Abstract
Fast and accurate replication of DNA is accomplished by the interactions of multiple proteins in the dynamic DNA replisome. The DNA replisome effectively coordinates the leading and lagging strand synthesis of DNA. These complex, yet elegantly organized, molecular machines have been studied extensively by kinetic and structural methods to provide an in-depth understanding of the mechanism of DNA replication. Owing to averaging of observables, unique dynamic information of the biochemical pathways and reactions are concealed in conventional ensemble methods. However, recent advances in the rapidly expanding field of single-molecule analyses to study single biomolecules offer opportunities to probe and understand the dynamic processes involved in large biomolecular complexes such as replisomes. This review will focus on the recent developments in the biochemistry and biophysics of DNA replication employing single-molecule techniques and the insights provided by these methods towards a better understanding of the intricate mechanisms of DNA replication.
Keywords: DNA replication, Polymerase, Replisome, Single-molecule Kinetics
[A] Introduction
Cell division in every organism involves the faithful duplication of its genetic information. The accurate replication of the genome is catalyzed by DNA polymerases aided by a multitude of accessory proteins. Together these proteins constitute the DNA replisome that is responsible for the rapid and accurate replication of genomic DNA [1, 2]. The replisome is conserved throughout all forms of life from viruses to higher order eukaryotes.
The DNA polymerase is a DNA-dependent polymerase that inserts deoxynucleotides in a 5′ to 3′ direction. Due to the bipolar nature of the duplex DNA (dsDNA), replication of DNA occurs simultaneously in a bidirectional manner and involves the coordinated synthesis of DNA strands with opposite polarity. As a result, synthesis of the leading strand is continuous whereas the lagging strand synthesis is discontinuous. The discontinuous nature of lagging strand synthesis generates shorter lengths of DNA called Okazaki fragments that are 1–3 kb long. Accessory factors such as the sliding clamp and clamp loading complexes not only increase the processivity of DNA polymerases but also assist in the coupling of the leading and lagging strand syntheses. The high processivity of DNA polymerases has been attributed to the clamp/clamp-loader complexes, where the processivity clamp is tethered to the polymerase to form the holoenzyme extending the interaction of the polymerase with DNA. Primosomes, a subcomplex of replisomes primarily composed of replicative helicases and primases, play an essential role in the progress of the replication fork by catalyzing DNA duplex unwinding and oligonucleotide primer synthesis necessary for Okazaki fragment formation on the lagging strand by helicases and primases, respectively. Another important protein involved in the replication of DNA is single-stranded DNA (ssDNA) binding protein. SsDNA binding proteins bind with high affinity to the ssDNA generated during various DNA metabolic processes such as DNA replication, repair and recombination [3]. These proteins are essential for DNA replication and are not only involved in coating the ssDNA generated during DNA replication, but are also involved in interactions with replication enzymes such as polymerases, primases and helicases and often stimulate their functions [3].
Ensemble studies of DNA polymerases have provided a great deal of information on the kinetic mechanism of the polymerization reaction. Extensive investigations of holoenzyme and ultimately replisome assembly have determined the order of addition of the accessory proteins necessary to form an active complex and led to the formulation of detailed kinetic models for holoenzyme and primosome assembly and the replisome [4]. These studies have been complemented by structural information obtained for the apo-form and various DNA polymerase complexes as well as accessory proteins [5–7]. In recent years, the rapidly growing field of single-molecule enzymology has paved the way for the analysis of details of DNA replication that cannot otherwise be determined.
Single-molecule techniques have been valuable tools in deciphering protein assemblies and the function of macromolecular complexes. Unlike ensemble methods that require synchronization and monitor average behavior, single-molecule techniques require no such synchronization [8]. This presents an advantage for detecting any transient intermediates involved in a reaction pathway, thus facilitating the investigation of enzymatic reaction mechanism manifolds [9]. Furthermore, analysis of a single time-trajectory can yield rate constants for conformational changes in both the forward and reverse directions [10]. Single-molecule methods have also been instrumental in analyzing the action of molecular motors such as kinesin [11], myosin [12] and helicases [13]. These approaches have also been employed to understand the assembly of the holoenzyme complex and the dynamic interactions between the proteins of the replisome. This review will focus on the recent advances in the field of DNA replication using single-molecule techniques to deduce and clarify the behavior of the replisome during DNA synthesis.
[B] Components of the replisome
As discussed above, several components are required to accomplish fast, accurate and coordinated DNA replication. A brief summary of the proteins that constitute the well-characterized replisome of three species: T7 bacteriophage, T4 bacteriophage and Escherichia coli and their respective functions within the replisome is presented below. The replisome architecture of higher order organisms such as Sacchromyces cerevisiae and Homo sapiens is still widely debated, because the identity of the replisome proteins is yet unambiguously defined. However, a brief discussion of the eukaryotic replisome will be presented in a context to compare conserved functions across replisomes and the corresponding accessory proteins.
The replisome of T7 phage
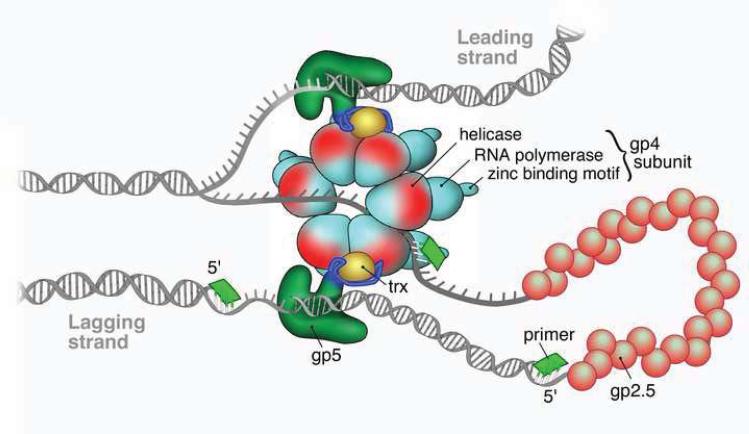
Bacteriophage T7 is the simplest of the three systems with its replisome composed of a polymerase (gp5), helicase/primase (gp4) and ssDNA binding protein (gp2.5) all encoded by the T7 genome. A schematic representation of the replisome of T7 phage is shown in Figure 1. The processivity factor thioredoxin (Trx) necessary for an efficient DNA synthesis is encoded by the E. coli host. The polymerase gp5 and the processivity factor Trx form a 1:1 stochiometric complex. In the absence of the processivity clamp, Trx, gp5 polymerase is a distributive enzyme incorporating 5–15 nucleotides before dissociation from the primer-template (P–T) junction. However, the presence of Trx increases the processivity by thousands of nucleotides in a single binding event [14]. Crystallographic structural information has revealed that the structure of the gp5-Trx complex resembles other polymerase structures with a palm, fingers and thumb of a right hand shape [15].
Figure 1.
Schematic representation of the T7 replisome (Courtesy of Nathan Tanner).
The primase and the helicase activities are carried out by the T7 gene 4 protein. The primase activity of gp4, responsible for the synthesis of tetraribonucleotide primers that are required for the lagging strand synthesis, is situated in the amino-terminus of the protein. The primase recognizes the DNA sequence 5′-GGGTC-3′, 5′-GTGTC-3′ or 5′-TGGTC-3′ and synthesizes the corresponding tetraribonucleotide primers, where the 3′- C is involved only in the recognition of the priming sequence, but is not copied by the primase [16–18]. The primase domain of gp4 is composed of a zinc-binding domain (ZBD) important for the recognition of the priming site and an RNA polymerization domain (RPD) that is the catalytic domain for the synthesis of ribonucleotide primers [19]. The helicase activity of gp4 responsible for unwinding the duplex DNA for the progression of replication fork is located at the carboxy-terminus of the protein. The functional form of helicase/primase gp4 is assembled as a hexameric ring as shown in Figure 1, which is loaded onto the lagging strand and unwinds the duplex DNA in a 5′ to 3′ direction. In the absence of the primase/helicase gp4, the polymerase is incapable of strand displacement synthesis. However, upon introduction of gp4, the rate of DNA synthesis by gp5 increases dramaticaly. The nascent ssDNA generated during the unwinding of duplex DNA by gp4 helicase translocating on the lagging strand is immediately coated with gp2.5, the ssDNA binding protein of T7 preventing the exposed ssDNA from nucleolytic degradation. Furthermore, it has been demonstrated that gp2.5 binds to gp4 and acts as a modulator of a multitude of its functions [20]. Although gp2.5 interacts with gp4, it does not have any direct effect on the DNA-dependent dTTP hydrolysis that is required for the helicase activity. The rate of polymerization by gp5 and the effect of accessory proteins on the polymerization reaction and processivity are listed in Table 1.
Table 1.
Rate of polymerization and processivities obtained from ensemble and single-molecule measurements:
| Ensemble studies | Single-molecule studies | |||
|---|---|---|---|---|
| Protein Complex | Rate of reaction (nt/s) | Processivity (kb) | Rate of reaction (nt/s) | Processivity (kb) |
| Phage T7: | ||||
| Gp4 | 15 [86] | 0.06 [86] | 29 (5.2 pN) [85] | 0.343 (5.2 pN) [85] |
| Gp5 | - | 0.015 [93] | 210 (exo-) [48] | - |
| Gp5+Trx | 110 [93] | 10 [93] | 100 [47] | 0.42 (15 pN) [47] |
| Gp5+Trx+gp4 | 120 [92] | - | 164 [52] | 17 [52] |
| Gp5+Trx+gp4+gp2.5 | 300 [105] | 10 [105] | 75 (22 °C) [61] 141 (37 °C)[61] |
25 ± 17 [61] 51 ± 9 [61] |
| Phage T4: | ||||
| Gp41 | 30 [106] | 0.65 [107] | 30 [49] | 0.8 [49] |
| Gp43 | 250 [108] | 0.85 [108] | - | - |
| Gp43+gp45 | 600 [108, 109] | 3 [108, 109] | - | - |
| Gp43+gp45+gp41+gp61+gp32 | 250 [108, 109] | > 20 [109] | - | - |
| E. coli: | ||||
| DnaB | 291 [110] | - | - | - |
| Klenow Fragment | 50 [67] | 0.188 [67] | 13 [48] | - |
| Pol III | 20 [27] | 0.01 [27] | - | - |
| Pol III + β | 750 [102, 103] | 1 [103] | 347 [51] | 1.40 [51] |
| Pol III+β+DnaB | - | - | 417 [51] | 10.5 [51] |
| Pol III + β+DnaG | - | - | 340 [51] | 2.9 [51] |
| Pol III+β+DnaB+SSB | 750 [101, 102] | >50 [101, 102] | 535 [61] | 85 [61] |
The replisome of T4 phage
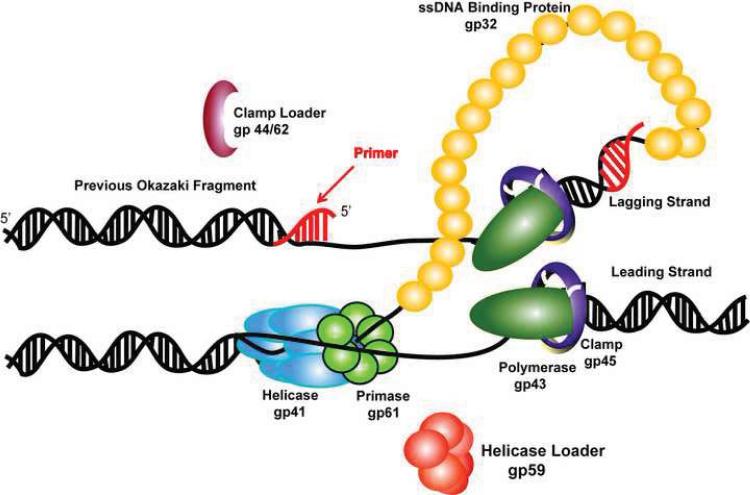
The bacteriophage T4 system provides all the proteins, except for the host cell RNA polymerase, necessary for the replication and repair of the 169-kb linear genome. It codes for the eight proteins necessary for DNA replication namely DNA polymerase (gp43), clamp (gp45), clamp loader (gp44/62), helicase (gp41), helicase loader (gp59), primase (gp61), and ssDNA binding protein (gp32). The assembly of the T4 replisome mediating the coordinated DNA synthesis is shown in Figure 2; the activity and processivity for the proteins and protein complexes are listed in Table 1.Gp43 is the replicative polymerase, which harbors 5′ to 3′ polymerase and 3′ to 5′ exonuclease proof reading activities. Like gp5 in the T7 replisome, gp43 extends both leading and lagging strand DNA. Although a crystal structure of gp43 from T4 has not been reported, the RB69 polymerase, which shares > 60% amino acid sequence identity with the T4 polymerase, has been crystallized as an apo-form, a binary complex with P-T, and a ternary complex with P-T and an incoming nucleotide [21–23]. These X-ray structures, in conjunction with kinetic studies of gp43, suggest that gp43 adapts the induced-fit mechanism to synthesize the DNA chain.
Figure 2.
Model of the replication fork of the T4 replisome.
