.
The structures of NrdIox-NrdF and NrdIhq-NrdF show that complexation results in continuation of the NrdF channel to the NrdI FMN cofactor, delineating an obvious path from the NrdI site of oxidant formation to Mn2. The NrdIhq-NrdFperox structure offers a crystallographic snapshot of a putative peroxide species within the channel. The lack of conformational changes at the MnII2 site in all three NrdI-NrdF complexes, when compared to MnII2-NrdF, suggests that the requirement for NrdI for MnII2-NrdF activation, if structural, is subtle. The structures strongly support the proposed mechanism of MnII2-NrdF activation by NrdIhq and O2 and afford new insight into how reactive small molecules are channeled within complex enzyme systems.
Supplementary Material
Footnotes
Note added in proof: In support of the in vivo relevance of our work, Auling, Lubitz, and co-workers have recently reported isolation of C. ammoniagenes NrdF from the native organism containing a MnIII2-Y• cofactor, characterization of the cofactor in detail by EPR spectroscopy, and structural determination of the protein to 1.36 Å resolution (33). Although the crystallized protein contained a MnIII2 cluster, the oxidation state of the observed Mn ions remains an open question because of possible photoreduction during data collection.
Supporting Online Material
www.sciencemag.org/cgi/content/full/science.1190187/DC1
Materials and Methods
Figs. S1 to S14
Tables S1 and S2
References
References and Notes
- 1.Nordlund P, Reichard P. Annu. Rev. Biochem. 2006;75:681. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 2.Gon S, Faulkner MJ, Beckwith J. Antioxid. Redox Signal. 2006;8:735. doi: 10.1089/ars.2006.8.735. [DOI] [PubMed] [Google Scholar]
- 3.McHugh JP, et al. J. Biol. Chem. 2003;278:29478. doi: 10.1074/jbc.M303381200. [DOI] [PubMed] [Google Scholar]
- 4.Monje-Casas F, Jurado J, Prieto-Alamo MJ, Holmgren A, Pueyo C. J. Biol. Chem. 2001;276:18031. doi: 10.1074/jbc.M011728200. [DOI] [PubMed] [Google Scholar]
- 5.Atkin CL, Thelander L, Reichard P, Lang G. J. Biol. Chem. 1973;248:7464. [PubMed] [Google Scholar]
- 6.Wu CH, Jiang W, Krebs C, Stubbe J. Biochemistry. 2007;46:11577. doi: 10.1021/bi7012454. [DOI] [PubMed] [Google Scholar]
- 7.Cotruvo JA, Jr, Stubbe J. Biochemistry. 2010;49:1297. doi: 10.1021/bi902106n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huque Y, et al. J. Biol. Chem. 2000;275:25365. doi: 10.1074/jbc.M002751200. [DOI] [PubMed] [Google Scholar]
- 9.Jordan A, et al. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12892. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willing A, Follmann H, Auling G. Eur. J. Biochem. 1988;170:603. doi: 10.1111/j.1432-1033.1988.tb13740.x. [DOI] [PubMed] [Google Scholar]
- 11.Cotruvo JA, Jr, Stubbe J. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14383. doi: 10.1073/pnas.0807348105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roca I, Torrents E, Sahlin M, Gibert I, Sjöberg BM. J. Bacteriol. 2008;190:4849. doi: 10.1128/JB.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hölgbom M, Huque Y, Sjöberg B-M, Nordlund P. Biochemistry. 2002;41:1381. doi: 10.1021/bi011429l. [DOI] [PubMed] [Google Scholar]
- 14.Voegtli WC, et al. J. Am. Chem. Soc. 2003;125:15822. doi: 10.1021/ja0370387. [DOI] [PubMed] [Google Scholar]
- 15.See supporting material on Science Online
- 16.Eklund H, Uhlin U, Färnegårdh M, Logan DT, Nordlund P. Prog. Biophys. Mol. Biol. 2001;77:177. doi: 10.1016/s0079-6107(01)00014-1. [DOI] [PubMed] [Google Scholar]
- 17.The metal site in the reported structure of the MnII2 form of C. ammoniagenes NrdF (13) differs from the E. coli MnII2-NrdF structure and is more similar to the FeII2-NrdF structures. Given that the C. ammoniagenes protein contained 1 Fe/β2 as compared to 0.03 Fe/β2 for the E. coli protein and the high sequence identity between C. ammoniagenes and E. coli NrdFs (66%), it is possible that the crystallized C. ammoniagenes protein did not contain two fully occupied MnII sites. Additionally, the presence of two coordinated solvent molecules in a MnII2 or FeII2 β2 as observed in E. coli MnII2-NrdF is unusual. A single solvent molecule is coordinated to the second metal site (Mn2 or Fe2) in class Ia E. coli MnII2-substituted β2 and class Ib S. typhimurium and M. tuberculosis FeII2-NrdF (18, 34, 35). However, the solvent molecule in the class Ib NrdF structures likely derives from preparation of FeII2-NrdF by chemical or photoreduction of the µ-oxo bridged diferric cluster.