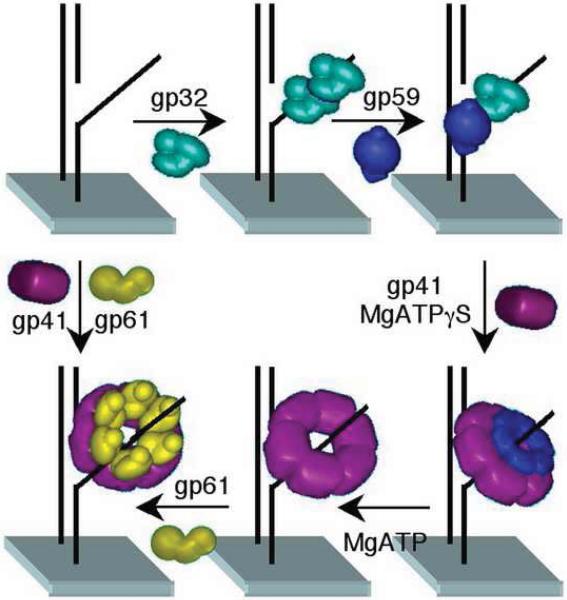
By itself gp43 is distributive and would not readily accomplish the genomic DNA replication. To achieve fast and accurate genomic synthesis, assembly of the T4 replisome is indispensable. When an origin of replication fork opens and a D-loop forms, gp32 binds to the transiently exposed ssDNA and the helicase loader gp59 is recruited to the site. Gp32-gp59 binary complexes encircle the ssDNA to form a helicase-loading complex (HLC). The sliding clamp gp45 is then loaded at the P-T junction by the clamp-loader complex gp44/gp62. The polymerase gp43 displaces the clamp loader, resulting in the formation of a holoenzyme that is inhibited by the interaction between gp59 and gp43 [24]. A homodimer of gp41 helicase is recruited to the replication site by HLC and is expanded to form a hexameric ring. Leading strand synthesis starts when gp41 unwinds dsDNA and releases the inhibition of gp43 by gp59. The lagging strand replication is initiated by the helicase-primase complex, formed by gp41 and gp61.
The sliding clamp protein gp45 plays an important role in stimulating the activity of gp43. The crystal structure of gp45 revealed that the homotrimeric gp45 forms a hollow circle with an inner diameter ca. 35 Å, large enough to accommodate duplex DNA [25]. The sliding clamp is loaded onto a primed template by the clamp loader composed of four gp44 subunits and one gp62 subunit.
The T4 primase, gp61, catalyzes the short RNA segment synthesis used to initiate leading strand replication and Okazaki fragment synthesis along the lagging strand. The priming recognition sequence is 5′-GTT-3′ in vivo; however, 5′-GCT-3′ is also recognized in vitro. At high concentrations, gp61 alone synthesizes short primers with varied lengths (mostly dimer and a small amount of 5 – 45 nt). The addition of gp41 to form the primosome shifts the preferred recognition sequence from 5′-GCT-3′ by gp61 alone to 5′-GTT-3′, greatly increases the overall primer synthesis rate, and favors the formation of 5nt RNA primers [26]. Given appropriate amounts of gp32, gp41 and gp59, each primosome produces one primer per second, which is sufficient for DNA replication in vivo [26].
The replisome of E. coli
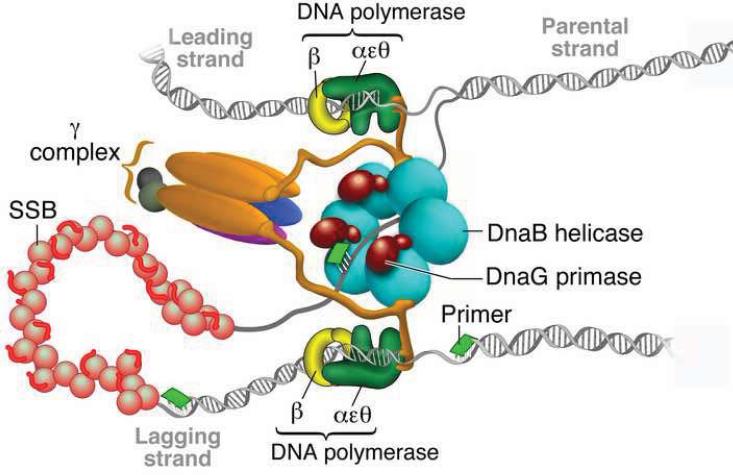
The organization of the E. coli replisome essential for the coordinated synthesis of DNA is represented in Figure 3. It requires more proteins to assemble the E. coli replisome compared with bacteriophage replisomes, although the proteins represent functional analogs found in T4: Pol III is the replicative polymerase; DnaB encodes the helicase; primase is the product of DnaG; β-clamp functions as the sliding clamp that is loaded onto a P-T junction by the clamp loader γ-complex; and SSB is the ssDNA binding protein. The holoenzyme of Pol III consists of ten subunits, including an isolable core polymerase composed of α, ε, and θ, among which α is the polymerase subunit and the ε subunit harbors an exonuclease activity. Both the α subunit and the core polymerase are poorly active, incorporating 8 and 20 nt/s respectively, and extend less than 10 bases per binding event [27]. The combination with β-clamp makes Pol III more active (750 nt/s) and processive (>50 kb). E. coli DNA primase (DnaG) initiates RNA primer synthesis on the recognition sequences 5′-CTG-3′ and 5′-CAG-3′ producing 8–12 nt primers for Okazaki fragment synthesis. The interaction between helicase and primase stimulates the activity of the latter by > 103 fold and favors 5′-CAG-3′ as the recognition sequence. The primosome (DnaG/DnaB) formation is important for the recruitment of primase to the replication fork. The homotetrameric SSB, χ subunit of POl III holoenzyme, and primase are crucial for coupled leading and lagging strand synthesis discussed below. The catalytic rates and processivity of polymerase alone or in combination with accessory protein(s) are provided in Table 1.
Figure 3.
Schematic representation of the replication fork of E. coli (Reproduced with permission from Ref. [51]).
The replisome of eukaryotes
As mentioned above, the exact composition of proteins that comprise a replisome in eukaryotes is not well defined; however, some of the proteins in such a complex are well characterized. These include the homotrimeric processivity clamp proliferating cell nuclear antigen (PCNA), the heteropentameric clamp-loader complex replication factor C (RFC), heterotrimeric ssDNA binding replication protein A (RPA), and polymerase α. Polymerase α, involved in the initiation of replication and Okazaki fragment synthesis on the lagging strand, possesses the primase activity. Furthermore, the discovery of two major replicative polymerases Pol δ and Pol ε has lead to the hypothesis that the labor of leading and lagging strand syntheses is divided between the two polymerases [28, 29].
This conclusion was based on a strategy developed by the Kunkel group to study mutagenesis during leading versus lagging strand replication [30]. They examined the spontaneous mutational specificity in the URA3 reporter gene placed adjacent to ARS306, an origin of replication on chromosome III that fires in early S phase in >90 % of cells in yeast strains that harbor these mutator alleles [30]. It is well known that several amino acid substitutions at or near the active site of DNA polymerases reduce the fidelity of DNA synthesis [31]. These mutants are thus error-prone and selectively introduce mismatches, for example, E. coli Klenow fragment E710A generates A-C mismatches selectively over T-G mismatches. These two mismatches give rise to an A-T to G-C mutation in vivo, depending upon which of the two template strands is being replicated when the error is generated. Using this strategy, a replicative polymerase with the property to generate such selective mismatches could be used to determine the strand it copies in vivo. Thus polymerase ε is suggested to be the leading strand polymerase [32] and polymerase δ is thought to be the lagging strand polymerase of the eukaryotic replisome [33].
The identity of the replicative helicase in eukaryotes is still debated. There are some suggestions that a minichromosocal maintenance (MCM) helicase is the replicative helicase; however, the question remains to be unambiguously resolved [34].
[C] Coordination of replication of leading and lagging strands
How the leading and lagging strand syntheses are coordinated has been long debated; however, the question still remains largely unanswered. A likely solution to coordinate leading and lagging strands synthesis employs a DNA looping mechanism as shown in Figures 1, 2 & 3. Both leading and lagging strands form a replication complex and these two complexes are linked together by accessory proteins: Trx-gp4 complex in T7, clamp-loader in E.coli and a putative dimer of leading and lagging strand polymerases in T4 phage. The lagging strand folds back to form a loop that ensures that both parallel polymerizations in the 5′ to 3′ direction can proceed simultaneously on both strands and the replication fork can move forward unhindered. The DNA looping, referred to as the trombone model, has been directly observed and verified by electron microscopy [35].
Whereas leading strand replication needs only one initiation event; replication of the lagging strand needs multiple initiation events that require the assembly of the holoenzyme repeatedly along the lagging strand. Generally dilution of the DNA polymerase did not change the Okazaki fragment pattern supporting a mechanism to recycle the DNA polymerase on the lagging strand to a newly synthesized RNA primer in order to synthesize another Okazaki fragment [35]. Two schemes were proposed to explain the trigger for the release of the polymerase from the lagging strand. In the collision model, when replication on the lagging strand reaches the end of the previous Okazaki fragment, a “collision” triggers the release of the DNA polymerase and synthesis of a new RNA primer. In the signaling model, the DNA polymerase recycling is associated with RNA primer synthesis and successful trapping of the primer by clamp/clamp-loader complex signals the polymerase recycling. This does not necessarily require a “collision”, so that the result is lagging strand synthesis with gapped DNA [36]. Mechanistic details of the primer handoff remain to be elucidated. For example, is there pausing of the replisome on the replication fork during priming?
[D] Ensemble studies on DNA polymerase
A general summary of the ensemble studies on various DNA polymerase catalyzed reactions is presented here for comparison to the single-molecules studies. Various DNA polymerases have been extensively studied to understand the kinetic, structural and stereochemical aspects of the polymerization reaction. The primary focus has been to understand the source of the high nucleotide specificity involved in selecting the correct nucleotides that are complementary to the templating bases contributing to the fidelity of the replicative polymerases [37]. However, exceptions to this include the Y-family of translesion synthesis (TLS) polymerases, examples of which include pol η, pol ι and pol κ in eukaryotes. TLS polymerases are capable of synthesizing DNA past lesions in the templating strand. The active site of the Y-family polymerases is organized to accommodate bulky lesions such as thymine dimers, 8-oxoguanine and cis-platin with a concomitant loss in specificity but appears to retain the overall kinetic sequence of nucleotide incorporation [38].
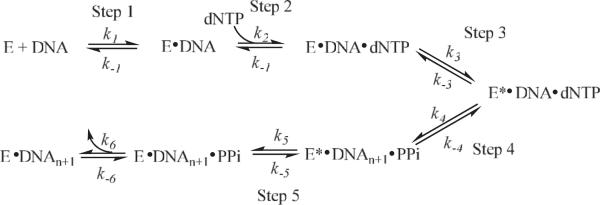
Scheme I shows the derived kinetic mechanism for the incorporation of the incoming dexoynucleotide triphosphate into the growing DNA chain in the DNA polymerization reaction by various polymerases based on detailed studies [39–42]. The fidelity of DNA polymerases and the kinetic and structural factors contributing to the fidelity have been reviewed elsewhere [37]. However, there are questions regarding the fidelity that remain unanswered. How does the polymerase regulate the selection of the correct incoming nucleotide for incorporation over an incorrect nucleotide? Does the rate-limiting step in the polymerase reaction pathway vary, particularly during misincorporations? If so, how general is this rate-limiting step variation? What are the factors that determine the switch between the polymerase mode and exonuclease mode? Questions of this nature as well as those previously mentioned can be investigated by single-molecule techniques to understand the details of DNA replication that otherwise would be difficult to determine.
Scheme 1.
Kinetic Scheme for the DNA chain elongation catalyzed by DNA polymerases.
[E] Single-molecule studies of DNA polymerase
On a sufficiently long time scale, a molecule stochastically visits all possible energetic or conformational states at a probability determined by the Boltzmann distribution. This dynamic aspect of individual molecules is concealed in ensemble measurements, because the fluctuation in the collective properties of the large number of molecules of the system is negligible on any experimental time scale. The ability to observe individual molecules opens the door to a wealth of information on their dynamic motions. Technological advances in the fields of optical and magnetic tweezers and fluorescence spectroscopy have paved the way for the development of novel methods to allow manipulations of molecules at the single-molecule level. These methods include: the use of single fluorophores on a macromolecule to probe the fluctuations in a local environment; the creation of single Fluorescence Resonance Energy Transfer (FRET) pairs to follow intermolecular distance changes; the manipulation of an individual molecule by Atomic Force Microscope (AFM), and the attachment of single molecules to micro-sized beads subject to forces generated by optical and magnetic tweezers, electric field or laminar flow [8, 43–46]. Apropos to our discussion, these methods have been employed to study the assembly and functions of complex macromolecular structures such as the DNA replisome composed of its multiple protein partners. The details of the operational principles of these single-molecule techniques are reviewed elsewhere [8, 43–46].