- 18.Eriksson M, Jordan A, Eklund H. Biochemistry. 1998;37:13359. doi: 10.1021/bi981380s. [DOI] [PubMed] [Google Scholar]
- 19.The E. coli FeII2-NrdF structure was obtained by crystallization of apo-NrdF followed by soaking with FeII ions in cryoprotectant solution. Nonheme FeII2 protein structures are routinely obtained in a similar manner (13, 14) or by crystallizing the FeIII2 form followed by chemical or photoreduction (18). As with the MnII2-NrdF structure reported here, FeII2-NrdF crystallized with one NrdF monomer per asymmetric unit with the second monomer in the functional β2 subunit related by crystallographic symmetry. Although a fully occupied class Ia FeII2-β2 structure was obtained by this method (14), the FeII sites in the FeII2-NrdF structure reported here are best modeled at 0.5 occupancy. Consequently, the model may represent an average of apo, partially loaded, and fully loaded states. The C. ammoniagenes FeII2-NrdF structure (13), obtained similarly, contains two β2 dimers in the asymmetric unit with the FeII2 sites fully occupied in one of the four NrdF molecules with 0.3 to 0.6 occupancy for the other three FeII2 sites. In the immediate vicinity of the metal site, the E. coli FeII2-NrdF structure is identical to that of the fully occupied C. ammoniagenes FeII2-NrdF subunit. This observation supports the conclusion that the unusual features of the E. coli MnII2-NrdF coordination environment result from MnII in the active site and are not an anomaly of the E. coli NrdF protein.
- 20.Sazinsky MH, Lippard SJ. J. Am. Chem. Soc. 2005;127:5814. doi: 10.1021/ja044099b. [DOI] [PubMed] [Google Scholar]
- 21.Sazinsky MH, Lippard SJ. Acc. Chem. Res. 2006;39:558. doi: 10.1021/ar030204v. [DOI] [PubMed] [Google Scholar]
- 22.The overall charge at the MnII2 site is neutral based on the structure if the Mn-bound solvent molecules are H2O. If H2O2 is the oxidant, the charge neutrality of the cluster would be conserved, with a proton being shuttled to one of the ligands upon oxidant binding to Mn2 (36). If the oxidant is HO2−, there is no obvious proton source near the active site, which may or may not be required to compensate for HO2− binding.