The recent decade has witnessed a rapid growth in the application of single-molecule techniques to study the biochemistry of DNA replication. In 2000, in the first report on the use of a single-molecule technique to study a DNA polymerase, the Bustamante group elegantly examined a single T7 polymerase carrying out DNA synthesis by monitoring the total length of primed ssDNA where one end was attached to a bead held by a micro pipette and the other end was attached to a bead trapped by a focused laser beam [47]. Extension by the DNA polymerase increases the double-stranded content of the primed DNA substrate leading to a decrease in the total DNA length under stretching forces greater than 6 pN. The same strategy has been exploited in other techniques that use different methods to stretch a single DNA molecule. For example, the Croquette group used magnetic force exerted on the DNA substrate through a magnetic bead attached to the free end of the primed ssDNA substrate that was immobilized on a microscope coverslip [48, 49]. Additionally, the van Oijen group employed laminar flow to produce a drag force on a bead attached to the free end of a surface immobilized ssDNA molecule in order to stretch the DNA [50–54] and Kim et al. reported a multiplexed version of this technique with improved flow stability [55].
Fluorescence based single-molecule methods have also been applied in the study of replisome assembly. The Benkovic/Hammes collaboration used single-molecule FRET (smFRET) to study the assembly of the T4 primosome [56] and polymerase holoenzyme [57]. SmFRET has also been successfully employed by the Zhuang group to investigate how HIV reverse transcriptase, an RNA-dependent DNA polymerase, switches its function between DNA polymerase and RNase H activities [58, 59]. Luo et al. probed T7 DNA polymerase conformational changes by monitoring the single-molecule fluorescence intensity of a Cy3-labeled primer-template DNA substrate [60]. In addition to the above methods, the van Oijen group reported a direct visualization of the elongation of double-stranded DNA with the aid of a fluorescent intercalating dye [61]. All these single-molecule studies of the DNA replisome will be examined in greater detail below to understand deeper several aspects of DNA replication. Before proceeding to the details of these single-molecule studies of DNA replication enzymes, a brief discussion of the elastic properties of ssDNA and dsDNA is presented below.
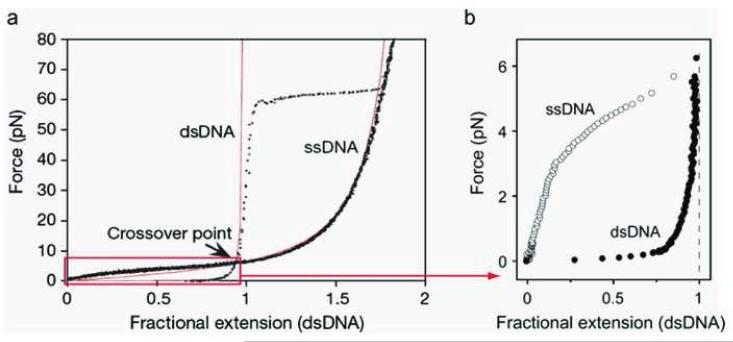
[F] DNA end-to-end distance as a function of stretching force
Several single-molecule techniques in the study of DNA enzymes are based on the differential elastic properties of ssDNA and dsDNA [47–49, 51–55, 62–66], and hence it is important to briefly describe the elasiticity of ssDNA and dsDNA. The elastic properties of ssDNA and dsDNA have been exploited in the study of enyzmes that generate either form of these DNA substrates during their function. For example, helicases produce ssDNA upon unwinding the duplex DNA and the ssDNA thus available is used as a template during replication by polymerases. With Watson-Crick base pairing and π-π stacking interactions, helical dsDNA is much more rigid than ssDNA. Consequently, dsDNA and ssDNA have distinct characteristic stretching curves with the end-to-end distance as a function of the stretching force, as shown in Figure 4. For dsDNA, a large stretching force is needed to stretch the dsDNA a small fraction longer than its full contour length. In contrast, ssDNA is much more elastic and can be stretched almost twice as long as its fully extended contour length. When the stretching force is ~ 60 pN, the helical structure of dsDNA is broken, and the molecule is denatured and overstretched to the length of corresponding ssDNA. A persistence length of ~ 50 nm for dsDNA and 0.8–3 nm for ssDNA was obtained by fitting these curves with the Worm-Like-Chain model. At a specific force, ssDNA and dsDNA have a fixed specific length of ( ) and ( ) respectively, where the length corresponds to one nucleotide and hence, the total length of a primed ssDNA substrate is calculated as ( ) = ( ) + ( ) where and are the number of nucleotides in the template strand that are single-stranded and double-stranded respectively. Therefore, the change in the total DNA length is expressed as Δ = Δ ( ) − ( ), where Δ is the change in the number of single-stranded nucleotides converted into double-stranded nucleotides. Consequently the change in the total length of stretched DNA can be used to measure the rate at which DNA polymerase extends the primer. Various DNA replication enzymes studied at the single-molecule level are described in detail below.
Figure 4.
Force-extension profile for dsDNA and ssDNA. The experimental data (dotted curves) can be well fit with the Worm-Like-Chain model (solid red curves) using 0.7 nm and 53 nm as the persistence length for ssDNA and dsDNA respectively. (Reproduced with permission from Ref. [50]).
[G] Single-molecule analysis of DNA polymerase activity
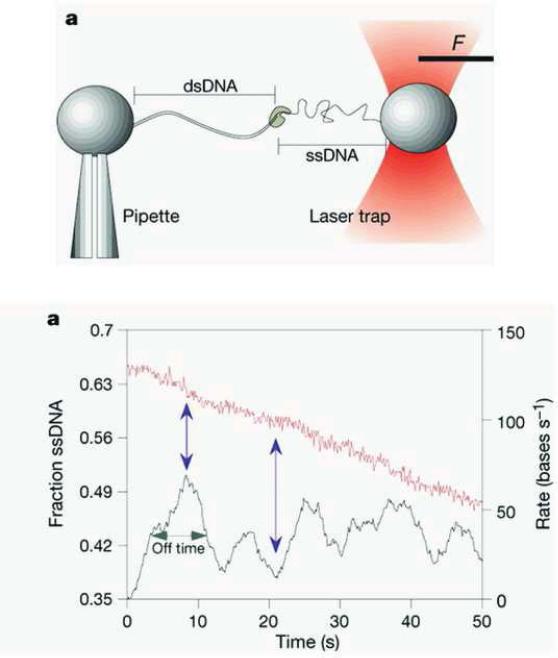
The activity of the T7 DNA polymerase tethered to Trx clamp in a 1:1 ratio was studied on a primed ssDNA substrate stretched by an optical trap setup shown in Figure 5a [47]. The experimental setup is briefly discussed next. A 10 kb plasmid DNA fragment was attached between two beads, where one end was held on the tip of a glass pipette and the other end in an optical trap. SsDNA was obtained by using the force-induced exonuclease activity of T7 DNA polymerase to remove the desired length of the non-template strand. The bead positions were imaged to obtain the end-to-end length of the DNA and the change in the light momentum of the bead which exits the dual beam trap was used to measure the force [47]. The replication rate was obtained from the change in the length of the ssDNA in the presence of polymerase and deoxynucleotides at any given force. Replication rates at various enzyme concentrations and various applied forces were measured. At a mean force of 34 ± 8 pN the enzyme stalled during DNA synthesis; however, occasionally polymerization occurred briefly above 50 pN.
Figure 5.
(a) Optical trap setup (b) Decrease of ssDNA fraction during DNA replication (red curve) and the DNA replication rate obtained by taking the derivative of the ssDNA fraction with respect to time. (Reproduced with permission from Ref. [47]).
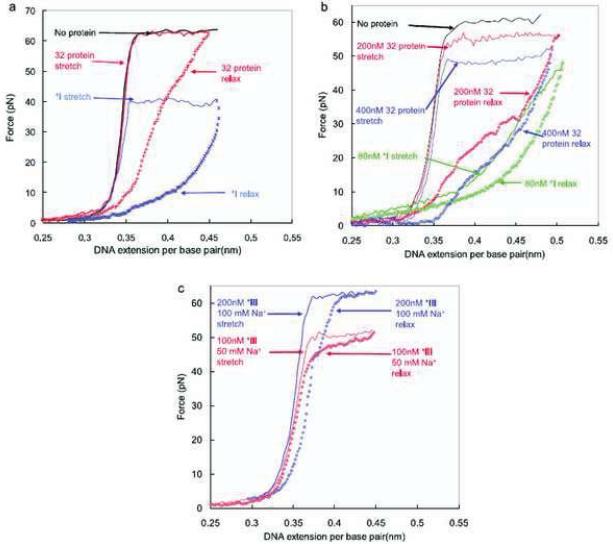
Unlike the constant polymerization activity obtained from ensemble measurements, the enzyme showed bursts of activity where the mean width of the burst corresponds to an off-rate of 0.13 ± 0.1 s−1 that was independent of the enzyme concentration and DNA stretching force (Figure 5b). The gap width between the adjacent bursts is negatively-dependent on the polymerase concentration. Therefore, the burst of activity is attributed to the enzyme loading onto and dissociating from the DNA substrate. The height of the burst varies between 26 and 60 bases at a force of 20 pN suggesting that each individual enzyme molecule behaves differently. The replication rate is found to be force-dependent, increasing from ~ 100 nt/s at zero force to a maximum of ~200 nt/s at 6.5 pN and then decreasing with increased force until it halts at 34 ± 8 pN, indicating the presence of a force dependent rate-limiting step in the polymerase reaction. In light of an induced-fit mechanism, the force dependence of the reaction was attributed to the effect of tension on the entropic term of the active energy of the rate equation. Fitting the data to an energetic model led to the conclusion that the finger domain organizes two nucleotides at the active site during each catalytic cycle.
Surprisingly, it was also observed that a force-induced burst of exonuclease activity is observed at high forces. No exonuclease activity was reported below a force of 34 pN where polymerization was predominant; however, when the template tension was greater than 40 pN the exonuclease activity increased to about 30 ± 11 nt/s. The exonucleolytic rate became independent of force above 42 pN and is two orders of magnitude faster than that observed at zero force on dsDNA. The mechanism of the tension induced exonuclease activity is not clear and may be characteristic of the T7 polymerase rather than a general property of all polymerases. It should be noted here that the rate of the reaction was dependent on the applied force, and hence the processivity measured also was dependent on the force. A measure of the processivity of 400 bases was obtained at a low force of 15 pN. It should be interesting to see whether other DNA polymerases behave identically at the single-molecule level.
An analogous set of measurements was carried out by Maier et al. on two polymerases, the Klenow fragment of E. coli Pol I and T7 Sequenase, on a primed ssDNA substrate stretched with magnetic forces in a magnetic tweezers apparatus. The Klenow fragment is produced from E. coli DNA polymerase I by the protease subtilisin and retains the polymerase and 3′ to 5′-exonuclease activities of the parent enzyme and replicates DNA with high fidelity. T7 Sequenase derived from DNA polymerase gp5 lacks the exonuclease activity of the wild-type polymerase. The behavior of the Klenow fragment at the single-molecule level is consistent with that noted above for the wild-type T7 polymerase and displays a burst of replication activity with a pause between each burst, as well as, a force-dependent rate of replication. The replication rate of 13 nt/s and 200 nt/s for Klenow fragment and T7 Sequenase respectively at low forces is comparable to 7 nt/s for Klenow fragment and 230 nt/s for T7 Sequenase obtained from ensemble measurement [39, 41, 67, 68]. Based on fitting the force dependent replication rate to a mechanochemical model, it was concluded that T7 Sequanase and Klenow fragment organize two and four template nucleotides respectively in their cognate active sites. This, however, raises a question of whether there is a correlation between the number of bases involved in the rate-limiting step of the polymerization reaction and the number of template bases within a polymerase active site.
The number of template nucleotides implicated in the active site - two for T7 polymerase and four for Klenow fragment - contradicts the observations from crystal structures of polymerases. For example, the ternary complex structure of T7 DNA polymerase/P–T junction DNA/dNTP shows that this polymerase has a compact active site and holds only one template base in its active site [15]. The rest of the template is located outside of the catalytic site almost perpendicular to the duplex region of the growing DNA chain. To reconcile this contradiction, the Herschbach group has employed a model involving a local instead of a global end-to-end profile to evaluate the mechanical work done on DNA by the enzyme [62]. This model assumed that the polymerase changes the geometry only of the ssDNA segment that is close to the active site and provided a satisfactory proof of the force-dependence of the replication rate for both enzymes.
The single-molecule methods above focused on the rate of replication and the processivity of single DNA polymerases under an applied force. The other important feature of polymerases is their high fidelity. The initial dNTP binding step of polymerization contributes to base selectivity for high-fidelity polymerases such as the T7 and T4 polymerases and the E. coli Klenow fragment. The conformational change that occurs in these polymerases upon binding the dNTP prior to the chemistry step has been proposed to be the rate-determining step for complementary dNTPs [39, 41, 68]. Crystal structures of several DNA polymerases have shown that the fingers subdomain of the polymerases completely encloses the incoming dNTP in the ternary complex structures [15, 69–72]. Fluorescence studies with either a fluorescent nucleotide or a FRET pair have indicated that there is a rapid equilibrium between the open and closed conformational states and that the closing of the fingers is a fast step [69, 73–76]. However, with fluorescently labeled T7 DNA polymerase this step could be partially rate-limiting for the base selectivity [77]. Hence, it is of great interest to understand how the enzyme's conformational dynamics contribute to the fidelity of base selection.
Luo et al. reported a single-molecule fluorescence study on the conformational dynamics of the active T7 polymerase/P–T/dNTP complex by taking advantage of the environmental sensitivity of Cy3 fluorescence [60]. This assay probed the conformational change associated with the dNTP binding. Binding of T7 polymerase to a Cy3 labeled primer-template enhanced the fluorescence of Cy3 by reducing the relaxation rate of the dye from its excited state. Furthermore, binding of a complementary dNTP induced a conformational change in T7 polymerase that led to a further enhancement of the Cy3 fluorescence. By using a polymerization incompetent dideoxy-terminated primer with an exonuclease resistant phosphorothioate linkage between the penultimate and 3′ terminal nucleotide, the conformational change associated with the active ternary complex formation was obtained from the Cy3 fluorescence response upon introduction of dNTP in the presence of Mg2+. The rate associated with this conformational change when measured for the complementary dNTP was much higher than the DNA replication rate, while the rate associated with a non-complementary dNTP was significantly slower than the DNA replication rate. The relationship between the rates of the conformation change and the correctness of the incoming nucleotide led to the conclusion that the conformational change step upon dNTP binding contributes to the high fidelity of DNA synthesis by the T7 polymerase. Thus Watson-Crick base pairing contribution to fidelity was augmented by the molecular interactions between the polymerase-DNA binary complex and the incoming dNTP.
One of the important developments based on single-molecule techniques is real-time DNA sequencing. The traditional and commercial method of DNA sequencing uses a DNA polymerase to incorporate 3′-dideoxynucleotides that terminate the DNA synthesis, but does not exploit the high catalytic rates or the processivity of the polymerase [78]. There are reports of methods that enhance the performance of DNA sequencing, that are still limited by the necessity for chain termination and short sequence read out (~400 nucleotides) [79–82]. The Turner group has provided a proof-of-concept for an approach to highly multiplexed single-molecule, real-time DNA sequencing based on the observation of the temporal order of fluorescently labeled nucleotide incorporations during uninterrupted DNA synthesis by a single DNA polymerase molecule [83]. Using this approach, they obtained with fifteen reads, a consensus sequence with 99.3% accuracy and no errors beyond those that arise from the fluorophore of the nucleotides. This method might prove to be a great tool for sequencing especially highly repetitive DNA, where challenges remain for de novo assembly or alignment.
[H] Characterization of ssDNA binding protein using single-molecule force spectroscopy
SsDNA binding protein (SSB) plays an essential part in DNA replication as it binds to the nascent ssDNA segment cooperatively and selectively to prevent the ssDNA from forming a secondary structure, as depicted in Figures 1, 2 &3. Its C-terminal domain is responsible for interaction with other proteins and the N-terminal domain is responsible for cooperative binding [1]. The preference of gp32 to bind ssDNA over dsDNA often leads one to expect this protein to destabilize dsDNA. However, wild-type gp32 does not affect the dsDNA melting temperature whereas its C-terminal truncated form does [84]. The origin of the ability of gp32 to specifically bind to ssDNA without destabilizing dsDNA was addressed by using single-molecule force spectroscopy [63–66]. In this technique, force is applied to the dsDNA using an optical tweezers setup similar to the one shown in Figure 5a. The stretching curve is recorded during the stretching phase in which dsDNA is stretched until it is denatured and overstretched, and in the relaxation phase in which the pulling force is gradually released. In the absence of a DNA binding protein, such as SSB, the stretching curve of the stretching phase overlaps perfectly with that of the relaxation phase as shown in Figure 6a. In the presence of a protein-DNA interaction, a hysteresis usually appears in the stretching curves, i.e. the stretching curve of the stretching phase does not overlap with that of the relaxation phase, and also the force needed to overstretch dsDNA may differ from that in its absence. The magnitude of the overstretching force and hysteresis consequently is dependent on the thermodynamic and kinetic aspects of the DNA-protein interaction.
Figure 6.
Stretch-force profile of a single dsDNA in the presence of wt gp32, Truncate I or Truncate III (Reproduced with Permission from Ref. [65]).
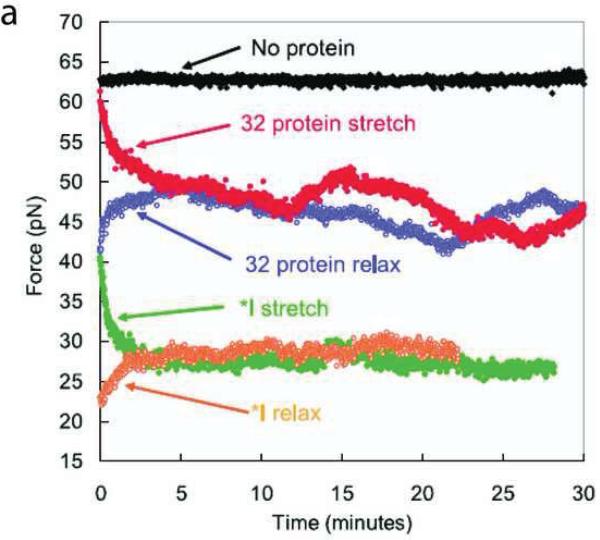
Three types of single-molecule force spectroscopic measurements have been performed to understand the gp32-DNA interaction. In the first type of experiment stretching curves were recorded for the stretching and relaxation phases for wt gp32, Truncate I in which the C-terminus of gp32 was removed, and Truncate III in which both the C- and N-termini were removed leaving only the ssDNA binding core. These proteins exhibited different effects in the stretching-force profile of a single dsDNA as shown in Figure 6. In the case of 200 nM Truncate I, the dsDNA overstretching force was reduced significantly indicating that the dsDNA was significantly destabilized (Figure 6a). In contrast, wt gp32 showed no effect on the overstretching force, but caused a hysteresis in the stretching curve that suggested gp32 remians associated with the denatured DNA. The destabilization effect of gp32 on dsDNA appeared at a high concentration as shown in Figure 6b. In the presence of 200 nM and 400 nM wt gp32, the force needed to overstretch dsDNA was reduced significantly with the effect increasing with increasing protein concentration. Destabilization of dsDNA by wt gp32 was not observed in ensemble measurements. In the presence of 80 nM Truncate I, the stretching curve resembled that of ssDNA (Figure 6b and 4a). The large hysteresis in the stretching curves for full-length gp32 and Truncate I implicate formation of a stable protein-DNA complex upon denaturation of the dsDNA. In contrast Truncate III exerted a significant effect on the dsDNA overstretching force only at a low ionic strength of 50 mM NaCl, with a corresponding smaller hysteresis in the stretching curve (Figure 6c), suggesting that its binding to DNA is weakened by removing the N-terminus. The differential effects of these three proteins revealed that the N-terminal domain primarily is responsible for the cooperative binding of gp32 to ssDNA through stabilizing the protein/ssDNA complex offset by the C-terminal domain's preferred interaction with dsDNA [65].
The second type of measurement involved monitoring the stretching force as a function of time. As shown in Figure 6, a larger force is needed to overstretch dsDNA without SSB than in the presence of SSB. If overstretching dsDNA to a fixed length is complete on a shorter timescale than that needed for dsDNA binding by SSB, the effective concentration of bound SSB is zero at the instant of stretching and the overstretching force will be comparable to that without SSB. At later times when the association of SSB with DNA reaches equilibrium, the force needed to maintain the DNA length equals the overstretching force in the presence of SSB. For the same reason, after a rapid relaxation of stretched SSB coated dsDNA, the force increases at longer times because of the dissociation of excessive SSB molecules from the dsDNA that occurred in the relaxation. The rates at which the force increases and decreases corresponds to the rates of dsDNA melting and re-annealing by SSB respectively. The dsDNA melting rate and re-annealing rate for gp32 and Truncate I were measured using this force-time experiment (Figure 7). In the presence of full length gp32 and Truncate I, the force decreased exponentially to an equilibrium value in the stretching phase and increased exponentially to the same equilibrium value in the relaxation phase, providing a denaturing rate of 18 bp/s and a reannealing rate of 57 bp/s for the full-length gp32 and denaturing and annealing rates of 80 bp/s and 107 bp/s respectively for Truncate I. These results suggest that the C-terminus of gp32 kinetically down regulates the ssDNA binding properties of gp32 protein.
Figure 7.
Stretching force as a function of time when dsDNA is stretched and then relaxed in the presence of wt gp32 and Truncate I (Reproduced with permission from Ref. [65]).
The third type of measurement determined the overstretching force as a function of pulling rate. Both the stretching force and the binding of SSB act to destabilize dsDNA. Intuitively, when pulling dsDNA slowly, SSB has time to find and associate with the nascent binding site. The availability of SSB-DNA binding energy demands less mechanical work from the pulling force to stretch the DNA molecule; therefore, the slower the pulling rate, the less overstretching force is necessary. A theoretical framework was proposed to relate the pulling rate to the overstretching force. Briefly, the binding rate of SSB to stretched DNA is determined by the time SSB takes to find a binding site on an ssDNA segment of efficient length and the probability at which such a binding site is produced. The probability is a function of the pulling force; the larger the pulling force, the greater the probability of revealing a binding site. For a rigorous derivation, one should refer to the original paper [64].
This method was applied to measure the cooperative binding and non-cooperative binding rates, binding size and non-specific binding rate of gp32 to ssDNA, as well as, the non-specific binding and sliding rates of gp32 on dsDNA. The cooperative binding rates at various protein concentrations were fit to this theoretical model assuming that the cooperative binding of gp32 to a nascent ssDNA segment generated during unwinding of duplex DNA was enhanced by 1-D diffusion of gp32 on dsDNA. The measured cooperative binding rate constant for full length gp32 was 32 s−1 at 200 nM, much lower than 28 000 s−1 for Truncate I. The binding site size for gp32 was 7 nt. Thus the ability of gp32 to specifically bind ssDNA without destabilizing dsDNA could be attributed to its slow cooperative binding rate which prevents binding of gp32 to spontaneously frayed dsDNA.
Salt effects on the equilibrium binding of SSB (T4 gp32 and T7 gp2.5) to either ssDNA or dsDNA were measured by the stretching force method and confirmed that the C-terminal truncated proteins had high ssDNA binding affinity and salt sensitivity [63–66].
[I] Real-time observation of dsDNA unwinding by replicative helicases
Helicases use the energy of NTP hydrolysis to unwind dsDNA or translocate on ssDNA. One intriguing question is how helicases couple NTP hydrolysis to the unwinding of dsDNA. Two competing mechanisms have been suggested. The simplest scenario is the passive unwinding mechanism in which ATP hydrolysis is used to generate directed ssDNA translocation of the enzyme to prevent reannealing of the transient thermal fraying of dsDNA at a fork junction. In the alternative active unwinding mechanism, the enzyme actively destabilizes DNA basepairs at a fork junction, shifting the equilibrium of the fork toward opening. Single-molecule experiments have been used to answer this question for two replicative helicases, gp4 from T7 and gp41 from T4.
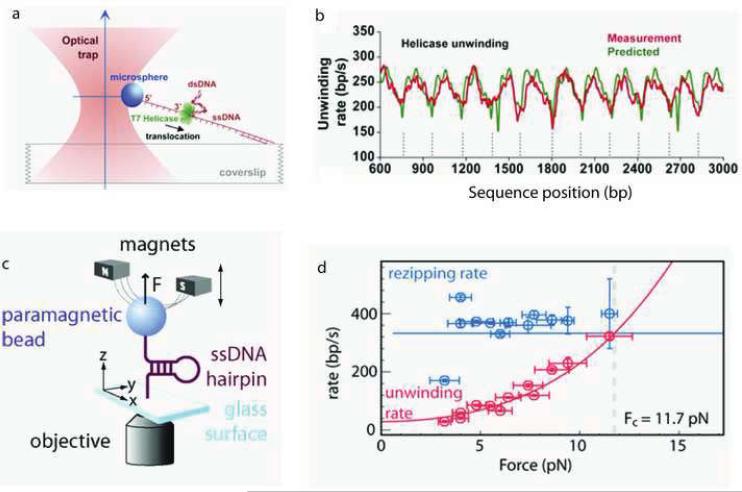
The Wang group has studied the ssDNA translocation and DNA unwinding properties of the T7 helicase [85]. The experimental configuration consisted of a dsDNA substrate with one strand attached to the surface of a microscope coverslip and the other strand attached to a microsphere held in a feedback-enhanced optical trap so that its position and the force on it could be measured (Figure 8a). In order to measure the ssDNA translocation rate of gp4, the dsDNA was mechanically unwound by moving the coverslip to generate a segment of ssDNA of known length and observing the time necessary for the helicase to reach the fork junction detected as a drop in the tension on the microsphere. In this manner, a ssDNA translocation rate of 322 ± 62 nt/s was measured at 14 pN. Helicase unwinding activity was monitored as an increase in the ssDNA length detected as a change in the microsphere position under constant tension. The unwinding rate showed a distinctive DNA sequence-dependence consistent with the periodicity of pseudorepeats in the DNA substrate (Figure 8b), indicating that it is harder for the helicase to unwind the DNA when the affinity between the two DNA strands is stronger, i.e. the unwinding rate was slower in GC-rich regions than in AT-rich regions of the DNA. The unwinding rate was also found to depend on the force exerted to separate the dsDNA strands thereby assisting helicase motion. The average unwinding rate increased nonlinearly with respect to force; at low force (5.2 pN) the unwinding rate of 29 bp/s was similar to that measured in bulk (10–15 bp/s; [86]), while at higher force (11.2 pN) the unwinding rate of 220 bp/s approached the ssDNA translocation rate. The average processivity of DNA unwinding was also dependent on force; at 11.2 pN, most helicase molecules completely unwound the remaining 3.6 kb DNA, while at 5.2 pN, the average length was only 343 bp.