- 23.Barynin VV, et al. Structure. 2001;9:725. doi: 10.1016/s0969-2126(01)00628-1. [DOI] [PubMed] [Google Scholar]
- 24.Uppsten M, Färnegårdh M, Domkin V, Uhlin U. J. Mol. Biol. 2006;359:365. doi: 10.1016/j.jmb.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig ML, Luschinsky CL. In: Chemistry and Biochemistry of Flavoenzymes. Müller F, editor. Boca Raton, FL: CRC Press; 1992. pp. 427–466. [Google Scholar]
- 26.Drennan CL, et al. J. Mol. Biol. 1999;294:711. doi: 10.1006/jmbi.1999.3151. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z, Swenson RP. Biochemistry. 1995;34:3183. doi: 10.1021/bi00010a007. [DOI] [PubMed] [Google Scholar]
- 28.Mattevi A. Trends Biochem. Sci. 2006;31:276. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Recent structures of Bacillus cereus NrdI in the absence of NrdF exhibit much less drastic conformational changes of the 50s loop upon reduction (37). Although most NrdI homologs possess 50s loops of 7 to 13 residues, typified by E. coli NrdI, some contain a short, three-residue 50s loop (usually Gly-Phe-Gly), typified by B. cereus NrdI. The shorter, less flexible 50s loop likely contributes to the more flavodoxin-like redox properties of B. cereus NrdI, and sequence alignments also reveal that the NrdFs from organisms containing NrdIs with short 50s loops (mainly Bacillus and Staphylococcus) also contain Gln at the equivalent position of Arg25 in E. coli. Both of these characteristics suggest that NrdI may play a different role in cofactor biosynthesis and maintenance in the former class Ib systems than in E. coli (6, 11).
- 30.Dixon M. Biochim. Biophys. Acta. 1971;226:241. doi: 10.1016/0005-2728(71)90092-2. [DOI] [PubMed] [Google Scholar]
- 31.Reaction of dithionite with O2 can also produce O2•−, but the electron density observed is unlikely to be this species. Analysis of the surrounding hydrogen bonding network (Fig. 4D) strongly suggests that at least one oxygen atom is protonated, and trapping of HO2 is unlikely (pKa ~ 4.8). Similarly, we have demonstrated (7) that reaction of NrdIhq with O2 does not generate appreciable amounts of O2•−. Because high concentrations of dithionite (100 mM) were required to bleach the crystals, we also considered the possibility that the density derives from dithionite itself. Anaerobic soaking with 100 mM dithionite does not produce electron density at this position, however (fig. S14).
- 32.Whereas connection of the channel to bulk solvent via a hydrogen bonding network would result in rapid protonation of HO2− through a proton conduction mechanism, analysis of the hydrogen bonding interactions at possible solvent access routes to the channel suggests that no direct connection exists (figs. S7 and S13). Thus, the structural analysis cannot distinguish between HO2− and H2O2.
- 33.Cox N, et al. J. Am. Chem. Soc. 2010;132:11197. doi: 10.1021/ja1036995. [DOI] [PubMed] [Google Scholar]
- 34.Atta M, Nordlund P, Aberg A, Eklund H, Fontecave M. J. Biol. Chem. 1992;267:20682. [PubMed] [Google Scholar]
- 35.Uppsten M, Davis J, Rubin H, Uhlin U. FEBS Lett. 2004;569:117. doi: 10.1016/j.febslet.2004.05.059. [DOI] [PubMed] [Google Scholar]
- 36.Do LH, Hayashi T, Moënne-Loccoz P, Lippard SJ. J. Am. Chem. Soc. 2010;132:1273. doi: 10.1021/ja909718f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Røhr AK, Hersleth H-P, Andersson KK. Angew. Chem. Int. Ed. 2010;49:2324. doi: 10.1002/anie.200907143. [DOI] [PubMed] [Google Scholar]
- 38.This work was supported by NIH grants GM58518 (A.C.R.) and GM81393 (J.S.), American Cancer Society grant PF-10-148-01-DMC (A.K.B.), and a National Defense Science and Engineering Graduate fellowship to J.A.C. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the Life Sciences Collaborative Access Team (LS-CAT) Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). Some data were collected at General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT), which has been funded in whole or in part with federal funds from the National Cancer Institute (grant Y1-CO-1020) and the National Institute of General Medical Science (grant Y1-GM-1104). We thank Z. Wawrzak for assistance with data processing and S. Lippard for helpful discussions. Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 3N37 (MnII2-NrdF), 3N38 (FeII2-NrdF), 3N39 (NrdIox-NrdF), 3N3A (NrdIhq-NrdF), and 3N3B (NrdIhq-NrdFperox).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.