Figure 8.
Real-time observation of dsDNA unwinding by replicative helicases. (a) Experimental configuration involving an optical trap. (b) Measured and predicted instantaneous velocity of T7 helicase along dsDNA template positions. (c) Experimental configuration of magnetic tweezers apparatus. (d) The unwinding rate (red circles) and rezipping rate (blue circles) for T4 helicase as a function of applied force. (Figures a and b reproduced with permission from Ref. [85].
The ssDNA translocation and DNA unwinding properties of the T4 helicase have been studied by the Croquette and Benkovic groups [49]. Experiments were carried out by tethering the two ends of a DNA hairpin between the surface of a microscope slide and a magnetic bead with force being applied to the DNA molecule through magnets positioned above the sample (Figure 8c). Helicase unwinding activity was monitored as an increase in the end-to-end distance of the DNA substrate in the presence of gp41 and varying concentrations of ATP over forces ranging from 3 to 11.5 pN. Following the complete unwinding of the hairpin by a helicase molecule, the ssDNA translocation velocity was measured from the gradual decrease in the end-to-end distance of the DNA substrate as the hairpin reannealed in the wake of the helicase translocating on ssDNA over the same ATP concentration and force ranges. In the case of the T4 helicase, the ssDNA translocation rate was 314 ± 15 nt/s independent of the applied force, while the unwinding rate was highly dependent on the force, approaching the ssDNA translocation rate at high force and extrapolating to 30 bp/s at zero force in a nonlinear fashion (Figure 8d). As expected, the helicase activity was dependent on the concentration of ATP following Michaelis-Menten kinetics with a Km of 1.1 ± 0.1 mM ATP.
Both studies used models to determine the passive/active nature of the helicase from the force dependent unwinding velocities. In both cases a helicase was considered a passive helicase if the fork junction was required to open n basepairs, the step size of the helicase, before translocation could take place; alternatively, an active helicase directly destabilized the dsDNA near the fork junction to facilitate unwinding. In the case of the T7 helicase, a strictly passive helicase model did not fit the data. Instead the Wang group used three parameters to describe the active nature of the T7 helicase: the step size, the interaction range of the helicase with the fork junction, and the interaction potential increase per base ΔGd. A good agreement between the measured unwinding velocity dependence on force and simulated data was obtained with a step size of 2 bp, an interaction range of 6 bp, and an interaction potential increase per base of 1.2 kBT. Considering that the free energy required to open a single GC basepair is ~3.4 kBT, their results support an active unwinding mechanism for T7 helicase, although the model also indicates that the helicase is not optimally active. In the case of the T4 helicase, the passive/active nature of the helicase is introduced in to the model through the fork opening and closing rates, thus the unwinding rate was equal to the maximum helicase ssDNA translocation rate times the probability that the next n basepairs are opened. The measured unwinding velocity dependence on force and ATP concentration was best fit by a model with a step size of 1.4 bp and a helicase destabilization energy equal to 0.05 kBT. In other words, the destabilization of the double helix by gp41 was very small (<15% of the base-pairing energy), consistent with the T4 helicase unwinding DNA by an essentially passive mechanism. Based on the helicase destabilization energy values, the T4 helicase is closer to the ideal passive helicase, whereas T7 helicase stands somewhere between the purely passive unwinding model and the purely active unwinding model. It should be noted that the passive/active classification distinguishes between two extreme cases that do not reflect the continuum of behaviors probably existing in nature. Indeed, both studies comment on the relatively slow unwinding velocity of each helicase at low force suggesting that the helicase needs to be coupled with the DNA polymerase or other replisome protein in order to achieve the maximum unwinding rate observed in DNA replication. This coupling between helicase and DNA polymerase may provide additional helix destabilization energy thereby shifting the unwinding mechanism from passive or moderately active to an optimally active unwinding mechanism.
[J] Primosome and holoenzyme assembly studied by single-molecule FRET
The single-molecule studies investigating the functions of replication proteins individually have been discussed so far. In the context of in vitro and in vivo DNA replication, it is important to look at the assembly of these proteins as they essentially form the active complexes for the synthesis of leading and lagging DNA strands. The primosome, comprised of helicase and primase, plays essential roles in DNA replication. The assembly of the T4 primosome from gp41 helicase, gp61primase, gp59 helicase loader and gp32 was studied with smFRET by the Benkovic and Hammes groups [56]. In these experiments, a fluorescence microscope equipped with a multi-wavelength Ar-ion laser and three filter sets was used. The first filter set was coupled to a 488 nm laser line to excite the donor fluorophore Alexa Fluor-488 and record the FRET signal of the donor. The second filter set was coupled with a 488 nm laser line to collect the fluorescence signal from the acceptor Alexa Fluor-555 or Cy3. The third filter set was coupled to a 514 nm laser line to directly excite and detect the acceptor fluorescence signal, which is useful to verify the presence of the acceptor modified molecule.
The interactions between proteins and the surface immobilized forked-DNA were probed by attaching fluorescent dyes to the proteins. Each of these four proteins alone was able to bind to the forked-DNA from solution. The protein-protein complex formation on the forked-DNA was investigated systematically by smFRET in the presence of ATP or ATPγS, a non-hydrolyzable analog of ATP. It was demonstrated that an ordered process of primosome assembly begins with gp32 and gp59 forming a tight 1:1 complex on the forked-DNA substrate agreeing well with earlier reports from ensemble measurements [24, 87]. This complex formation involves gp32 binding to the forked DNA with subsequent or simultaneous binding of the gp59 helicase-loader complex. In the presence of ATPγS, gp41 forms a weak complex with the DNA coated with gp32 and gp59, whereas in the presence of ATP, gp41 binds to gp59 and is loaded onto the DNA. With ATP, the unwinding activity of gp41 displaces gp32 and gp59 probably by translocation. In the presence of ATP, gp61 unlike gp41 could not be loaded onto the DNA/gp32/gp59 complex, but gp41/61 could be loaded onto a DNA.
From these single-molecule results a primosome assembly pathway was devised as shown in Figure 9. Gp32 and gp59 bind to forked-DNA, and then gp41 binds to gp59 that in the presence of nucleotide and ATP hydrolysis leads to dissociation of gp32 and gp59 from the DNA. Finally gp61 binds to gp41 to form the active primosome complex. In the context of the replication fork, the polymerase and the processivity clamp are present in the functional replisome and the above pathway for the primosome assembly may be modified. Results from electron microscopy on DNA/replisome complexes reveal a bobbin-like structure on lagging strand ssDNA composed of gp32 and gp59. The results from this work, however, suggest that the active primosome is composed only of the gp41 and gp61 complex. Since gp61alone does not bind at the interface of gp32 and gp59, it is suggested that gp59 is imbedded at other loci in the bobbin and may reflect an abortive complex or act in the coordination of the action of the leading and lagging strand holoenzymes [88].
Figure 9.
Assembly of the T4 primosome, gp41(helicase)/61(primase), onto surface immobilized forked-DNA promoted by gp32 and gp59 in the presence of ATP. (Reproduced with permission from Ref. [56]).
A strategy analogous to the above experiment was used to investigate the formation of holoenzyme from T4 gp43 polymerase and processivity clamp gp45 in the presence of the clamp loader complex gp44/62 [57]. In this case, the clamp gp45 and the polymerase gp43 were labeled with the acceptor, Alexa Fluor-555 and the donor Alexa Fluor-488 respectively. The smFRET signals between the donor and the acceptor in these experiments suggested that there are four pathways for the assembly of gp43/gp45 holoenzyme on forked-DNA. The assembly pathways involve: (a) binding of the gp45-gp44/62 complex to DNA followed by gp43 binding; (b) binding of gp44/62 to DNA followed by sequential binding of gp45 and gp43; (c) binding of gp45 to DNA followed by sequential binding of gp44/62 and gp43; and (d) binding of gp43 to DNA and then binding of the gp45-gp44/62 complex. Pathway c is possible because gp45 is partially open in solution unlike other clamp proteins [89, 90].
The existence of the multiple pathways for T4 holoenzyme assembly provides a mechanism to effectively recycle lagging-strand polymerase during DNA replication. It has been shown that there are multiple pathways for the initiation of DNA replication in bacteriophage T4 [91]. The early stages of T4 phage infection involve initiation of the leading strand DNA synthesis from an RNA primer that forms an R-loop structure. In the late stages of T4 phage infection, DNA replication involves priming from DNA recombination intermediates such as D-loop structures that are formed by the invasion of a partially unreplicated 3′-terminus derived from the homologous region of another segment of chromosomal DNA. Thus, the holoenzyme is assembled on either the D-loop or R-loop structures to initiate the processive leading strand synthesis. However, the lagging strand synthesis requires the repetitive assembly of the holoenzyme on a short RNA primer. Hence, the nature of specific DNA structures might influence the choice of the assembly pathway of the holoenzyme. Several of the assembly pathways for the primosome and the holoenzyme of T4 phage might be general and possibly be conserved among other replisomes.
[K] Single-molecule observation of coordinated leading and lagging strand synthesis
How the leading and lagging strand syntheses are coordinated at the same pace is both remarkable and puzzling. The leading strand DNA synthesis by the T7 replication machinery composed of gp5, Trx and gp4 was studied by the van Oijen group on a long λ, phage DNA molecule that was immobilized onto a functionalized surface through the 5′-end of the lagging strand and stretched by a drag force on a microbead attached to the 3′-end of the same strand created by flow as shown in Figure 10b [52]. A replication complex without the lagging strand polymerase was preassembled on a primed λ, DNA fork. The leading strand DNA synthesis was initiated by introducing Mg2+ copying the leading strand template generated by the gp4 helicase. The leading strand synthesis was detected by the decrease in the DNA length; the lagging strand will remain single-stranded due to the absence of a lagging strand polymerase. The difference between the lengths of ssDNA and dsDNA was employed to calculate the number of nucleotides incorporated by the leading strand polymerase. The rate of nucleotide incorporation by the leading strand polymerase was found to be 164 ± 20 bp/s and corresponds well with the helicase unwinding rate obtained for the T7 helicase of 130 bp/s [92].
Figure 10.
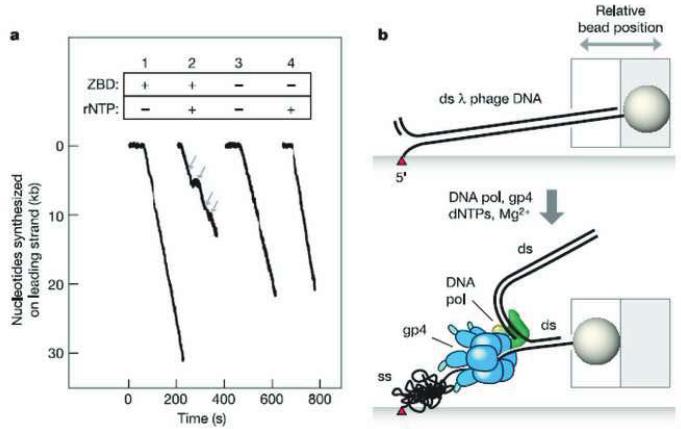
Observation of leading strand synthesis by T7 DNA replication proteins using single-molecule method. (a) DNA length decreases during leading strand synthesis; ZBD, zinc-binding domain is necessary for primase activity; rNTP is material for primer; gray arrows denote pauses. (b) The single-molecule event corresponding to the time trace in Panel a. (Reproduced with permission from Ref. [52]).
The processivity obtained in this single-molecule study was 17 ± 3 kb, which is dramatically different from the processivity of 1 kb for gp5 polymerase alone [93] or the 60 bp unwinding processivity of gp4 alone [86]. The high processivity most likely originates from the stabilization of the replisome complex by the interaction of the thioredoxin binding domain of gp5 polymerase and the acidic C-terminal tails of gp4 and gp2.5 [94]. In the presence of rNTP necessary for primer synthesis by gp4, pauses approximately 6 s long were observed in the leading strand DNA synthesis as shown in Figure 10a (Trace 2). When a gp4 enzyme lacking the zinc-binding domain that forms the active hexameric helicase but defective in primase activity was used, no such pauses were observed even when rNTPs were present (Figure 10a, Trace 4). Therefore, the pauses in the leading strand synthesis were attributed to DNA-templated RNA primer synthesis by the replisome.
The inclusion of additional DNA polymerase in this assay ensured the formation of both the leading and lagging strand holoenzymes. In this case a gradual shortening of the DNA associated with a pause is followed by a sudden increase in the length of the DNA. This is attributed to the replication loop formation and its release during lagging strand DNA synthesis. The average pause of 6 s agrees well with ensemble measurement of the rate of primase catalyzed primer synthesis of 1–3 nt/s. The pauses reveal that leading strand synthesis is halted until primer synthesis on the lagging strand is completed allowing the leading and lagging strand holoenzymes to resume coordinated DNA synthesis. Primer synthesis, therefore, acts as a brake on leading strand DNA synthesis, i.e. initiation of primer synthesis interrupts the leading strand DNA synthesis allowing time for primer synthesis and other probably slow steps in Okazaki fragment initiation to be completed before resuming synthesis on the leading strand. The molecular mechanism responsible for the pauses observed in the leading strand DNA synthesis by the primase activity is not well defined at this time [53]. The possibility of primase-RNA-DNA interactions as the cause of stalling has been ruled out due to the low affinity of the primase for the P-T junction [95]. Among other possibilities is an allosteric modulation of the helicase activity by the primase that arises during primer synthesis.
It had been previously demonstrated from bulk biochemical assays that the C-terminal tail of gp4 is required for maintaining the interaction of gp4 with gp5/Trx for effective strand-displacement synthesis [96, 97]. Furthermore, the crystal structure of gp5/Trx revealed the presence of two basic loops (residues 275–285 and residues 299–314) [15]. Plausible electrostatic interactions of the acidic C-terminus of gp4 and the basic loops of gp5 were probed by mutational analysis of these proteins, where the C-terminal acidic tail of gp4 containing 17 amino acids was removed and the basic residues of gp5 were mutated to alanines. A measure of the rate and processivity of these proteins at the single-molecule level showed that the mutation of either of these proteins led to a drastic reduction in the processivity of leading strand DNA synthesis from 17 kb for the wild-type enzyme to 6 kb for the gp5 mutant and 5 kb for the gp4 C-terminal deletion mutant. It should be noted that the rate of DNA synthesis was not significantly reduced. Thus, it is evident that there exists an electrostatic interaction between the C-terminal tail of gp4 and the basic loops of gp5 confirming an earlier suggestion [96, 97].
As discussed above, the primase and helicase of the T4 phage, E. coli and eukaryotic replisomes exist as two separate polypeptides and not as a covalently fused primase and helicase protein like the T7 phage. Although the primase and helicase proteins from these species are not covalently linked, they do form tightly coupled complexes during in vitro DNA replication [98, 99]. It is possible for such an electrostatic interaction to extend to these replisomes as well.
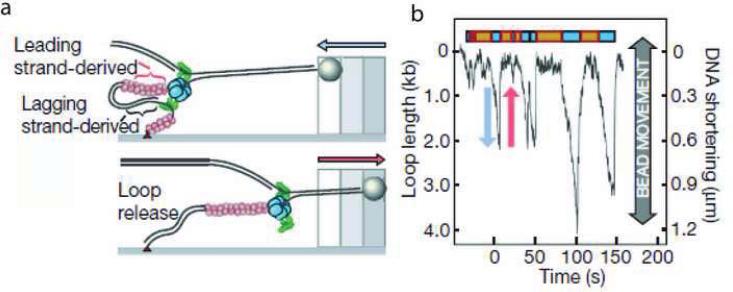
In the above single-molecule study of the leading strand synthesis from T7, the replication loop formation and release in the lagging strand was observed in the presence of the gp4 protein. However, ssDNA binding protein, an important protein involved in the lagging strand synthesis was not included. The lagging strand DNA synthesis was studied in the presence of T7 SSB gp2.5 and the lagging strand polymerase employing the single-molecule system shown in Figure 11a [54]. The polymerization reaction was monitored by the change in the DNA length of the substrate indicated by the movement of the bead attached to the DNA substrate. The time trace of the DNA length contains a series of valleys, as shown in Figure 11b, attributed to the loop formation due to lagging strand synthesis and subsequent loop release.
Figure 11.
(a) Schematic depiction of loop formation during lagging strand synthesis; the red circles are gp2.5; the blue objects are the gp4 helicase/primase and the green objects are the polymerase/Trx complex. (b) Time course of the change in DNA length during DNA synthesis of a single DNA molecule. The change in the DNA length is monitored by observing the beadmovement. The gradual decrease in the length of the DNA is due to the replication loop formation during lagging strand DNA synthesis and the rapid increase in the DNA length arises from the release of the replication loop as shown in panel (a) (Reproduced with permission from Ref. [54]).
The factors that contribute to triggering loop release need to be identified in order to understand how the coordination of leading and lagging synthesis is achieved. The loop length was measured to be 1.4 kb, corresponding to twice the Okazaki fragment size of 0.8 kb. Furthermore, the rate of 146 bp/s for DNA shortening during the replication loop growth is nearly twice that of 80 bp/s observed for leading strand polymerization alone [52]. This indicates that the loop is formed during coordinated leading and lagging stand DNA synthesis with a net rate contributed by both polymerases. It was also found that there is always a lag time, 12.0 s on average, between loop release and the formation of a new loop. Both the loop length and the lag time are dependent on the concentration of ATP and CTP; reducing their concentration decreases the loop length and increases the lag time. This effect on loop size can be compensated for by adding a pAC dinucleotide that can be utilized for primer synthesis; however, the lag time is only a function of the ATP and CTP concentrations. These data indicate that initiation of primer synthesis triggers loop release before the completion of the previous Okazaki fragment. Statistical analysis of the relative length of a pair of successive loops implicated the completion of an Okazaki fragment as another trigger for releasing the loop. The existence of these two modes for triggering loop release guarantees that the lagging-strand polymerase is efficiently recycled. Nevertheless, observation of replication fork dynamics by measurement of the conversion of ssDNA to dsDNA is indirect. Whether the observed events of loop expansion and collapse as well as replisome pausing are events tightly coupled to primer synthesis will require assays that measure the extent of primer synthesis at a given pause.
A single-molecule experiment similar to the one discussed above was performed using the E. coli replication machinery with the Pol III holoenzyme composed of the core polymerase along with DnaB helicase and DnaG primase [51]. The substrate was a preassembled replication fork and the experimental setup was identical to that used for the T7 single-molecule assay [52]. However, primer extension by Pol III holoenzyme was monitored as the lengthening of the DNA as the ssDNA was converted into dsDNA. The Pol III holoenzyme with a clamp-loader complex lacking χ and ψ subunits extends a primer with a rate of 347 nt/s with a processivity of 1.4 kb. For comparison the processivity obtained from ensemble measurements is reported to range from one hundered to several hundred nucleotides [100]. The authors have argued that the discrepancy between the single-molecule and ensemble measurements arises from multiple-binding events and difficulties in obtaining precise values from ensemble measurements. Such a variation is also evident in the processivity value for the T7 polymerase obtained by the Bustamante group, where the single-molecule measurement yielded a value of 420 bases compared with 15–20 bases obtained from ensemble experiments. In addition, enzyme dependent pauses were observed that reflect a slow association of the enzyme on the DNA substrate and further comfirm single-enzyme event measurements.
Next the effect of the DnaB helicase and the DnaG primase on leading strand synthesis was assayed. To efficiently load the helicase onto the lagging strand of the substrate, the helicase-loader protein DnaC was also included in the experiment. Thus, DnaB and Pol III holoenzyme lacking the χ and ψ subunits of the clamp-loader complex replicated the leading strand at a rate of 417 bp/s with a processivity of 10.5 kb. This observed processivity is significantly lower compared to the value obtained from bulk biochemical assays of >50 kb [101]. The authors attribute this deviation to the differences in single and multiple binding events between the single-molecule and the ensemble assays. However, in the previous case where DnaB was absent in the assay, the processivity obtained from the single-molecule measurements was greater than the ensemble case. One possible explanation is that the movement of the helicase is challenged by a surface immobilized fork substrate leading to a lower probability of reinitiation of leading strand synthesis upon dissociation of any one of the replisome components. Upon introduction of the primase and under conditions of active priming, leading strand synthesis proceeded with a similar rate, but with a significantly reduced processivity of 2.9 kb and no observable pausing. This is in sharp contrast to the results on leading strand synthesis for the T7 replisome described earlier. How the interaction between primase and helicase accelerates the departure of helicase from the replisome thereby decreasing its processivity requires further clarification especially whether the reduction in processivity is indeed the mechanism by which the E. coli replisome achieves coordination of leading and lagging strand synthesis.
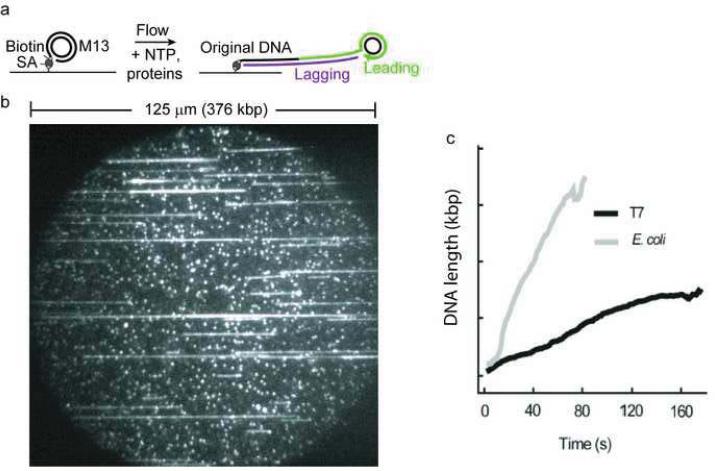
Recently a promising single-molecule method was presented by Tanner et al. for measuring coordinated DNA replication [61]. The real-time observation of coordinated DNA synthesis by the T7 and E. coli replisomes at the single-molecule level was obtained through the direct visualization of the growing DNA chain (Figure 12). The authors employed a well established rolling-circle DNA substrate capable of highly processive DNA synthesis. The polymerization of the substrate attached to the surface by a biotin-streptavidin conjugate was initiated by introducing all the replication proteins responsible for both leading and lagging strand synthesis. The growing chain of DNA was stretched by constant laminar flow and the increasing dsDNA length was monitored directly with the aid of an intercalating dye, SYTOX Orange. This provided a real-time measure of the growing DNA molecules and furnished replication rates by plotting the endpoint trajectories versus time as shown in Figure 12c. A replication rate of 75.9 ± 4.8 bp/s and a processivity of 25.3 ± 1.7 kb at 22 °C were obtained for the T7 replisome. When the temperature was raised to 37 °C, the authors observed a slight increase in the rate of the reaction and the processivity doubled to 51 ± 9 kb. The replication rate and processivity for the E. coli system was observed to be 535.5 ± 39 bp/s and 85.3 ± 6.1 kb, respectively. The replication rate observed here is in good agreement with other single-molecule and ensemble measurements. Although the processivity of 85 kb is much larger than the 10 kb observed in other single-molecule observations that did not include SSB, the value is in accord with ensemble assays where a processivity of 50 kb has been reported [102, 103]. This underscores the importance of SSB in the coordination of leading and lagging strand DNA synthesis.
Figure 12.
(a) Schematic depiction of single-molecule measurement using a rolling-circle substrate. (b) A picture of the image field. (c) Length of DNA as a function of time (Reproduced with permission from Ref. [61]).
[L] Study of the dynamic behavior of HIV reverse transcriptase by smFRET
Another type of DNA polymerase, a reverse transcriptase falls within the context of DNA polymerases. The reverse transcriptases are RNA-dependent DNA polymerases that synthesize DNA in a 5′ to 3′ direction employing RNA templates. The reverse transcriptase (RT) of human immunodeficiency virus (HIV), responsible for the cause of Acquired-Immune Deficiency Syndrome (AIDS) in humans, has been an extensively studied member of this group of enzymes. The HIV RT has been reviewed in detail by Le Grice in this issue; however, in order to discuss the single-molecule studies on this class of RNA-dependent DNA polymerases, a brief summary of this enzyme is presented below.
The HIV RT is a multifunctional enzyme with DNA polymerase and RNase H activities. This enzyme catalyzes the synthesis of dsDNA from its RNA genome in three successive steps. The first involves the synthesis of the minus strand of the DNA employing the viral RNA as the template to form a RNA-DNA hybrid. This hybrid becomes the substrate for the RNase H activity of the enzyme leading to partially degraded RNA fragments that bear purine-rich sequences known as polypurine tracts (PPTs). These tracts are used as primers for the synthesis of the plus strand of the dsDNA completing the replication of the viral genome.
Crystal structures of the enzyme in complex with RNA-DNA or DNA-DNA duplexes with or without dNTP, in conjunction with biochemical studies, revealed that RT interacts with either nucleic acid duplex and the incoming nucleotide for polymerization. However, when RT is acting as an RNase, the enzyme interacts with the RNA-DNA complex positioning the RNA strand within a second active site. It is, thus, important for the activities of RT to be well regulated in order to process the substrate correctly.
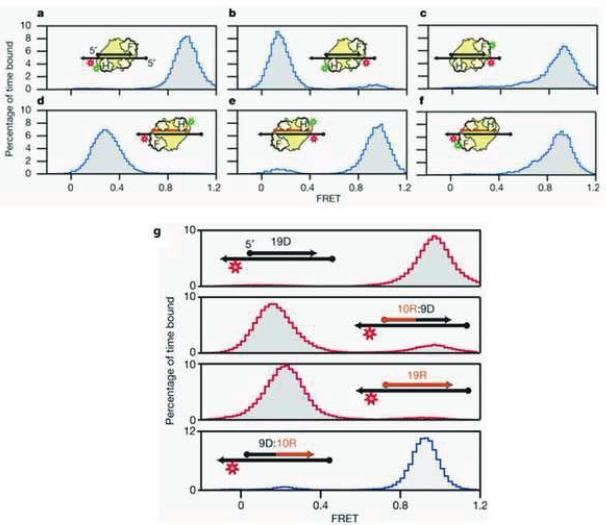
The Zhuang group, through a series of elaborately designed smFRET experiments, was able to observe RT undergoing a dynamic switch between its DNA polymerase and RNase H activities [58]. In these experiments, the position of fluorescent probes on the DNA/DNA substrate and RT were systematically varied to precisely determine the orientation of RT binding to the substrate by measuring the smFRET, as illustrated in Figure 13. In these experiments, the protein and the DNA substrate were labeled with a fluorescent donor and acceptor, respectively. In Figure 13a, the fluorescent donor was attached to the RNase H domain of RT and the acceptor was conjugated to the 3′-terminus of the template. When RT binds to the P-T substrate in the polymerization orientation, the donor and the acceptor are proximal so that a FRET with a high efficiency of 0.9 was observed; accordingly, when the 5′-terminus was labeled with the acceptor as showed in Figure 13b, the distance between the donor and the acceptor was increased due to the change in the position of the acceptor so that the FRET efficiency dropped to 0.3. Movement of the donor probe to the RT finger domain generated a high efficiency FRET (Figure 13c). Data from Figure 13a–c clearly demonstrate that when the template is primed with a DNA primer, RT binds to the P-T DNA/DNA junction in a polymerase active mode.
Figure 13.
(a–f) SmFRET experimental design for probing the binding orientation of HIV RT on DNA/DNA or DNA/RNA substrates. Red and green indicate the position of the Cy5 and Cy3 probes, respectively. Letter H represents the RNase H domain; Letter F represents the finger domain. (g) Binding orientation as a function of the backbone composition of the 5′-end of the primer. Letter xR:yD means a primer containing from 5′ to 3′ x ribonucleotides and y deoxyribonucleotides (Reproduced with permission from Ref. [58]).
With the same experimental design but using RNA as the primer, RT bound to the RNA/DNA substrate in an RNase mode as revealed in Figure 13d–f. With a series of RNA/DNA hybrid primers and the RT probe in the RNaseH domain two separate FRET efficiency peaks were observed (Figure 13g) indicating that RT binds to its substrate with different orientations depending upon the backbone composition of the 5′-end of the primer. RT binds to a substrate with a DNA primer or a hybrid primer containing DNA at the 5′-end with its DNA polymerase domain close to the 3′-end of the DNA primer competent for polymerase activity. In contrast RT binds to a substrate with an RNA primer or a hybrid primer containing RNA at the 5′-end RT binds with its RNase H domain close to the 3′-end of the primer in a conformation competent for RNase activity. These results indicate that the substrate binding orientation is the determining factor for polymerase or RNase activity. However, when an RNA PPT and its derivatives were used as primers, a portion of RT bound in its polymerase mode consistent with previous reports that PPTs serve as the primers for DNA-dependent DNA synthesis in vivo. Additional experiments showed that a correct incoming dNTP increased the probability of RT binding in the polymerase-competent orientation, whereas, the anti-HIV drug nevirapine, a non-nucleoside RT inhibitor that allosterically inhibits DNA synthesis by binding in a hydrophobic pocket near the polymerase active site, increased the probability of RT binding in the RNase-competent orientation. These smFRET experiments strongly support the hypothesis that the substrate composition determines the activity of RT.
The distributive nature of RT is reflective of the dissociation from its DNA extension site. How the enzyme finds the 3′-end of the primer and resumes DNA synthesis was addressed by Liu et al. using smFRET experiments [59]. By varying the length of the primer labeled with a dye as shown in Figure 14 a–c, smFRET revealed how RT moves along the substrate. When a 19mer DNA primer with an RNA template was used, RT binds to the P-T substrate with a FRET efficiency of 0.9. When the primer length was increased to 38 or 56 nucleotides, two FRET populations were observed suggesting that RT moves between the 5′-end of the primer and the 3′-P-T junction (Figure 14b, c). The temporal dependence of the sliding was tracked: no energy transfer was observed until 20 s, after which a moderate but constant FRET signal was recorded (Figure 14d) suggesting that RT moved 5′ to ′ the duplex as depicted in scenario A of Figure 14f. Surprisingly, Liu et al. also observed a short but drastic change in the FRET signal (Figure 14e). This observation suggested that occasionally RT binds incorrectly and must flip to a correct orientation (Scenario B). On such substrates RT binds randomly followed by sliding in either direction along the DNA primed duplex until the 3′-end of the primer is identified. The presence of a correct dNTP increased the frequecny of RT binding to the 3′-end. In summary, the smFRET experimental results show that RT searches for its synthesis site through a binding and sliding mechanism.
Figure 14.
(a–c) Sliding of RT along DNA primed substrates with the finger domain facing the 3′-end of the DNA primer and the observed FRET distribution. (d) Donor and acceptor fluorescence time traces corresponding to the binding scenario in Panel f (e) A quick change in FRET corresponding to a flip in the binding orientation at the 3′-end of the DNA primer. (Reproduced from Ref. [59]).
[M] Comparison of ensemble and single-molecule measurements of the replisome
The results obtained from the single-molecule observations of the activities of replisome eznymes individually or in complex with other proteins are tabulated in Table 1 for comparison with the corresponding values from ensemble methods. The single-molecule experiments generally involve indirect measurement of the DNA polymerization from changes in DNA length. Almost all reports of single-molecule studies involve the use of a force to stretch the DNA or have the reaction initiated on force-stretched DNA substrates. Furthermore, these reactions are force-dependent. The single-molecule studies provide a more precise processivity value often not attainable in ensemble measurements owing to multiple binding events but in reasonable agreement. One exception appears to be the the processivities measured with the intercalating dye that are significantly higher than the other values, for example, 25 kb (22 °C) or 51 kb (37 °C) compared to 17 kb for the T7 replisome and 85 kb compared to 10.5 kb for the E. coli replisome. This might arise from inaccuracy in measuring the signal intensity of the intercalating dye or its effect on the replication process itself.
Reaction rates of the polymerases and helicases obtained from both the ensemble and single-molecule studies also are in good agreement so that gratifyingly the behavior of the enzymes is identical under both conditions (Table 1). In the holoenzymes examined thus far the rates of replication were similar for both the single-molecule and ensemble levels, the exception being the T7 holoenzyme whose rate of 164 nt/s in the absence of rNTPs was two-fold faster than ~80 nt/s.
[N] Conclusions
In summary, single-molecule methods can address a variety of important questions on DNA replication such as: protein/DNA interactions; the movements of polymerase, primase and helicase during DNA synthesis; the assembly pathway of a replisome at the replication fork from its constitutional components; the coordination of leading and lagging strand DNA synthesis; and the sliding and switching of an enzyme to capture its substrate. Although limited so far in number, these successful single-molecule studies of DNA replication are encouraging and demonstrate the power of the methods. Single-molecule kinetics of DNA polymerase made it possible to “visualize” the dynamic movement of polymerases and replisome components on the DNA chain by tracking the movement of a bead tethered to the macromolecule, by the change in mechanical tension or by the energy transfer between dyes attached to DNA and proteins. All measurements such as the rate of DNA polymerase extension, its processivity, and the rates of helicase unwinding correspond to similar measurements using ensemble experiments.
The advantage of single-molecule experiments is in detecting the transient states that are invisible due to the “averaging effect” in ensemble experiments. For example, the switch between the polymerase active mode and exonuclease active mode for the T7 polymerase depends on the applied mechanical tension on the DNA chain, which is difficult, if not impossible, to demonstrate by ensemble experiment since it is not feasible to apply mechanical tension on millions or billions of DNA molecules simultaneously. Another example is the observation of leading strand synthesis pausing during primer synthesis required for initiation of lagging strand synthesis in T7 DNA replication in single-molecule experiments providing evidence for the coupling between leading and lagging strands synthesis although tempered by the cautionary note expressed earlier. These transient but important events monitored at the single-molecule level increase our understanding about the DNA replisome and underscore the potential utilization of the techniques in the system to probe other unanswered questions.
However, compared to ensemble experiments, the manipulation of biomolecules for single-molecule experiments is much more complicated. There are several factors that need to be taken into consideration when designing single-molecule experiments. Although the effect of modification of macromolecules are contributing factors in these and ensemble experiments, a difficulty general to single-molecule experiments is the concentrations employed in the assays [104]. In ensemble experiments, the concentration of protein or substrate are sub-micromolar or even higher; however, in single-molecule experiments, in order to reduce the background signal, the concentrations of enzyme and substrate are typically in the nanomolar and sometimes the sub-nanomolar range. The reduced concentration may shift the equilibrium dynamics from a productive enzyme-substrate complex to dissociated individual molecules and result in few events being observed. For instance, with a Kd equal to 50 nM between a DNA polymerase and a DNA substrate, in ensemble experiments containing 100 nM polymerase and DNA, the concentration of polymerase-DNA complex formed is 50 nM (50% of total enzyme); however, if the polymerase and substrate are reduced to 10 nM, the concentration of polymerase-DNA complex formed would be only 1.5 nM (15% of total enzyme). This applies to a higher order replisome where loss of auxiliary proteins may not be recognized. The rates of extension by DNA polymerases in ensemble studies are systematically higher than those of single-molecule studies and the reason might be due to one or more of these factors.
Almost all of the complexes involved in DNA metabolism such as replication, repair, recombination and transcription involve multiple pathways and many interacting protein partners. The nature of the complexes and their respective functions can be studied in the future employing single-molecule techniques with further improvements. With the exception of FRET based analyses of individual enzyme function or replisome assembly, investigation of replisome DNA synthesis have relied primarily on length changes in the DNA. Because these cannot be verified by actual product analysis, these conclusions are inferential. Future studies should aspire to direct measurements. The real challenge lies in our ability to extend these techniques to monitor the dynamics of enzymes in vivo in their natural environment.
[O] Acknowledgements
This work is supported by National Institute of Health Grant GM013306 to S.J.B. We would like to thank Nathan Tanner and Prof. Antoine van Oijen for providing us with their high-resolution figures.
[P] References
- [1].Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- [2].Hamdan SM, Richardson CC. Motors, Switches, and Contacts in a Replisome. Annu. Rev. Biochem. 2009 doi: 10.1146/annurev.biochem.78.072407.103248. doi:10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- [3].Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sexton DJ, Kaboord BF, Berdis AJ, Carver TE, Benkovic SJ. Dissecting the order of bacteriophage T4 DNA polymerase holoenzyme assembly. Biochemistry. 1998;37:7749–7756. doi: 10.1021/bi980088h. [DOI] [PubMed] [Google Scholar]
- [5].Jeruzalmi D, O'Donnell M, Kuriyan J. Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol. 2002;12:217–224. doi: 10.1016/s0959-440x(02)00313-5. [DOI] [PubMed] [Google Scholar]
- [6].O'Donnell M, Jeruzalmi D, Kuriyan J. Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr. Biol. 2001;11:R935–946. doi: 10.1016/s0960-9822(01)00559-0. [DOI] [PubMed] [Google Scholar]
- [7].Patel PH, Suzuki M, Adman E, Shinkai A, Loeb LA. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- [8].Cornish PV, Ha T. A survey of single-molecule techniques in chemical biology. ACS Chem. Biol. 2007;2:53–61. doi: 10.1021/cb600342a. [DOI] [PubMed] [Google Scholar]
- [9].Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- [10].Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- [11].Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- [12].Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- [13].Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr., Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huber HE, Tabor S, Richardson CC. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- [15].Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- [16].Mendelman LV, Richardson CC. Requirements for primer synthesis by bacteriophage T7 63-kDa gene 4 protein. Roles of template sequence and T7 56-kDa gene 4 protein. J. Biol. Chem. 1991;266:23240–23250. [PubMed] [Google Scholar]
- [17].Tabor S, Richardson CC. Template recognition sequence for RNA primer synthesis by gene 4 protein of bacteriophage T7. Proc. Natl. Acad. Sci. U. S. A. 1981;78:205–209. doi: 10.1073/pnas.78.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kusakabe T, Richardson CC. Template recognition and ribonucleotide specificity of the DNA primase of bacteriophage T7. J. Biol. Chem. 1997;272:5943–5951. doi: 10.1074/jbc.272.9.5943. [DOI] [PubMed] [Google Scholar]
- [19].Kato M, Ito T, Wagner G, Richardson CC, Ellenberger T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell. 2003;11:1349–1360. doi: 10.1016/s1097-2765(03)00195-3. [DOI] [PubMed] [Google Scholar]
- [20].He ZG, Richardson CC. Effect of single-stranded DNA-binding proteins on the helicase and primase activities of the bacteriophage T7 gene 4 protein. J. Biol. Chem. 2004;279:22190–22197. doi: 10.1074/jbc.M401100200. [DOI] [PubMed] [Google Scholar]
- [21].Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- [22].Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- [23].Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc. Natl. Acad. Sci. U. S. A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ma Y, Wang T, Villemain JL, Giedroc DP, Morrical SW. Dual functions of single-stranded DNA-binding protein in helicase loading at the bacteriophage T4 DNA replication fork. J. Biol. Chem. 2004;279:19035–19045. doi: 10.1074/jbc.M311738200. [DOI] [PubMed] [Google Scholar]
- [25].Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- [26].Valentine AM, Ishmael FT, Shier VK, Benkovic SJ. A zinc ribbon protein in DNA replication: primer synthesis and macromolecular interactions by the bacteriophage T4 primase. Biochemistry. 2001;40:15074–15085. doi: 10.1021/bi0108554. [DOI] [PubMed] [Google Scholar]
- [27].Maki H, Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J. Biol. Chem. 1985;260:12987–12992. [PubMed] [Google Scholar]
- [28].Fukui T, Yamauchi K, Muroya T, Akiyama M, Maki H, Sugino A, Waga S. Distinct roles of DNA polymerases delta and epsilon at the replication fork in Xenopus egg extracts. Genes. Cells. 2004;9:179–191. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- [29].Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends. Cell. Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- [31].Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- [32].Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Costa A, Onesti S. The MCM complex: (just) a replicative helicase? Biochem. Soc. Trans. 2008;36:136–140. doi: 10.1042/BST0360136. [DOI] [PubMed] [Google Scholar]
- [35].Alberts BM, Barry J, Bedinger P, Formosa T, Jongeneel CV, Kreuzer KN. Studies on DNA replication in the bacteriophage T4 in vitro system. Cold Spring Harb. Symp. Quant .Biol. 1983;47(Pt 2):655–668. doi: 10.1101/sqb.1983.047.01.077. [DOI] [PubMed] [Google Scholar]
- [36].Yang J, Nelson SW, Benkovic SJ. The control mechanism for lagging strand polymerase recycling during bacteriophage T4 DNA replication. Mol. Cell. 2006;21:153–164. doi: 10.1016/j.molcel.2005.11.029. [DOI] [PubMed] [Google Scholar]
- [37].Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- [38].Yang W. Damage repair DNA polymerases Y. Curr. Opin. Struct. Biol. 2003;13:23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- [39].Dahlberg ME, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow fragment): identification of a second conformational change and evaluation of the internal equilibrium constant. Biochemistry. 1991;30:4835–4843. doi: 10.1021/bi00234a002. [DOI] [PubMed] [Google Scholar]
- [40].Eger BT, Benkovic SJ. Minimal kinetic mechanism for misincorporation by DNA polymerase I (Klenow fragment) Biochemistry. 1992;31:9227–9236. doi: 10.1021/bi00153a016. [DOI] [PubMed] [Google Scholar]
- [41].Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- [42].Wong I, Patel SS, Johnson KA. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- [43].Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annu. Rev. Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- [44].van Oijen AM. Single-molecule studies of complex systems: the replisome. Mol Biosyst. 2007;3:117–125. doi: 10.1039/b612545j. [DOI] [PubMed] [Google Scholar]
- [45].Walter NG, Huang CY, Manzo AJ, Sobhy MA. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–106. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- [48].Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 2000;97:12002–12007. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].van Oijen AM. Honey, I shrunk the DNA: DNA length as a probe for nucleic-acid enzyme activity. Biopolymers. 2007;85:144–153. doi: 10.1002/bip.20624. [DOI] [PubMed] [Google Scholar]
- [51].Tanner NA, Hamdan SM, Jergic S, Loscha KV, Schaeffer PM, Dixon NE, van Oijen AM. Single-molecule studies of fork dynamics in Escherichia coli DNA replication. Nat. Struct. Mol. Biol. 2008;15:998. doi: 10.1038/nsmb0908-998a. [DOI] [PubMed] [Google Scholar]
- [52].Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- [53].Hamdan SM, Johnson DE, Tanner NA, Lee JB, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol. Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- [54].Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457:336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kim S, Blainey PC, Schroeder CM, Xie XS. Multiplexed single-molecule assay for enzymatic activity on flow-stretched DNA. Nat. Methods. 2007;4:397–399. doi: 10.1038/nmeth1037. [DOI] [PubMed] [Google Scholar]
- [56].Zhang Z, Spiering MM, Trakselis MA, Ishmael FT, Xi J, Benkovic SJ, Hammes GG. Assembly of the bacteriophage T4 primosome: single-molecule and ensemble studies. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3254–3259. doi: 10.1073/pnas.0500327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Smiley RD, Zhuang Z, Benkovic SJ, Hammes GG. Single-molecule investigation of the T4 bacteriophage DNA polymerase holoenzyme: multiple pathways of holoenzyme formation. Biochemistry. 2006;45:7990–7997. doi: 10.1021/bi0603322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Abbondanzieri EA, Bokinsky G, Rausch JW, Zhang JX, Le Grice SF, Zhuang X. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Luo G, Wang M, Konigsberg WH, Xie XS. Single-molecule and ensemble fluorescence assays for a functionally important conformational change in T7 DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12610–12615. doi: 10.1073/pnas.0700920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Tanner NA, Loparo JJ, Hamdan SM, Jergic S, Dixon NE, van Oijen AM. Real-time single-molecule observation of rolling-circle DNA replication. Nucleic Acids Res. 2009;37:e27. doi: 10.1093/nar/gkp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Goel A, Frank-Kamenetskii MD, Ellenberger T, Herschbach D. Tuning DNA “strings”: modulating the rate of DNA replication with mechanical tension. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8485–8489. doi: 10.1073/pnas.151261198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shokri L, Marintcheva B, Richardson CC, Rouzina I, Williams MC. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J. Biol. Chem. 2006;281:38689–38696. doi: 10.1074/jbc.M608460200. [DOI] [PubMed] [Google Scholar]
- [64].Pant K, Karpel RL, Rouzina I, Williams MC. Mechanical measurement of single-molecule binding rates: kinetics of DNA helix-destabilization by T4 gene 32 protein. J. Mol. Biol. 2004;336:851–870. doi: 10.1016/j.jmb.2003.12.025. [DOI] [PubMed] [Google Scholar]
- [65].Pant K, Karpel RL, Williams MC. Kinetic regulation of single DNA molecule denaturation by T4 gene 32 protein structural domains. J. Mol. Biol. 2003;327:571–578. doi: 10.1016/s0022-2836(03)00153-0. [DOI] [PubMed] [Google Scholar]
- [66].Pant K, Karpel RL, Rouzina I, Williams MC. Salt dependent binding of T4 gene 32 protein to single and double-stranded DNA: single molecule force spectroscopy measurements. J. Mol. Biol. 2005;349:317–330. doi: 10.1016/j.jmb.2005.03.065. [DOI] [PubMed] [Google Scholar]
- [67].Carroll SS, Benkovic SJ. Mechanistic aspects of DNA polymerases: Escherichia coli DNA polymerase I (Klenow fragment) as a paradigm. 1990;90:1291–1307. [Google Scholar]
- [68].Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- [69].Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- [71].Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- [72].Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- [74].Purohit V, Grindley ND, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- [75].Dunlap CA, Tsai MD. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase beta. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- [76].Fidalgo da Silva E, Mandal SS, Reha-Krantz LJ. Using 2-aminopurine fluorescence to measure incorporation of incorrect nucleotides by wild type and mutant bacteriophage T4 DNA polymerases. J. Biol. Chem. 2002;277:40640–40649. doi: 10.1074/jbc.M203315200. [DOI] [PubMed] [Google Scholar]
- [77].Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Balasubramanian S, Bentley DR. 2001.
- [80].Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- [82].Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- [83].Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- [84].Gold L, O'Farrell PZ, Russel M. Regulation of gene 32 expression during bacteriophage T4 infection of Escherichia coli. J. Biol. Chem. 1976;251:7251–7262. [PubMed] [Google Scholar]
- [85].Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jeong YJ, Levin MK, Patel SS. The DNA-unwinding mechanism of the ring helicase of bacteriophage T7. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7264–7269. doi: 10.1073/pnas.0400372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lefebvre SD, Wong ML, Morrical SW. Simultaneous interactions of bacteriophage T4 DNA replication proteins gp59 and gp32 with single-stranded (ss) DNA. Co-modulation of ssDNA binding activities in a DNA helicase assembly intermediate. J. Biol. Chem. 1999;274:22830–22838. doi: 10.1074/jbc.274.32.22830. [DOI] [PubMed] [Google Scholar]
- [88].Jones CE, Mueser TC, Nossal NG. Bacteriophage T4 32 protein is required for helicase-dependent leading strand synthesis when the helicase is loaded by the T4 59 helicase-loading protein. J. Biol. Chem. 2004;279:12067–12075. doi: 10.1074/jbc.M313840200. [DOI] [PubMed] [Google Scholar]
- [89].Alley SC, Shier VK, Abel-Santos E, Sexton DJ, Soumillion P, Benkovic SJ. Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry. 1999;38:7696–7709. doi: 10.1021/bi9827971. [DOI] [PubMed] [Google Scholar]
- [90].Millar D, Trakselis MA, Benkovic SJ. On the solution structure of the T4 sliding clamp (gp45) Biochemistry. 2004;43:12723–12727. doi: 10.1021/bi048349c. [DOI] [PubMed] [Google Scholar]
- [91].Mosig G, Colowick N, Gruidl ME, Chang A, Harvey AJ. Multiple initiation mechanisms adapt phage T4 DNA replication to physiological changes during T4's development. FEMS Microbiol. Rev. 1995;17:83–98. doi: 10.1111/j.1574-6976.1995.tb00190.x. [DOI] [PubMed] [Google Scholar]
- [92].Stano NM, Jeong YJ, Donmez I, Tummalapalli P, Levin MK, Patel SS. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Tabor S, Huber HE, Richardson CC. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- [94].Hamdan SM, Marintcheva B, Cook T, Lee SJ, Tabor S, Richardson CC. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Frick DN, Richardson CC. Interaction of bacteriophage T7 gene 4 primase with its template recognition site. J. Biol. Chem. 1999;274:35889–35898. doi: 10.1074/jbc.274.50.35889. [DOI] [PubMed] [Google Scholar]
- [96].Lee SJ, Marintcheva B, Hamdan SM, Richardson CC. The C-terminal residues of bacteriophage T7 gene 4 helicase-primase coordinate helicase and DNA polymerase activities. J. Biol. Chem. 2006;281:25841–25849. doi: 10.1074/jbc.M604602200. [DOI] [PubMed] [Google Scholar]
- [97].Notarnicola SM, Mulcahy HL, Lee J, Richardson CC. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J. Biol. Chem. 1997;272:18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- [98].Yang J, Xi J, Zhuang Z, Benkovic SJ. The oligomeric T4 primase is the functional form during replication. J. Biol. Chem. 2005;280:25416–25423. doi: 10.1074/jbc.M501847200. [DOI] [PubMed] [Google Scholar]
- [99].Mitkova AV, Khopde SM, Biswas SB. Mechanism and stoichiometry of interaction of DnaG primase with DnaB helicase of Escherichia coli in RNA primer synthesis. J. Biol. Chem. 2003;278:52253–52261. doi: 10.1074/jbc.M308956200. [DOI] [PubMed] [Google Scholar]
- [100].Fay PJ, Johanson KO, McHenry CS, Bambara RA. Size classes of products synthesized processively by DNA polymerase III and DNA polymerase III holoenzyme of Escherichia coli. J. Biol. Chem. 1981;256:976–983. [PubMed] [Google Scholar]
- [101].Mok M, Marians KJ. The Escherichia coli preprimosome and DNA B helicase can form replication forks that move at the same rate. J. Biol. Chem. 1987;262:16644–16654. [PubMed] [Google Scholar]
- [102].Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- [103].Studwell PS, O'Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J. Biol. Chem. 1990;265:1171–1178. [PubMed] [Google Scholar]
- [104].van Oijen AM. Cutting the forest to see a single tree? Nat. Chem. Biol. 2008;4:440–443. doi: 10.1038/nchembio0808-440. [DOI] [PubMed] [Google Scholar]
- [105].Lechner RL, Richardson CC. A preformed, topologically stable replication fork. Characterization of leading strand DNA synthesis catalyzed by T7 DNA polymerase and T7 gene 4 protein. J. Biol. Chem. 1983;258:11185–11196. [PubMed] [Google Scholar]
- [106].Raney KD, Carver TE, Benkovic SJ. Stoichiometry and DNA unwinding by the bacteriophage T4 41:59 helicase. J. Biol. Chem. 1996;271:14074–14081. doi: 10.1074/jbc.271.24.14074. [DOI] [PubMed] [Google Scholar]
- [107].Richardson RW, Nossal NG. Characterization of the bacteriophage T4 gene 41 DNA helicase. J. Biol. Chem. 1989;264:4725–4731. [PubMed] [Google Scholar]
- [108].Mace DC, Alberts BM. T4 DNA polymerase. Rates and processivity on single-stranded DNA templates. J. Mol. Biol. 1984;177:295–311. doi: 10.1016/0022-2836(84)90458-3. [DOI] [PubMed] [Google Scholar]
- [109].Cha TA, Alberts BM. The bacteriophage T4 DNA replication fork. Only DNA helicase is required for leading strand DNA synthesis by the DNA polymerase holoenzyme. J. Biol. Chem. 1989;264:12220–12225. [PubMed] [Google Scholar]
- [110].Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J. Mol. Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]